Abstract
Molecular chaperones are known to be involved in many cellular functions, however, a detailed and comprehensive overview of the interactions between chaperones and their cofactors and substrates is still absent. Systematic analysis of physical TAP-tag based protein–protein interactions of all known 63 chaperones in Saccharomyces cerevisiae has been carried out. These chaperones include seven small heat-shock proteins, three members of the AAA+ family, eight members of the CCT/TRiC complex, six members of the prefoldin/GimC complex, 22 Hsp40s, 1 Hsp60, 14 Hsp70s, and 2 Hsp90s. Our analysis provides a clear distinction between chaperones that are functionally promiscuous and chaperones that are functionally specific. We found that a given protein can interact with up to 25 different chaperones during its lifetime in the cell. The number of interacting chaperones was found to increase with the average number of hydrophobic stretches of length between one and five in a given protein. Importantly, cellular hot spots of chaperone interactions are elucidated. Our data suggest the presence of endogenous multicomponent chaperone modules in the cell.
Keywords: chaperone modules, chaperone networks, protein folding, TAP-tag
Introduction
Molecular chaperones are defined as a group of highly interactive proteins that modulate the folding and unfolding of other proteins, or the assembly and disassembly of protein–protein, protein–DNA, and protein–RNA complexes (Hartl and Hayer-Hartl, 2002; Deuerling and Bukau, 2004; Saibil, 2008). In addition, chaperones are known to be involved in many cellular processes and pathways such as protein translocation across membranes, ribosomal RNA processing, and endoplasmic reticulum associated protein degradation (ERAD). Most chaperones are members of the heat-shock regulon and this seems to be a generally conserved feature for most known molecular chaperones. Chaperones are usually categorized into distinct groups according to their sequence similarity, which also reflects their distinct functions. A well-studied and well-defined model organism like Saccharomyces cerevisiae (budding yeast) has seven small heat-shock proteins, three chaperones of the AAA+ family, eight proteins of the CCT/TRiC complex, six proteins of the prefoldin/GimC complex, 22 Hsp40s, one Hsp60, 14 Hsp70s, and two Hsp90s (Supplementary Table S1) (Sghaier et al, 2004). Hsp10 acts mainly as a cofactor for Hsp60 and, hence, is not considered as a chaperone in our analysis (but its interactors are listed in Supplementary Table S2).
Earlier studies on molecular chaperones have largely focused on the detailed biochemical and biophysical analysis of a single or a group of closely related chaperones. Some efforts have also recently been made toward elucidating a system's view for some chaperones (for example refer to (Kerner et al, 2005; Albanese et al, 2006; Dekker et al, 2008)). To this end, we recently presented a comprehensive physical and genetic analysis of the Hsp90 chaperone interaction network, showing the broad role for Hsp90 in many cellular pathways and complexes (Zhao et al, 2005). To obtain a global view of the entire chaperone network and to gain insights into the rules that govern chaperone-mediated protein folding processes inside the cell, we undertook a further comprehensive analysis of the physical interaction network of all 63 chaperones in yeast. This study showed the interplay of specificity and promiscuity of chaperone interactions with their substrates and elucidated the modular organization of the yeast chaperoning system.
Results
Overview of chaperones of S. cerevisiae and their interactors
The types, names, subcellular localization, essentiality, and molecular weight of all 63 yeast chaperones are listed in Supplementary Table S1 and are also charted in Figure 1. The collected information is mainly derived from the Saccharomyces Genome Database (SGD) (Nash et al, 2007) and published literature (Supplementary Table S1). In the subsequent analysis, we grouped together the chaperones that are functionally identical, namely Hsp90 (Hsp82 and Hsc82) or that form well-established stable complexes, namely the prefoldins (PFD, with subunits Yke2, Gim3, Gim4, Gim5, Pac19, and Pfd1) (Vainberg et al, 1998) and CCT (with subunits Tcp1 and Cct2–8) (Tang et al, 2007) (Supplementary Figure S1), hence, resulting in 50 chaperones/chaperone complexes.
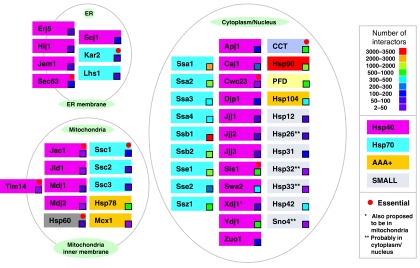
Figure 1.
Overview of chaperones and their interactor numbers. 50 chaperones/chaperone complexes in yeast are shown arranged according to their subcellular localization. The essentiality and types of chaperones are indicated together with the number of identified interactors.
The presence of molecular chaperones in complexes obtained from the tandem affinity purification of 4562 different, endogenously TAP-tagged proteins in yeast cells was determined by mass spectrometry. Each preparation was analyzed by both liquid chromatography tandem mass spectrometry (LC-MS/MS) and by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF). Confidence scores for the interactions were calculated as described earlier (Krogan et al, 2006) (see Materials and methods and Supplementary Figure S2). A total of 21 687 unique pairs of interactions were identified as high confidence (Supplementary Table S2). These interactions are between 63 chaperones and a total of 4340 other proteins; in addition, there are 259 chaperone–chaperone interactions. There are 711 pairs of reciprocal interactions; 653 are chaperone–protein interactions and 58 are chaperone–chaperone interactions. All of our data is deposited in a publicly available and searchable database that we created and termed ChaperoneDB (http://chaperonedb.ccbr.utoronto.ca/).
We expect that most of the chaperone interactors represent putative substrates whose conformational stability is modulated by molecular chaperones although a very small proportion represent cofactors or co-chaperones that modulate chaperone activity. It should be emphasized that the interactions presented are indirect TAP-tag based interactions and not direct binary interactions.
For the 63 yeast chaperones, the numbers of non-chaperone protein interactors vary greatly, ranging from a minimum of two for Hsp32 to a maximum of 3269 for Ssb1 (Figure 1 and Supplementary Table S1). On the basis of this, we divided the yeast chaperones into two groups: specific chaperones, which have <200 non-chaperone interactors per chaperone, and promiscuous chaperones with more than 200 non-chaperone interactors per chaperone. According to our classification, the specific chaperones are (ordered from low to high number of interactors): Hsp32, Sno4, Hsp33, Tim14, Mdj2, Jac1, Cwc23, Jid1, Mcx1, Mdj1, Jjj2, Hlj1, Jjj3, Erj5, Ssc3, Kar2, Lhs1, Hsp26, Jem1, Jjj1, Xdj1, Hsp12, Hsp60, Ssc2, Hsp31, Zuo1, Sec63, Apj1, Scj1, Djp1, and Ssc1. The promiscuous chaperones are: Caj1, Sse2, Swa2, Ssa3, Hsp104, Hsp42, Ssa4, Sis1, Ssz1, Hsp78, Ydj1, CCT, PFD, Ssb2, Hsp90, Ssa2, Sse1, Ssa1, and Ssb1. Interestingly the specific chaperones are mostly present in the ER and mitochondria with the exception of Hsp78, whereas promiscuous chaperones are mostly found in cytoplasm/nucleus. It should be emphasized that more abundant chaperones, whose expression levels have been experimentally determined (Ghaemmaghami et al, 2003; Newman et al, 2006), do not necessarily interact with more proteins as no such correlation was observed (data not shown).
