Abstract
In vitro selection is a key component of efforts to discover functional nucleic acids and small molecules from libraries of DNA, RNA, and DNA-encoded small molecules. Such selections have been widely used to evolve RNA and DNA catalysts and, more recently, to discover new reactions from DNA-encoded libraries of potential substrates. While effective, current strategies for selections of bond-forming and bond-cleaving reactivity are generally indirect, require the synthesis of biotin-linked substrates, and involve multiple solution-phase and solid-phase manipulations. In this work we report the successful development and validation of reactivity-dependent PCR (RDPCR), a new method that more directly links bond formation or bond cleavage with the amplification of desired sequences and that obviates the need for solid-phase capture, washing, and elution steps. We show that RDPCR can be used to select for bond formation in the context of reaction discovery and for bond cleavage in the context of protease activity profiling.
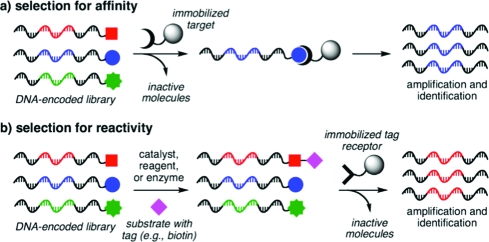
In vitro selection is a key component of efforts to discover functional nucleic acids and small molecules from libraries of DNA, RNA, and DNA-encoded small molecules.(1) When the desired activity is binding affinity, as is the case for aptamer evolution(2) or for the discovery of DNA-linked small molecules that bind to protein targets,(3) a direct selection is possible; the library is typically incubated with immobilized target molecules, and bound library members are washed and eluted before being subjected to PCR amplification (Figure 1a).
Figure 1.
Traditional approaches to in vitro selection.
In vitro selections have also been applied to evolve RNA and DNA catalysts(4) and, more recently, to discover new reactions from DNA-encoded libraries of potential substrates.(5) For these applications, desired library members undergo bond formation or bond cleavage. Selections for reactivity are significantly more complicated than selections for binding affinity. Typically, libraries are incubated with biotinylated substrates or potential reaction partners. Bond formation results in the attachment of biotin to a library member, which in turn enables its capture by immobilized avidin (Figure 1b).(6) For bond cleavage, an inverse approach is commonly used in which immobilized, biotinylated library members liberate themselves upon bond scission.(7) While effective, such selections for reactivity are indirect, require the synthesis of biotin-linked substrates, and involve multiple solution-phase and solid-phase manipulations.
Motivated by our ongoing interest in applying in vitro selection to discovery problems that involve chemical reactivity,(8) we sought to develop a new method that more directly links bond formation or bond cleavage with the amplification of desired sequences(9) and that obviates the need for solid-phase capture, washing, and elution steps. Here we report the development and validation of such a method, reactivity-dependent PCR (RDPCR).
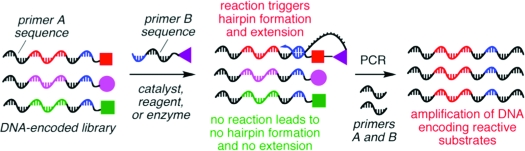
RDPCR is based on the well-established observation that the melting temperatures (Tm) of double-stranded nucleic acids are substantially higher when hybridization occurs intramolecularly as opposed to intermolecularly.(10) For example, the DNA hairpin 1 with an 8 bp stem is predicted(11) to exhibit a Tm of 48 °C, while the intermolecular hybridization of two DNA strands of the same sequence (2 and 3) is predicted to be far less favorable, with a Tm of only 11 °C (Figure 2). We hypothesized that the significant difference in intramolecular versus intermolecular duplex stability could enable a new type of in vitro selection, wherein bond formation or bond cleavage is transduced into the formation of a self-priming DNA hairpin. This hairpin enables the selective PCR amplification of those DNA sequences that encode the reactive species (Figure 2b).(12)
Figure 2.
Principles underlying reactivity-dependent PCR (RDPCR). Conditions in (a): 10 nM DNA, 2 mM Mg2+, 100 mM NaCl.