Physical, structural, and functional properties of chaperone interactors
We found that many proteins (828) interact with only one chaperone and this occurs for 33 chaperones/chaperone complexes (Supplementary Table S3). In other words, these proteins have very high specificity for particular chaperones whereas other proteins interact with multiple chaperones (Supplementary Table S3). For example, in the most extreme case Hca4, a putative nucleolar DEAD box RNA helicase, and Rrp5, an RNA binding protein component of both the ribosomal small subunit processosome and the 90S preribosome, interact with 25 chaperones (Supplementary Table S3).
To determine if there are any physical characteristics that influence the number of chaperones that a protein interacts with, we investigated the relationship between the number of interactions and the following molecular properties: molecular weight (MW), peptide length, isoelectric point (pI), GRAVY score, aromaticity score, and the percentage of a given amino acid in the protein sequence. Protein interactors were grouped according to the number of interacting chaperones. For each group, the average value of a given physical property was calculated and plotted against the number of interacting chaperones. The Spearman's rank correlation coefficient (SCC) was used to describe the relationship between the physical property and the number of interacting chaperones. The trends observed are shown in Supplementary Figure S3. Proteins that have the following properties tend to interact with more chaperones (SCC >0.1, red frame): having greater molecular weight or sequence length, or being enriched with Asp, Glu, or Lys residues. However, proteins sharing the following properties tend to interact with fewer chaperones (SCC <−0.1, green frame): having greater aromaticity or GRAVY scores, or being enriched with Cys or Phe residues. However, if the length of a hydrophobic stretch is considered and the numbers of such stretches are counted, then it is found that, per protein, the average number of hydrophobic stretches of length between one and five increases with the number of interacting chaperones (Supplementary Figure S4). This correlation is statistically significant (SCC >0.1, red frame). We also investigated whether structurally disordered proteins are more likely to interact with more chaperones by using the disorder prediction software DisEMBL (Linding et al, 2003), but no significant trends were found.
Longer proteins are expected to consist of multiple structural domains and, therefore, either interact with more chaperones or require more chaperones to assist in their folding. Our experimental observation is consistent with a recent bioinformatic analysis of published pair-wise interaction data that reached a similar conclusion (Hegyi and Tompa, 2008). On the other hand, proteins with high average hydrophobicity were found to interact with fewer chaperones, however, more chaperones are found to interact with proteins that have a larger number of hydrophobic stretches of length between one and five. This seems to indicate that a larger number of hydrophobic stretch in a given protein is a better determinant for chaperone interaction rather than the overall hydrophobicity of the protein. This might also reflect the possibility that proteins with more overall hydrophobicity require specific chaperones rather than a larger number of chaperones or that such proteins tend to be membrane associated/integrated and, hence, could be sequestered from chaperone interactions. However, it should be noted that the total number of integral membrane proteins in our dataset is only 410 out of 4340 proteins.
Potential specificity was observed for chaperone interactions with certain structural protein domains using the SCOP database (Andreeva et al, 2008). For example, proteins in the structure class ‘membrane and cell surface proteins and peptides' are enriched in interactors of few chaperones (one or two), whereas proteins in other categories are enriched in interactors of seven and more chaperones (Supplementary Figure S5A). Enrichment in SCOP folds is given in Supplementary Table S4. The most highly enriched folds that interact with 17 or more chaperones seem to be primarily related to DNA binding and modulation such as: ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase, OB-fold, F-box domain, Type II DNA topoisomerase, DNA breaking-rejoining enzymes, Eukaryotic DNA topoisomerase I or N-terminal DNA-binding fragment (Supplementary Table S4).
Consistent with the finding that more chaperones interact with longer proteins (Supplementary Figure S3), the number of interacting chaperones was found to increase with the number of Pfam domains (Finn et al, 2006) in a given protein (Supplementary Figure S5B).
Supplementary Table S5 provides an overall view of the average molecular weight, pI, length, GRAVY score, and aromaticity score for the interactors of each chaperone/chaperone complex. The top ten Pfam domains and GO biological processes are also listed. Inspection of Supplementary Table S5 indicates a wide variability in the properties of interactors of the different chaperone systems.
The stability of chaperone interactors
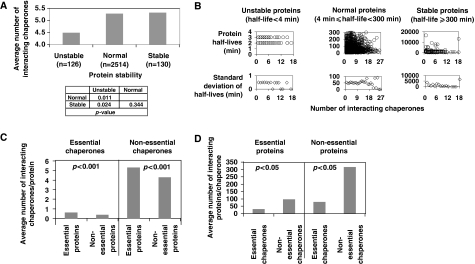
We next investigated whether there is any potential correlation between protein stability, as measured by protein half-life in vivo, and the number of interacting chaperones. For this purpose, a recently published dataset of protein half-lives of 3751 yeast proteins measured in vivo in log phase at 30°C was used (Belle et al, 2006). Proteins were grouped into three categories based on their half-lives: ‘stable' proteins (half-lives ⩾300 min), ‘normal' proteins (half-lives ⩾4 and <300 min), and ‘unstable' proteins (half-lives <4 min). A fourth group of proteins with negative detected half-lives and artificially assigned a half-life of 300 min by Belle et al was not used in our analysis. The number of interacting chaperones for each protein in these three groups were counted and compared, and, as the distribution is not normal, we used the non-parametric Mann–Whitney test to calculate the statistical significance. Figure 2A shows that, on average, proteins of the ‘normal' and ‘stable' groups interact with more chaperones than those of the ‘unstable' group. However, as shown in Figure 2B, there is no direct correlation between protein half-lives and the number of interacting chaperones whether using the ‘stable', ‘normal', or ‘unstable' group of proteins. This might be an important indication that, although chaperones could affect the in vivo stability of proteins, the degradation machinery or chaperone cofactors might play a more prominent role in dictating the determination of protein half-lives in the cell.
Figure 2.
Characterization of chaperone interactors. (A) The average number of chaperones interacting with proteins of different half-lives. (B) Top panels show a plot of protein half-lives versus the number of interacting chaperones for the ‘unstable', ‘stable', and ‘normal' groups of proteins. Lower panels show the s.d. of protein half-lives versus the number of interacting chaperones. (C) A comparison of the average number of interacting chaperones per protein on the basis of essentiality. (D) A comparison of the average number of interacting proteins per chaperone on the basis of essentiality. In (A, C, D) statistical significance is on the basis of the Mann–Whitney test.
It is interesting to note that, on average, essential yeast proteins interact with more essential or nonessential chaperones than nonessential proteins (Figure 2C). This observation suggests that more chaperone resources are spent to insure the proper folding of essential proteins. However, nonessential chaperones have more essential or nonessential protein interactors than essential chaperones (Figure 2D). This observation suggests that essential chaperones might work as specialists as they are required to assist the folding of specific proteins, whereas non-essential chaperones are generalists that interact with a large number of proteins as suggested by the number of interactors in Figure 1.
Cellular hot spots of chaperone interactions
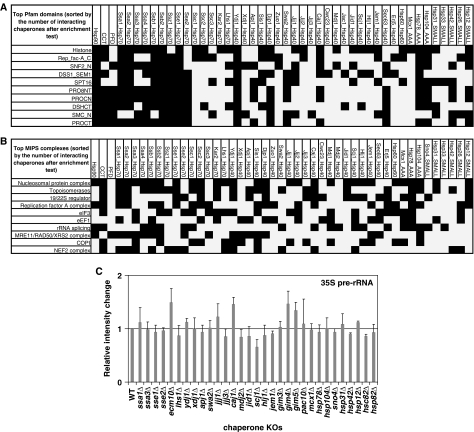
Functional analysis of chaperone interactors was carried out using Gene Ontology annotations GO slim tools (Ashburner et al, 2000) for all three ontology categories, namely: cellular component, molecular function, and biological process. Strikingly, we found a clear trend that the further the proteins are located from the nucleus, the fewer the number of chaperones they interact with (Supplementary Figure S6A). For example, a protein found in ER or near plasma membrane interacts, on average, with one or two chaperones whereas a protein localized in the nucleus or nucleolus interacts, on average, with more than seven chaperones (Supplementary Figure S6A). This observation also holds true for molecular functions (Supplementary Figure S6B) and biological processes (Supplementary Figure S6C) related to different cellular compartments. This seems to indicate that the cell spends more chaperone resources in maintaining the conformational integrity of proteins near or close to the nucleus or that such proteins encounter more stresses during their lifetime in the cell as many cellular processes including ribosome biogenesis, DNA metabolism, and protein transport occur in or near the nucleus. Consistent with these observations, we found that Pfam domains related to nuclear activities are highly enriched for chaperone interactions (Figure 3A and Supplementary Table S6) calculated based on the enrichment test as described in Materials and methods. Significantly, we found domains corresponding to the N-terminus (PRO8NT), central (PROCN), and C-terminus (PROCT) of the nucleolar pre-mRNA splicing factors of the PRO8 family (Staub et al, 2004) are among the top ten Pfam domains enriched for chaperone interactors.
Figure 3.
Interaction of chaperones with Pfam domains and protein complexes. (A) The top ten Pfam domains enriched in chaperone interactors. (B) The top ten MIPS complexes enriched in chaperone interaction. In (A) and (B), a hypergeometric distribution was assumed to assess enrichment (refer to Materials and Methods). (C) Results of the northern blot analyses for 35S pre-rRNA from 31 different chaperone knockout strains grown at 30°C in YPD media for 3 days. Data shown are averages of three repeats. Bands were normalized to the band corresponding to U2 RNA and then divided by the corresponding value for the wildtype (Zhao et al, 2008).
We also determined whether any protein complexes are ‘hot spots' for chaperone interactions using the manually curated yeast complexes from the MIPS database (Mewes et al, 2006). Among the 215 MIPS complexes containing at least two distinct subunits, 199 interacted with at least one chaperone (Supplementary Table S7). Figure 3B shows the top ten complexes interacting with the maximum number of chaperones based on the enrichment test (see Materials and Methods). As discussed above, there is an over-representation of complexes related to nuclear functions such as nucleosomal protein complex, topoisomerases, and rRNA splicing (Figure 3B).
We had recently shown that Hsp90 affects box C/D snoRNA accumulation and maintenance, especially in stationary phase cells, resulting in an effect of the chaperone on rRNA processing (Zhao et al, 2008). As the data in the current analysis suggest that the rRNA splicing complex is a hot spot for chaperone interactions, northern blot experiments were carried out to determine the effect of chaperone deletion on 35S rRNA processing in stationary-phase cells grown at 30°C. As shown in Figure 3C, the single deletion of several molecular chaperones affects rRNA processing resulting in the accumulation of 35S rRNA.
Interactions between chaperones
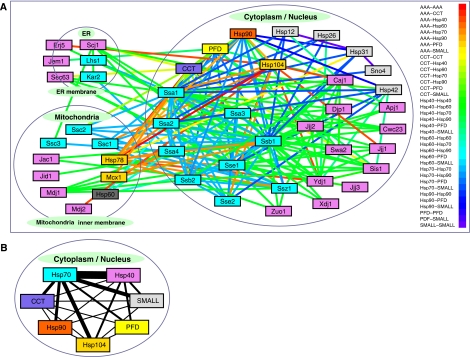
In addition to interactions of chaperones with other proteins, we also detected extensive TAP-tag protein–protein interactions between different chaperones (Figure 4A and Supplementary Table S2), suggesting the prevalence of functional cooperation, physical association, or functional redundancy between individual chaperones. There are 195 such chaperone–chaperone interactions between the 50 chaperones/chaperone complexes. Figure 4B shows the network for cytoplasmic/nuclear chaperones after grouping the chaperones into their different families, with the thickness of the edges indicating the number of interactions between any two groups. The large number of interactions between the Hsp70s and the Hsp40s reflects the large number of chaperones belonging to these two families (Figure 1 and Supplementary Table S1) that interact with each other. However, the Hsp70s have a larger number of interactions with Hsp104, Hsp90, CCT, PFD, and SMALL than the Hsp40s. If we assume that the frequency of interactions between two groups of chaperones indicates the degree of functional overlap, then the networks shown in Figure 4A and B are generally consistent with the current understanding of how molecular chaperones cooperate in maintaining the proper folding of substrate proteins (Young et al, 2004). The Hsp70s are thought to be able to mediate the transfer of substrates to other chaperone systems, whereas Hsp40s are generally thought to act together with Hsp70s, but not directly with other chaperones.
Figure 4.
Chaperone interaction network on the basis of TAP-tag interactions between chaperones. (A) Chaperone interaction network on the basis of chaperone–chaperone TAP-based interactions. (B) Same as the network shown in (A) but for cytoplasmic chaperones grouped into seven families. The thickness of the edges represents the number of interactions between the members of the families. Source data is available for this figure at www.nature.com/msb.