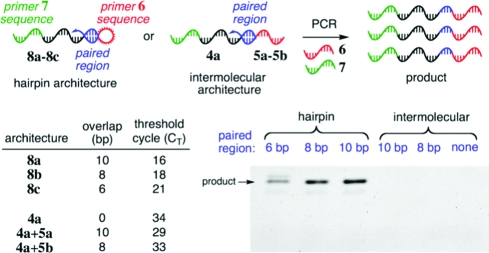
We first assessed the ability of intramolecular self-priming to result in preferential DNA amplification. We synthesized a series of oligonucleotide pairs (4a+5), each predicted to hybridize intermolecularly at their 3′ ends to form a short duplex region of 8 or 10 bp (Figure 3). The 5′ end of each DNA oligonucleotide in the pair contained a sequence identical to either primer 6 or primer 7. PCR amplification cannot occur until after DNA hybridization and 3′ extension take place to generate a single-stranded DNA molecule containing both primer 6 and a sequence complementary to primer 7. This 3′-extended species can then hybridize with primer 7 and initiate PCR amplification.
Figure 3.
Comparison of PCR efficiency of intramolecularly primed versus intermolecularly primed DNA templates. PCR conditions for PAGE samples: 19 fmol of 8 or 19 fmol of 4a+5 in 30 μL, 25 cycles.
We also prepared an analogous series of DNA oligonucleotides capable of hybridizing intramolecularly to form hairpin structures with 10-, 8-, or 6-bp stems (8a−8c). As with the intermolecularly hybridizing oligonucleotides, PCR amplification must be initiated by primer extension of the 3′ end. Quantitative, real-time PCR(13) (qPCR) was used to compare the ability of these oligonucleotides to undergo PCR amplification.
Consistent with our initial hypothesis, under identical PCR conditions and with equal starting concentrations of DNA, the intramolecularly hybridizing templates were amplified much more efficiently than their intermolecular counterparts (8a vs 4a+5a, 8b vs 4a+5b). The intramolecularly hybridizing templates reached a threshold level of amplified product 13 to 15 PCR cycles (CT) earlier than the intermolecular templates, corresponding to a > 2(13)-fold (>8000-fold) difference in effective initial template abundance. These qPCR results were corroborated by PAGE analysis; after 25 cycles of PCR, amplified product was only detected in reactions containing hairpin DNA. Collectively, these findings demonstrate that intramolecularly hybridizing templates can be amplified to abundant levels under conditions that fail to appreciably amplify the corresponding intermolecularly hybridizing templates. Subsequent experiments in this work were carried out with an 8-base stem, which was found to optimally balance robust intramolecular priming and poor intermolecular priming (see Supporting Information).
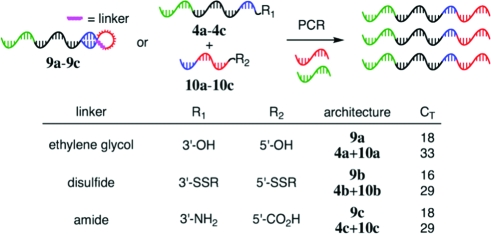
To use RDPCR in a general selection for bond formation, the covalently linked functional groups in the hairpin loop must not interfere with the required hybridization and 3′-extension events. To test the compatibility of non-natural linkers with the preferential amplification of self-priming templates, we repeated the qPCR experiment in Figure 3 with a series of non-natural linker structures, including ether, disulfide, and amide hairpin linkers. In all cases tested, DNA templates containing non-natural linkers (9a−9c) were far more efficiently amplified than the analogous intermolecularly hybridizing templates (4+10) (Figure 4).
Figure 4.
Non-natural hairpin linkers support self-priming PCR. R = (CH2)6OH.
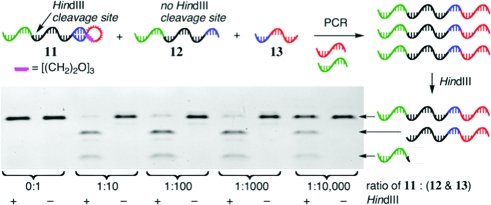
Since RDPCR will ultimately be applied to mixtures of both active (resulting in DNA hairpins) and inactive (resulting in separate linear oligonucleotides) library members, we next tested the ability of hairpin templates to undergo preferential amplification in the presence of large excesses of corresponding linear molecules (Figure 5). For these library-format experiments, the hairpin (11) and linear (12) template sequences vary only by a single base, such that DNA amplified from the hairpin contains a cleavage site for the restriction enzyme HindIII, while DNA arising from 12 does not.
Figure 5.
Selectivity of RDPCR in a library-format mock selection. PCR conditions: 19 fmol of 12 and 13 in 60 μL, 25−35 cycles.