Shared interactors between chaperones
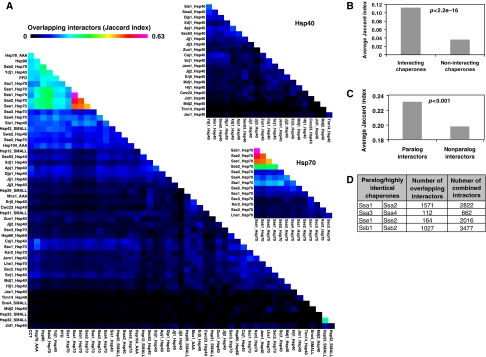
The numbers of shared interactors between chaperones were calculated to determine the functional coordination or redundancy between different chaperone systems. Figure 5A shows the overlap in interactors between chaperones as represented by the Jaccard indices (Tan et al, 2005) (refer to Materials and Methods). CCT, PFD, Hsp90, and several Hsp70s share many common interactors, suggesting functional collaboration among these chaperones. Indeed, a functional collaboration between mammalian CCT and Hsp70 has been suggested recently on the basis of the structural analysis (Cuellar et al, 2008). Furthermore, there is a strong overlap between interactors of the Hsp70 chaperones, namely: Ssa1, Ssa2, Ssb1, Ssb2, and Sse1, with the strongest overlap between Ssa1 and Ssb1 (Figure 5A), suggesting a very strong cooperativity among Hsp70 members (James et al, 1997). Figure 5B shows that interacting chaperones share more interactors than non-interacting chaperones further supporting the functional overlap or redundancy of chaperones found in the same complexes.
Figure 5.
Interactor overlap among chaperones. (A) A heat map highlighting the overlap of non-chaperone interactors among the 50 chaperones/chaperone complexes using the Jaccard indices. (B) Comparison of the number of interacting chaperones versus non-interacting chaperones on the basis of the Jaccard index derived from several non-chaperone interactors. (C) The relationship between paralog and non-paralog interactors on the basis of chaperone overlap. (D) Shown are the numbers of interactor overlap between paralog/highly identical chaperones. The identities between chaperones in each pair are as follows: Ssa1–Ssa2 98.2%, Ssa3–Ssa4 86.9%, Sse1–Sse2 77.2%, and Ssb1–Ssb2 99.4%. In (B and C) statistical significance is on the basis of the Mann–Whitney test.
Many yeast genes have duplicated paralogs, which are thought to be the result of whole genome or small-scale duplication events (Kellis et al, 2004). Paralogs can be identified on the basis of either genomic synteny or sequence similarity. It was reported earlier that about half of the yeast paralogs tend to be co-clustered into the same protein complexes (Musso et al, 2007). Consistent with this, paralog proteins share more common chaperones than non-paralog proteins (Figure 5C). The numbers of interactors overlapping between paralog or highly identical chaperones are given in Figure 5D. Ssa1/Ssa2 and Ssb1/Ssb2 share the highest percentage of interactors probably indicating functional redundancy or perhaps functional cooperativity.
We next asked whether subcellular localization of the chaperones or of the protein substrates can have any influence on their interactions. We found, in general, chaperones in the cytoplasm and nucleus share many more interactors than those in the ER and mitochondria (Supplementary Figure S7). However, both Scj1 in the ER and Hsp78 in the mitochondria share a considerable number of interactors with cytoplasmic chaperones. For example, Scj1 shares 143 and 120 non-chaperone interactors with Ssa1 and Ssa2, respectively, whereas Hsp78 shares 135, 253, 351, 128, 725, and 550 non-chaperone interactors with Hsp42, CCT, Hsp90, Hsp104, Ssb1, and Ssa2, respectively. This suggests that Scj1 and Hsp78 might play a major role in mediating the translocation of proteins from the cytoplasm to the respective organelles.
Chaperone functional modules
The network shown in Figure 4, which is based on TAP-tag interactions between individual chaperones/chaperone complexes, is very similar to the networks shown in Figure 5A and Supplementary Figure S7, which are based on the shared interactors between individual chaperones. In fact, as already shown in Figure 5B, chaperones interacting with each other share, on average, significantly more protein interactors than non-interacting chaperones. This suggests that a TAP-tag interaction between two chaperones reflects a functional relationship or redundancy between them. Such two chaperones might form what we term a functional module.
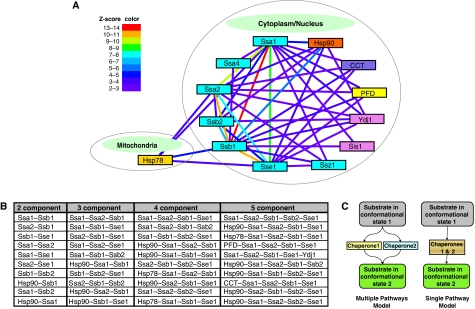
To first elucidate significantly important two-chaperone relationships based on interactor overlap, the numbers of shared interactors for each pair of chaperones were counted and a Z-score was calculated to determine enrichment for common interactors (see Materials and methods). Chaperone pairs with a Z-score greater than or equal to two were considered to form a statistically significant functional module. Between the 50 chaperones/chaperone complexes, 41 out of a total of 1079 chaperone pairs with overlapping interactors were found to be significant (Figure 6A, B and Supplementary Table S8). Most of the modules were found in the cytoplasm/nucleus, but four modules were found between Ssa1, Ssa2, Ssb1 and Sse1, which are located in the cytoplasm/nucleus, and the mitochondrial chaperone Hsp78. It is interesting to note that 38 out of the 41 chaperone modules are also found in the 195 chaperone–chaperone TAP-based interactions shown in Figure 4A.
Figure 6.
Chaperone modules based on interactor overlap. (A) Two-component chaperone modules are shown. Two chaperones in a module are linked with an edge. Edge color represents significance on the basis of the Z-score derived from interactor overlap. (B) A list of the top ten chaperone modules containing 2–5 components. The modules are sorted from top to bottom on the basis of Z-scores. For each module, chaperones are listed alphabetically. (C) The schematics for the single pathway and multiple pathways models. Source data is available for this figure at www.nature.com/msb.
As it is known that more than two chaperones might be involved in mediating the correct folding or stability of substrate proteins, significant modules composed of 3–5 individual chaperones were also derived using a similar procedure as for the two-component modules. The most frequently observed larger modules contain the chaperones of the Hsp40 and Hsp70 family members or of the Hsp90 and Hsp70 members (Figure 6B and Supplementary Table S9). This is consistent with our current detailed understanding of the biochemistry of chaperone function: Hsp40s are known to activate the ATPase of Hsp70s (Hennessy et al, 2005), whereas Hsp70s are known to transfer substrates to the Hsp90s (Pearl and Prodromou, 2006).
For two-component chaperone modules, we then addressed the question of whether the two chaperones in the module act along a single folding pathway of a given substrate protein or whether the substrate protein has two different folding pathways that the chaperones act on independently (Figure 6C). We call the former model, the ‘single pathway model', and we call the latter, the ‘multiple pathways model' (Figure 6C). Such an analysis would allow us to gain insights into whether chaperone modules evolved to facilitate multiple folding pathways for proteins. To do this, it is reasonable to assume that chaperone-mediated folding pathways have similar features as enzyme-mediated metabolic pathways. Hence, pathway information was extracted from the KEGG database (Kanehisa et al, 2008) and the gene co-expression data measured by microarrays were used to provide potential indicators of pathway relationship of two proteins (see Materials and Methods). Such an analysis was carried out only on two-component chaperone modules as gene co-expression can readily be computed for such cases. Three models were considered: single pathway, multiple pathways, and both single and multiple pathways models.
As shown in Supplementary Table S10, the probability for a two-component chaperone module to act on multiple folding pathways for a given substrate protein is the highest for all chaperone pairs (ranging from 0.44 to 0.58), whereas the probability of acting on a single folding pathway only is lower (ranging from 0.17 to 0.35). The probability of being both is relatively constant (about 0.2). Thus, in general, two-component chaperone modules are most likely to function in multiple pathways, but for several chaperone pairs, such as Ssa2–Hp90, Sse1–Hsp90, Ssa2–Sse1, Ssa1–Ssa2, Ssa1–Hsp90, Ydj1–Ssa2, Ssa1–Ssa4, Ssa1–Sse1, and Sis1–Ssa1, the probability of functioning in single pathway is also considerable (P>0.3). These results seem to indicate that although two-component chaperone modules have generally evolved to facilitate multiple folding pathways for proteins, two-component modules containing Hsp90, one of four Hsp70s (Ssa1, Ssa2, Ssa4, and Sse1), or one of two Hsp40s (Ydj1 and Sis1) are also important to ensure proper folding along a single pathway.
Discussion
Considerations
The data described in this study rely on a single proteomic approach based on TAP-tag pulldowns. In most cases, these interactions reflect the binding between a given chaperone and a protein complex, rather than a direct binary interaction. The TAP-tag protocol employed in our study involves two stringent washing steps in which the interacting proteins are purified on two different columns (refer to Materials and Methods). Subsequently, hits identified by mass spectrometry are further filtered using an experimentally determined confidence score for mass spectrometry data and checked for enrichment for known MIPS complexes (Mewes et al, 2006), which are manually curated from low-throughput experiments only. The data are also checked for enrichment in published interactors using the BioGRID database (Stark et al, 2006). The specificity of the interactions we detect for chaperones is highlighted by the fact that our findings are consistent with several general observations. For example, the Hsp40 chaperone Ydj1 (809 hits in our dataset) is thought to be a more general chaperone than a dedicated Hsp40 chaperone such as ribosome-bound Zuo1 (110 hits) (Sahi and Craig, 2007). Also, as would be expected, chaperones in the ER and mitochondria generally have a lower number of hits than chaperones in the cytoplasm/nucleus (Figure 1).
In some instances cytoplasmic chaperones are found to interact with ER or mitochondrial proteins (Supplementary Table S2) and this could reflect an interaction of the chaperone with the precursor protein as the protein is being translocated into the specific cellular compartment. In other cases, we also found ER or mitochondrial chaperones interacting with cytoplasmic proteins. This might reflect the multiple localization of the protein or chaperone; for example, Xdj1 has been reported to be a mitochondrial as well as a cytoplasmic protein (Figure 1) (Schwarz et al, 1994; Reinders et al, 2006; Sahi and Craig, 2007). Alternatively, the detected interaction might be a false positive hit, possibly occurring post-lysis.
Although it would be difficult to provide an accurate estimate of false positive hits, however, with the filtering approach described above, and based on our earlier, current, and ongoing chaperone interaction studies, we estimate that the fraction of false positives in our current dataset is comparable to that of other large-scale interaction studies reported. By combining the TAP-tag based proteomic approach with other screening approaches such as yeast two-hybrid (2H) methods and genetic interaction analyses, a high fidelity dataset of chaperone interactions should ultimately be obtained. Such an integrative proteomic approach has already been used by us to map the Hsp90 chaperone network (Zhao et al, 2005) and can be used to map the networks for all other yeast chaperones.
Comparison with other available yeast chaperone physical interactor datasets
We compared our chaperone–protein interaction data with interactions documented in publicly available databases such as BioGRID (Stark et al, 2006). Of the 4340 interactions identified in our study, 733 are also reported in BioGRID.
There are two other detailed experimental high throughput interaction datasets available for yeast chaperones. One dataset is from our own laboratory where we analyzed the yeast Hsp90 physical and genetic interactors (Zhao et al, 2005). In that study, we identified 112 and 88 non-chaperone Hsp90 interactors based on TAP-tag and 2H methods, respectively. TAP-tag pulldowns were carried out from WT C-terminally TAP-tagged strains used in the current study as well as from a strain in which yeast HSP82 gene was knocked out and the HSC82 gene was N-terminally TAP-tagged but kept under its endogenous promoter. The 2H screens were carried out using full-length as well as domains of Hsp90 screened against an ordered array of 6084 yeast colonies expressing activation domain fusions to individual ORFs (Zhao et al, 2005). In this study, which is solely based on the TAP-tag pulldown approach, we identify 1144 non-chaperone hits for Hsp90 (Supplementary Table S2). The overlap in interactors of yeast Hsp90 between the current study and our earlier study is 88 non-chaperone interactors (26 for 2H and 69 for TAP). Hence, 29.5 and 61.6% of the 2H and TAP hits that we reported earlier are verified again in this study. We would consider such reproducible hits to be of high fidelity and represent true substrates or cofactors of Hsp90.
Another high throughput interaction dataset is available for the eight-subunit CCT chaperone complex (Dekker et al, 2008). In that study, the authors also use physical and genetic methods to map interactors of the CCT complex. Dekker et al (2008) provide a supplementary table where they list 143 non-chaperone CCT interactors based on co-precipitation criteria. In this study, we identified 639 non-chaperone CCT interactors, no matter which subunit they interact with (Supplementary Table S2). The overlap between our dataset and that of Dekker et al is 36 non-chaperone interactors. Hence, 25.2% of the Dekker et al dataset is reproducible in our study. This is a significant overlap given that our approach is not specifically optimized for CCT pulldowns. The maximum number of newly-synthesized proteins estimated to interact with CCT is about 9–15% of all newly translated proteins (Thulasiraman et al, 1999). Our number of interactors of CCT (about 10% of total yeast proteins) is in line with this estimate.