The quantity of 11 spiked into an equimolar mixture of 12 and 13 was varied to determine the selectivity of RDPCR in a library format. As little as 1 attomol of 11 (600 000 molecules) could be selectively amplified in the presence of a 10 000-fold excess of 12 and 13. The use of larger quantities of hairpin (corresponding to a 10- to 1000-fold excess of 12 and 13) overwhelmingly provided the desired product. These results demonstrate the ability of hairpin templates to be preferentially amplified even in the presence of large excesses of linear templates and indicate that RDPCR can be applied to library-format selections.(14)
Following these foundational studies, RDPCR was applied to two model in vitro selections. First, RDPCR was validated as a bond-formation selection for DNA-encoded reaction discovery. Previously, DNA-encoded reaction discovery required the capture, washing, and elution of active library members on avidin-linked beads (Figure 1b).(5) In a RDPCR version of reaction discovery, pairs of functional groups are attached to encoding DNA strands (Figure 6). A disulfide linker temporarily joins each substrate pair (9b, 9d). Exposure to a set of reaction conditions and subsequent cleavage of the disulfide bond provide one of two possible outcomes. If a new covalent bond has formed between the functional groups, the hairpin-forming nucleotides remain tethered together through the reaction product, leaving a self-priming DNA hairpin (14). If no bond has formed, then only intermolecular hybridization is possible (15+16), resulting in inefficient PCR amplification. In contrast with previous reaction discovery selections, the RDPCR version requires no solid-phase steps and minimal manipulation.
Figure 6.
RDPCR-based DNA-encoded reaction discovery selection. PCR conditions for PAGE samples: 1 fmol of 9 in 20 μL, 23 cycles.
To test the ability of RDPCR to support reaction discovery, we synthesized a disulfide-linked substrate (9d) with pendant alkene and aryl iodide groups, which should undergo a Pd-mediated Heck-type reaction (Figure 6).(15) An unreactive control substrate (9b) containing an azide and an aldehyde was similarly generated. Each substrate was treated with 1 mM Na2PdCl4 (which is reduced to Pd(0) in situ) in aqueous pH 7.5 buffer for 30 min at 65 °C, followed by DTT to cleave the disulfide bond. The resulting material was subjected to PCR. The DNA attached to the alkene-aryl iodide pair (9d) amplified efficiently (Figure 6). Omission of Na2PdCl4 resulted in much less efficient amplification. Likewise, the unreactive substrate pair 9b did not undergo PCR amplification after identical treatment. Omission of DTT, however, enabled the disulfide-linked starting substrate to amplify efficiently. Collectively, these results indicate that RDPCR can selectively and efficiently amplify DNA templates that have undergone bond formation and that amplification is dependent on the intramolecularity of the resulting template-primer species.(16) These experiments were corroborated using an azide/alkyne substrate that undergoes a Cu(I)-catalyzed cycloaddition reaction (see Supporting Information).
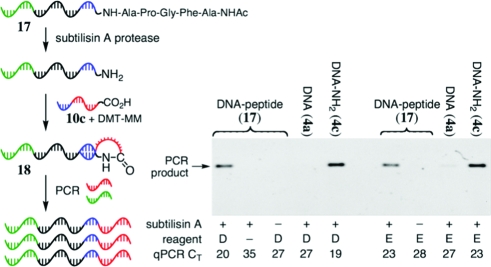
In principle, RDPCR can also enable efficient selection for bond cleavage, which has yet to be studied in a DNA-encoded context. To explore this possibility, we evaluated the ability of DNA-linked peptides to undergo cleavage mediated by a protease (Figure 7). We anticipated that protease-mediated cleavage of a DNA−peptide conjugate would expose a primary amine group, which would then undergo DNA-templated amide bond formation to generate a hairpin template for efficient PCR.(17) In contrast, the absence of proteolysis should result in no amide formation and thus inefficient PCR amplification.
Figure 7.
RDPCR-based protease-mediated peptide cleavage selection. PCR conditions for PAGE samples: 19 fmol of DNA in 30 μL, 23 cycles (lanes 1−5) or 25 cycles (lanes 6−9). D = DMT-MM; E = EDC + sNHS.