It is known that the N- and C-termini of the CCT subunits are buried when they are part of the CCT complex (Klumpp et al, 1997; Ditzel et al, 1998; Pappenberger et al, 2002, 2006). Hence, the presence of a C-terminal TAP-tag, although not lethal to the cells, might destabilize the complex. Consistent with this, we found that only when Cct4 was C-terminally TAP-tagged (i.e. Cct4 is used as bait), we recovered Cct2 and Cct6 (Supplementary Table S2). The C-terminal tagging of any of the other CCT subunits did not recover another CCT subunit. Indeed, Dekker et al placed an internal tag in an exposed region in Cct3 and Cct6 subunits to be able to pulldown the full CCT complex. Hence, the interactions that we detect in our procedure for C-terminally TAP-tagged CCT subunits most probably represent strong interactions between a given subunit and the interacting protein that persist even if the CCT complex is disrupted when the cells are lysed and the pulldown procedure is carried out. Furthermore, it has been experimentally found that a substrate protein bound to the CCT complex only engages a defined subset of the CCT subunits (Llorca et al, 1999, 2000); hence, if the C-terminally TAP-tagged CCT complex is unstable, we could expect that only the interaction between those subunits and the substrate protein might be preferentially detected using our pulldown and washing protocols. In this regard, of the most established substrates for CCT, actin (Act1) and tubulin (Tub1–4), only Tub1 (bait) was found to interact with Cct4 (prey), but the interaction did not meet our cutoff criteria and is not listed in Supplementary Table S2. Hence, actin and tubulin might require the full CCT chaperone complex for stable interaction.
Nevertheless, it should be emphasized that 355 out of the total 639 non-chaperone CCT interactions identified in our study are from experiments in which the interacting protein is C-terminally TAP-tagged and not a CCT subunit. Also, it has generally been observed that WD40 domain-containing proteins preferentially interact with CCT (Ho et al, 2002; Pappenberger et al, 2006). Consistent with this, we found that WD40 is the top domain enriched in CCT interactors as it is present in about 10% of those interactors (Supplementary Table S5).
In future pull-down experiments, the protocols have to be individually optimized for each chaperone system. However, the validity of our current generic approach relies on the fact that significant overlaps are obtained with datasets published earlier.
Overall properties of chaperone interactors
Our data provide the first global view of the chaperone interaction network in an eukaryotic model organism, the budding yeast S. cerevisiae. The network that we have built is based on physical interactions derived from TAP-tagging co-purification experiments and provides a comprehensive list of putative substrates and cofactors of all 63 chaperones of yeast. A correlation analysis with the number of interacting chaperones showed several protein properties that influence the propensity of proteins to interact with chaperones: (1) proteins that have a larger number of hydrophobic stretches of length between one and five interact with more chaperones (Supplementary Figure S4); (2) however, more hydrophobic proteins interact with fewer chaperones whereas more hydrophilic proteins interact with more chaperones (Supplementary Figure S3); (3) larger proteins and multi-domain proteins interact with more chaperones than smaller and simpler proteins (Supplementary Figures S3 and S5B); (4) proteins enriched in the charged residues Asp, Glu, and Lys also interact with more chaperones (Supplementary Figure S3).
Our correlation analysis showed that, in general, yeast proteins tend to interact more with non-essential chaperones than with essential chaperones (Figure 2D), which suggests extensive functional redundancy among these non-essential chaperones. Such a correlation could also imply that heavy dependence on the essential chaperones might be avoided during evolution. We also found that essential proteins have more chaperone interactors than nonessential proteins (Figure 2C), which suggests that yeast has evolved more chaperones to help to stabilize these proteins that are important for cell survival.
Analysis of the half-lives of chaperone interactors yielded an important observation (Figure 2B). Our data indicate that the number of chaperone interactors does not correlate with in vivo protein turnover. Hence, although chaperones might work to stabilize proteins, they do not directly affect their half-lives. Hence, the main determinant of protein half-life in the cell might reside with the degradation machinery.
Overall properties of the chaperone interaction networks
Chaperone networks can be drawn based on chaperone–chaperone TAP-tag interactions or based on interactor overlap between the different chaperones. Our analysis shows the presence of differences in the interaction capacity of different chaperone families. Particularly, many substrate-mediated and TAP-based interactions exist within the Hsp70 family members and between Hsp70 and Hsp40 family members, whereas very few interactions were found between the Hsp40 family members (Figures 4A and 5A). Hsp70 family members also interact with many chaperones of the SMALL, CCT, and Hsp90 families (Figure 4B). Most promiscuous chaperones are those present in the cytoplasm/nucleus, whereas most specific chaperones are those of the ER and mitochondria (Figure 1). However, Scj1 of the ER and Hsp78 of the mitochondria seem to play important direct or indirect roles in communicating with the chaperones of the cytoplasm/nucleus (Figure 4 and Supplementary Figure S7). Hence, our analysis suggests that Scj1 and Hsp78 would be the most promiscuous chaperones in their respective cellular compartments.
Molecular chaperones seem to be especially important for the maintenance of protein complexes and pathways that are closely associated with nuclear activities (Figure 3 and Supplementary Figure S6). This strongly indicates an important role for the chaperone systems in maintaining genomic stability and gene expression. Indeed, this might be the major cellular role of chaperones rather than in the folding of newly translated proteins, which is expected to mainly occur in the cell cytoplasm outside the nucleus. However, it should be pointed out that our data do not inform us of what part of the total fraction of a given protein interacts with a given chaperone. Hence, a stronger conclusion in this regard will require quantitative data describing the strength of the reported interactions.
Chaperone modules
Our analysis has revealed the presence of multi-component chaperone modules (Figure 6 and Supplementary Tables S8 and S9), which could either act on single or multiple folding pathways (Figure 6C). From a mechanistic perspective, the formation of a chaperone module could reflect one of three possible experimentally observed models, namely: functional similarity, functional dependence, and functional coupling. In the case of functional similarity, two or more chaperones have very similar or even redundant functions such as binding to the same group of substrates and folding them in a similar but independent fashion. This would correspond to ‘multiple pathways' chaperone modules. Functional dependence means that the chaperones have to form either a stable or transient complex or have to be in close physical proximity to collaborate on substrate folding, transport, or repair. Functional coupling is a situation where the substrate needs different chaperones at different stages of maturation. The latter two models would correspond to ‘single pathway' chaperone modules.
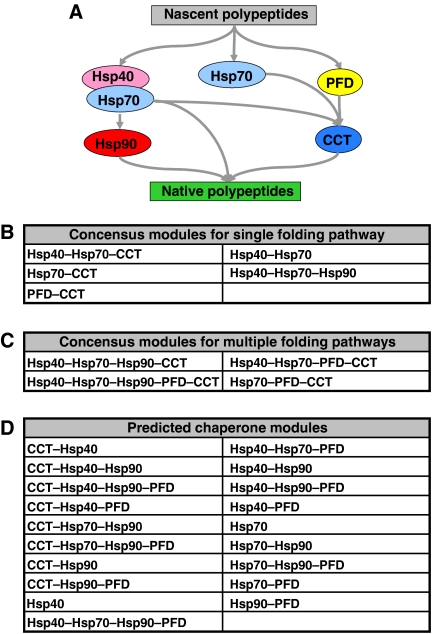
Figure 7A shows a current consensus model for the chaperone-mediated protein folding pathways of newly synthesized proteins in the cell cytoplasm (Young et al, 2004). The consensus model includes only five types of chaperones, namely, Hsp40, Hsp70, Hsp90, PFD, and CCT. From this model, five single pathway modules and four multiple pathways modules can be enumerated (Figure 7B and C). However, if we map our derived multicomponent modules (Supplementary Tables S8 and S9) to this consensus model (after removing Hsp60, AAA, and SMALL because these three types of chaperones were not included in the consensus model), then 28 unique modules are predicted to be present. These 28 modules include the nine modules that have been experimentally observed (Figure 7B and C). Hence, our data predict the presence of another 19 new functional chaperone modules that can be experimentally investigated (Figure 7D). The elucidation of these modules and the determination of the crosstalk between different chaperone systems is essential to understanding protein homeostasis in the cell.
Figure 7.
Chaperone modules in an experimental folding pathway model. (A) A generally accepted experimental model for chaperone-mediated protein folding pathway in the cell cytoplasm (Young et al, 2004). (B) 5 single pathway modules derived from the model. (C) Four multiple pathway modules derived from the model. (D) A list of the 19 new modules predicted to be present in the experimental model on the basis of our data. In (B), (C), and (D), the chaperones in each module are ordered from left to right on the basis of the model in (A).
In conclusion, our comprehensive analysis of the yeast chaperone physical interaction network reflects an attempt to gain a global view of protein folding pathways in the eukaryotic cell. Our described efforts have provided an emerging picture of the rules that govern chaperone-mediated cellular protein folding. On the basis of our data, molecular chaperones can best be described as carrying out a surveillance function aimed at maintaining the proper homeostasis of a multitude of cellular complexes and pathways.
Materials and methods
Biochemical protocols
The purification of endogenously TAP-tagged proteins from cells, grown in rich YPD medium at 30°C to an OD600 of about two, was carried out as described earlier (Zhao et al, 2005). Identification of proteins in the isolated complexes was done by MALDI-TOF or LC-MS/MS (Krogan et al, 2006).
Northern blot analysis of strains deleted for different chaperones was carried out on cells grown at 30°C in YPD as described earlier (Zhao et al, 2008). Bands were quantified using Quantity One (BioRad). All deletion strains were obtained from the yeast gene deletion-mutant collection constructed by the Yeast Deletion Consortium (Winzeler et al, 1999).
Filtering the interaction dataset
The MALDI-TOF dataset contains 38 387 total and 22 791 unique records, consisting of 1980 unique bait proteins (TAP-tagged) and 3394 unique prey proteins. The LC-MS/MS dataset contains 10 846 total and 9096 unique records, with 2745 unique baits and 954 unique preys. This study is part of our ongoing yeast interactome work. In this study, we provide the list of all interactors identified using the TAP-tag approach for all the 63 yeast chaperones.
The confidence scores for LC-MS/MS data were calculated from the mass spectrometry database search scores according to Krogan et al (2006), whereas the MALDI-TOF confidence scores are Z-scores based on the mass spectrometry database search scores. LC-MS/MS data with confidence scores higher than 70% were selected following the procedures published earlier (Krogan et al, 2006). To obtain cutoff of MALDI-TOF confidence scores for the protein–protein interactions involving chaperone proteins, the distributions of the confidence scores for the interactions between subunits in the MIPS complexes were used as a reference (Mewes et al, 2006). The MIPS protein complexes are manually curated from low-throughput experiments only, that is excluding data from high throughput methods such as TAP-tagging or yeast two-hybrid methods. MALDI-TOF data with confidence scores higher than 0 were selected, based on the fact that many interactions between subunits of MIPS complexes have a confidence score between 0 and 1 (Supplementary Figure S2A). Interactions were assigned following a ‘spoke model' (Bader and Hogue, 2002). Using these confidence score cutoffs, we derived a final set of 21 687 interactions from the combined data sets, involving 63 chaperones and 4340 other proteins; in addition, there are 259 chaperone–chaperone interactions. As a confirmation of the cutoffs, potential enrichment of known chaperone interactors as documented in BioGRID (Stark et al, 2006) was tested with hypergeometric distribution. Known interactors were enriched in the above dataset for 34 chaperones (Supplementary Figure S2B).
All our chaperone interaction data is deposited in a publicly searchable database that we termed ChaperoneDB (http://chaperonedb.ccbr.utoronto.ca/). The database has a friendly web interface allowing users to query by either chaperones or by substrates. Users can also bulk download the entire data set. Furthermore, all reciprocal interaction data have been deposited in BioGRID.
Bioinformatics and statistical analyses
Chaperone sequences were extracted from GenBank FTP website (ftp://ftp.ncbi.nih.gov/genbank/) and were compared using the BLAST program (Altschul et al, 1990). E-value cutoff was set to one in the BLAST searches (Supplementary Figure S1) and default values for other parameters were used. Chaperones with very similar sequences (E-value lower than 1e–45) are Hsc82 and Hsp82, CCT subunits TCP-1 and Cct2 through Cct8, and small heat-shock chaperones Hsp32, Sno4, and Hsp31. In addition, several chaperones of the Hsp70 family possess very high sequence similarity: Ssa1, Ssa2, Ssa3, Ssa4, Ssb1, Ssb2, Ssc1, Ssc2, Ssc3, and Kar2. The following pairs of chaperones have been shown to be paralogs following a whole-genome duplication event: Ssa3 and Ssa4, Sse1 and Sse2, Ssb1 and Ssb2, and Hsc82 and Hsp82 (Musso et al, 2007).
To measure interactor overlap between chaperones, the number of shared interactors was calculated and the Jaccard index was then derived:
 |
We used the Mann–Whitney rank–sum test to calculate the significance in the difference between two properties, as the data do not follow normal distribution. This was true for the data in Figures 2A, C, D and 5B, C. The statistical package R (http://www.r-project.org/) was used for most of the statistical analysis including the Spearman's rank correlation coefficient (SCC) for Supplementary Figures S3, S4 and S5B.
To assess the enrichment in nominal features of the interactors, such as Pfam domains (Figure 3A), MIPS complexes (Figure 3B), SCOP classes (Supplementary Figure S5A), and GO terms (Supplementary Figure S6), a hypergeometric distribution (Robinson et al, 2002) was assumed and the enrichment of the nominal features was calculated using the following formula:
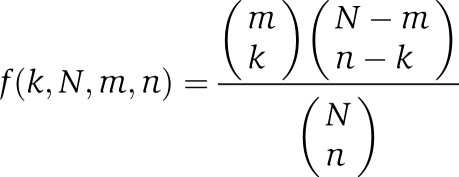 |
In the above equation, N is the number of all interactors, n is the number of all interactors possessing a nominal feature, m is the number of interactors of a group of chaperones, and k is the number of interactors in a group which possesses the nominal feature. Sum of the probability, p, for all i⩾k gives the significance of the interactors with the nominal feature. A p-value <0.05 indicates enrichment of the nominal feature in the group of interactors. We used R function Phyper (http://www.r-project.org/) for this calculation. False discovery rate (FDR) was controlled with adjusted p-value for multiple independent tests (Benjamini and Hochberg, 1995). The mean threshold for the multiple tests was used as a rough FDR (RFDR). The adjusted p-value, p′, was calculated as:
The variable t is the number of independent tests, and α is the threshold, which was set to 0.05.