A DNA−N-acetyl-pentapeptide conjugate (17), synthesized by solid phase cosynthesis, was exposed to subtilisin A. The peptide sequence (Ac-N-AFGPA) was designed to include cleavage sites for subtilisin A.(18) The enzyme-treated DNA was combined with a carboxylic acid-linked DNA primer (10c) under conditions (DMT-MM or sNHS+EDC) that support DNA-templated amide bond formation.(19)
Addition of the protease-digested and carboxylate-ligated DNA−peptide conjugate (18) to a PCR reaction resulted in efficient PCR amplification. In contrast, no PCR product was detected by PAGE when unfunctionalized DNA (4a) was used in place of the pentapeptide or when subtilisin A was omitted. Likewise, omission of the amide formation reagents also resulted in inefficient PCR amplification, consistent with the necessity of intramolecular primer hybridization for rapid amplification. These findings together demonstrate the ability of RDPCR to rapidly detect DNA-linked peptide substrates of protease enzymes.
In conclusion, we have developed and validated RDPCR as a new, entirely solution-phase method for the selective amplification of DNA sequences encoding molecules that undergo bond formation or bond cleavage.(9) By obviating the need to perform solid-phase capture, washing, and elution steps, RDPCR can greatly streamline the selection process for applications such as DNA-encoded reaction discovery and protease activity profiling. Compared with the performance characteristics of previous in vitro selection methods,3c,5a the data above suggest that RDPCR may also offer superior enrichment factors (signal:background ratios). In addition, RDPCR may be applicable to the evolution of ribozymes and DNAzymes that catalyze bond-forming or bond-cleaving reactions including those that do not generate nucleic acid-like products.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and the NIH/NIGMS (R01GM065865). D.J.G. gratefully acknowledges a joint American Cancer Society-Canary Foundation postdoctoral fellowship and research support from the Kavli Foundation. We thank Dr. Yinghua Shen for assistance with mass spectrometry.
Supporting Information Available
Experimental procedures and compound characterization data. This material is available free of charges at http://pubs.acs.orgThis material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Seminal reports:; a Tuerk C.; Gold L. Science 1990, 249, 505. [DOI] [PubMed] [Google Scholar]; b Ellington A. D.; Szostak J. W. Nature 1990, 346, 818. [DOI] [PubMed] [Google Scholar]; c Robertson D. L.; Joyce G. F. Nature 1990, 344, 467. [DOI] [PubMed] [Google Scholar]; For a general review, see:; d Wilson D. S.; Szostak J. W. Annu. Rev. Biochem. 1999, 68, 611. [DOI] [PubMed] [Google Scholar]
- Recent reviews of evolved DNA/RNA aptamers:; a Shamah S. M.; Healy J. M.; Cload S. T. Acc. Chem. Res. 2008, 41, 130. [DOI] [PubMed] [Google Scholar]; b Gopinath S. H. B. Anal. Bioanal. Chem. 2007, 387, 171. [DOI] [PubMed] [Google Scholar]
- In vitro selections of DNA-linked small molecules:; a Wrenn S. J.; Weisinger R. M.; Halpin D. R.; Harbury P. B. J. Am. Chem. Soc. 2007, 129, 13137. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Melkko S.; Zhang Y.; Dumelin C. E.; Scheuermann J.; Neri D. Angew. Chem., Int. Ed. 2007, 46, 4671. [DOI] [PubMed] [Google Scholar]; . Mock selections:; c Doyon J. B.; Snyder T. M.; Liu D. R. J. Am. Chem. Soc. 2003, 125, 12372. [DOI] [PubMed] [Google Scholar]; d Gartner Z. J.; Tse B. N.; Grubina R.; Doyon J. B.; Snyder T. M.; Liu D. R. Science 2004, 305, 1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Silverman S. Chem. Commun. 2008, 3467. [DOI] [PubMed] [Google Scholar]; b Joyce G. F. Annu. Rev. Biochem. 2004, 73, 791. [DOI] [PubMed] [Google Scholar]