Identification of chaperone functional modules
Chaperone functional modules were deduced based on the number of overlapping interactors shared between two or more chaperones. For a pair of chaperones, the number of overlapping interactors was counted and a Z-score was then calculated for each combination using the formula:
The variable x is the number of overlaps for two chaperones, $bar{x}$ is the average of all numbers, and σ is the s.d. calculated as:
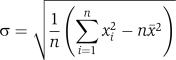 |
n is the total number of possible combinations. Combinations of chaperones with Z greater than two were considered as significant functional modules. This procedure was also applied to other combinations with up to five chaperones.
Analysis of pathway relationships for the two-component chaperone modules
We employed an integration and learning approach to determine whether two chaperones act on a single, multiple, or single and multiple folding pathways. For this analysis, the assumption is made that chaperone-mediated folding pathways have similar features as enzymatic pathways: chaperones working on single pathways have higher probability of being co-expressed, whereas chaperones working on multiple pathways have lower probability of being co-expressed. Hence, a training dataset derived from the biological pathways in KEGG was used, together with the microarray results from Cho et al (1998) (cell cycle) and Gasch et al (2000) (20 environmental conditions). The relationships between gene co-expression and single/multiple/both pathways models were found to be very similar across the experiments. When the Pearson correlation coefficient is lower than 0.6, the probability that the two chaperones act in multiple pathways is much higher than the probability for acting in a single pathway. For Pearson correlation coefficients higher than 0.6, the probability for multiple pathways decreases and the probability for single pathway increases. The probability for the two proteins to be in both remains relatively constant as the value of the Pearson correlation coefficient changes. Subsequently, the probabilities of pathway location for two chaperones across the experiments were integrated using the formula (Hon Nian Chua, National University of Singapore, personal communication):
Where Du,v is the set of expression data sources that contains both chaperones, P(k) is the probability that the two chaperones have a particular pathway relationship determined using KEGG pathways as training dataset, and P is the integrated probability that the two chaperones have a particular pathway relationship. Three probabilities were derived for single, multiple, or single and multiple folding pathways. The final probabilities were calculated by normalization, that is, each probability was divided by the total of the three.
Data visualization
Network graphs were created with CytoScape (Cline et al, 2007). Heat-map diagrams were created using in-house PHP scripts. For some heat-map diagrams, data were clustered beforehand in R.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S7
Supplementary Tables S1-10
Acknowledgments
The authors thank Dr Rongmin Zhao for helpful discussions and Mr Vincent Fong and Mr Ke Jin for assistance with data processing. YK is a postdoctoral fellow of the Canadian Institutes of Health Research Training Program Grant in Protein Folding: Principles and Diseases. This work was supported by grants from the Canadian Institutes of Health Research (MOP-81256) to AE and WAH, and Genome Canada through the Ontario Institute of Genomics to ZZ, JG, and AE.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG (2008) Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res 36: D419–D425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW (2002) Analyzing yeast protein-protein interaction data obtained from different sources. Nat Biotechnol 20: 991–997 [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA 103: 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, Davis RW (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar J, Martin-Benito J, Scheres SH, Sousa R, Moro F, Lopez-Vinas E, Gomez-Puertas P, Muga A, Carrascosa JL, Valpuesta JM (2008) The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat Struct Mol Biol 15: 858–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR (2008) The interaction network of the chaperonin CCT. EMBO J 27: 1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Bukau B (2004) Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol 39: 261–277 [DOI] [PubMed] [Google Scholar]
- Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S (1998) Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93: 125–138 [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34: D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hegyi H, Tompa P (2008) Intrinsically disordered proteins display no preference for chaperone binding in vivo. PLoS Comput Biol 4, e1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci 14: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA (1997) Functional specificity among Hsp70 molecular chaperones. Science 275: 387–389 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220 [DOI] [PubMed] [Google Scholar]
- Klumpp M, Baumeister W, Essen LO (1997) Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell 91: 263–270 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453–1459 [DOI] [PubMed] [Google Scholar]
- Llorca O, Martin-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J 19: 5971–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM (1999) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402: 693–696 [DOI] [PubMed] [Google Scholar]
- Mewes HW, Frishman D, Mayer KF, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V (2006) MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res 34: D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Zhang Z, Emili A (2007) Retention of protein complex membership by ancient duplicated gene products in budding yeast. Trends Genet 23: 266–269 [DOI] [PubMed] [Google Scholar]
- Nash R, Weng S, Hitz B, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Livstone MS, Oughtred R, Park J, Skrzypek M, Theesfeld CL, Binkley G, Dong Q, Lane C, Miyasato S et al. (2007) Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res 35: D468–D471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Pappenberger G, McCormack EA, Willison KR (2006) Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/gamma subunit. J Mol Biol 360: 484–496 [DOI] [PubMed] [Google Scholar]
- Pappenberger G, Wilsher JA, Roe SM, Counsell DJ, Willison KR, Pearl LH (2002) Crystal structure of the CCTgamma apical domain: implications for substrate binding to the eukaryotic cytosolic chaperonin. J Mol Biol 318: 1367–1379 [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271–294 [DOI] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5: 1543–1554 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR (2002) FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahi C, Craig EA (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA 104: 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil HR (2008) Chaperone machines in action. Curr Opin Struct Biol 18: 35–42 [DOI] [PubMed] [Google Scholar]
- Schwarz E, Westermann B, Caplan AJ, Ludwig G, Neupert W (1994) XDJ1, a gene encoding a novel non-essential DnaJ homologue from Saccharomyces cerevisiae. Gene 145: 121–124 [DOI] [PubMed] [Google Scholar]
- Sghaier H, Le Ai TH, Horiike T, Shinozawa T (2004) Molecular chaperones: proposal of a systematic computer-oriented nomenclature and construction of a centralized database. In Silico Biol 4: 311–322 [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Fiziev P, Rosenthal A, Hinzmann B (2004) Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays 26: 567–581 [DOI] [PubMed] [Google Scholar]
- Tan P-N, Steinbach M, Kumar V (2005) Introduction to Data Mining. Boston: Addison-Wesley [Google Scholar]
- Tang YC, Chang HC, Hayer-Hartl M, Hartl FU (2007) SnapShot: molecular chaperones, Part II. Cell 128: 412. [DOI] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J 18: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ (1998) Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93: 863–873 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727 [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, Yip CM, Houry WA (2008) Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol 180: 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S7
Supplementary Tables S1-10