- a Kanan M. W.; Rozenman M. M.; Sakurai K.; Snyder T. M.; Liu D. R. Nature 2004, 431, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Rozenman M. M.; Liu D. R. ChemBioChem 2006, 7, 253. [DOI] [PubMed] [Google Scholar]; c Rozenman M. M.; Kanan M. W.; Liu D. R. J. Am. Chem. Soc. 2007, 129, 14933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasow T. M.; Tarasow S. L.; Eaton B. E. Nature 1997, 389, 54. [DOI] [PubMed] [Google Scholar]; b Seelig B.; Jaschke A. Chem. Biol. 1999, 6, 167. [DOI] [PubMed] [Google Scholar]; Other tags may be used. For selected examples, see:; c Chandra M.; Silverman S. K. J. Am. Chem. Soc. 2008, 130, 2936. [DOI] [PubMed] [Google Scholar]; d Pradeepkumar P. I.; Hobartner C.; Baum D. A.; Silverman S. K. Angew Chem., Int. Ed. 2008, 47, 1753. [DOI] [PubMed] [Google Scholar]; e Johnston W. K.; Unrau P. J.; Lawrence M. S.; Glasner M. E.; Bartel D. P. Science 2001, 292, 1319. [DOI] [PubMed] [Google Scholar]
- a Breaker R. R.; Joyce G. F. Chem. Biol. 1994, 1, 223. [DOI] [PubMed] [Google Scholar]; b Sheppard T. L.; Ordoukhanian P.; Joyce G. F. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 7802. [DOI] [PMC free article] [PubMed] [Google Scholar]; For an alternative approach, see:; c Hobartner C.; Pradeepkumar P. I.; Silverman S. K. Chem. Commun. 2007, 2255. [DOI] [PubMed] [Google Scholar]
- a Rozenman M. M.; McNaughton B. R.; Liu D. R. Curr. Opin. Chem. Biol. 2007, 11, 259. [DOI] [PubMed] [Google Scholar]; See also:; b Wrenn S. J.; Harbury P. B. Annu. Rev. Biochem. 2007, 76, 331. [DOI] [PubMed] [Google Scholar]
- Note that in contrast with a conventional selection in which unfit library members are discarded and sequences encoding surviving members are replicated in a subsequent step, RDPCR achieves the selective replication of only those sequences encoding desired library members.
- Ogawa A.; Maeda M. Biorg. Med. Chem. Lett. 2007, 17, 3156. [DOI] [PubMed] [Google Scholar]
- Computation carried out using the Oligonucleotide Modeling Platform (OMP, DNA Software, Inc
- For examples of phosphodiester bond formation-dependent PCR, see:; a Bartel D. P.; Szostak J. W. Science 1993, 261, 1411. [DOI] [PubMed] [Google Scholar]; b Makrigiorgos G. M. Human Mut. 2004, 23, 406. [Google Scholar]; c Troutt A. B.; McHeyzer-Williams M. G.; Pulendran B.; Nossal G. J. V. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For a general review, see:Kubista M.; Andrade J. M.; Bengtsson M.; Forootan A.; Jonák J.; Lind K.; Sindelka R.; Sjöback R.; Sjögreen B.; Strömbom L.; Ståhlberg A.; Zoric N. Mol. Asp. Med. 2006, 27, 95. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Liu D. R. J. Am. Chem. Soc. 2001, 123, 6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck R. F. Org. React. 1982, 27, 345.See also ref 5a. [Google Scholar]
- As little as 10 fmol of 9d and 9b could be carried through the process with similar results, suggesting that selections can be performed on a very small scale.
- For approaches to protease substrate profiling by derivitization of free α-amines, see:; a Mahrus S.; Trinidad J. C.; Barkan D. T.; Sali A.; Burlingame A. L.; Wells J. A. Cell 2008, 134, 866. [DOI] [PMC free article] [PubMed] [Google Scholar]; b McDonald L.; Robertson D. H. L.; Hurst J. L.; Beynon R. J. Nat. Methods 2005, 2, 955. [DOI] [PubMed] [Google Scholar]; c Gevaert K.; Goethals M.; Martens, Van Damme J.; Staes A.; Thomas G. R.; Vandekerckhove J. Nat. Biotechnol. 2003, 21, 566. [DOI] [PubMed] [Google Scholar]
- Substrate designed in consultation with Sigma-Aldrich Protease finder (http://sigma-aldrich.com/proteasefinder
- DNA-templated amide bond formation with DMT-MM:; a Li X. Y.; Gartner Z. J.; Tse B. N.; Liu D. R. J. Am. Chem. Soc. 2004, 126, 5090. [DOI] [PubMed] [Google Scholar]; With EDC/sNHS:; b Gartner Z. J.; Kanan M. W.; Liu D. R. Angew. Chem., Int. Ed. 2002, 41, 1796. [DOI] [PubMed] [Google Scholar]; For a review of DTS, see:; c Li X. Y.; Liu D. R. Angew. Chem., Int. Ed. 2004, 43, 4848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.