Abstract
Isobutanol has emerged as a potential biofuel due to recent metabolic engineering efforts. Here we used gene expression and transcription network connectivity data, genetic knockouts, and network component analysis (NCA) to map the initial isobutanol response network of Escherichia coli under aerobic conditions. NCA revealed profound perturbations to respiration. Further investigation showed ArcA as an important mediator of this response. Quinone/quinol malfunction was postulated to activate ArcA, Fur, and PhoB in this study. In support of this hypothesis, quinone-linked ArcA and Fur target expressions were significantly less perturbed by isobutanol under fermentative growth whereas quinol-linked PhoB target expressions remained activated, and isobutanol impeded growth on glycerol, which requires quinones, more than on glucose. In addition, ethanol, n-butanol, and isobutanol response networks were compared. n-Butanol and isobutanol responses were qualitatively similar, whereas ethanol had notable induction differences of pspABCDE and ndh, whose gene products manage proton motive force. The network described here could aid design and comprehension of alcohol tolerance, whereas the approach provides a general framework to characterize complex phenomena at the systems level.
Keywords: biofuel, network component analysis, quinone, respiration, transcriptional regulation
Introduction
The recent metabolic engineering of Escherichia coli for higher branched-chain alcohol production has enabled isobutanol to emerge as a potential biofuel (Atsumi et al, 2008). However, this compound is toxic to microorganisms, causing growth retardation at concentrations as low as 1% vol/vol in E. coli. Although production continues long after growth stops (Atsumi et al, 2008), the toxicity is important for the final titer of the product. As this compound was previously considered a minor microbial fermentation product, isobutanol cytotoxicity remains largely uncharacterized. Thus, to facilitate improved isobutanol production, an understanding of how microorganisms respond to isobutanol stress is desirable.
A considerable amount of work has focused on the effects of similar alcohols on microorganisms; namely, ethanol on E. coli and n-butanol on Clostridium acetobutylicum (Ingram, 1976; Dombek and Ingram, 1984; Sikkema et al, 1995; Yomano et al, 1998; Gonzalez et al, 2003; Tomas et al, 2003, 2004; Alsaker et al, 2004; Borden and Papoutsakis, 2007). In general, the cytotoxicity of these alcohols has been attributed to membrane disruption, which is thought to occur by direct insertion of lipophilic side chain into the cellular membrane (Ingram, 1976; Borden and Papoutsakis, 2007). Physiological changes associated with this chaotropic action include increased membrane fluidity (Ingram, 1976; Borden and Papoutsakis, 2007), dissipation of proton motive force (PMF) (Tomas et al, 2004; Jovanovic et al, 2006; Borden and Papoutsakis, 2007; Kobayashi et al, 2007), disruption of protein–lipid interactions (Borden and Papoutsakis, 2007), and inhibition of glucose and nutrient transport (Bowles and Ellefson, 1985). To understand solvent stress, a number of genomic techniques, including gene-expression profiling and genomic libraries, have been used. Using DNA microarray to identify transcriptional differences between an ethanol-tolerant E.coli mutant and its parent, Gonzalez et al (2003) identified three metabolites (glycine, betaine, and serine) and one non-functional transcription factor (FNR) as important for ethanol tolerance in anaerobic E. coli cultures. In C. acetobutylicum, multiple studies involving a genomic library and/or DNA microarray were used to investigate gene overexpressions that conferred increased n-butanol tolerance (Tomas et al, 2003, 2004; Alsaker et al, 2004; Borden and Papoutsakis, 2007). These studies provide useful foundations to analyze solvent toxicity, and highlight the complexity and breadth of the solvent-stress phenotype. Owing to this inherent complexity, we used a systems-biology approach to investigate isobutanol stress.
Here we mapped the initial response network of E. coli to isobutanol using gene expression and transcription factor (TF)–gene interaction data (Gama-Castro et al, 2008), genetic knockouts (Baba et al, 2006), and network component analysis (NCA) (Liao et al, 2003; Tran et al, 2005; Yang et al, 2005; Galbraith et al, 2006). ArcA was identified as the most significantly perturbed TF by NCA, and verified by knockout to be a major regulator of respiratory changes induced by isobutanol. We postulated that ArcA activation resulted from quinone malfunction, and linked the activations of Fur and PhoB, two other verified regulators of the isobutanol response in this study, to these important electron carriers. To support this hypothesis, quinone-linked ArcA and Fur target gene expressions were shown to be comparatively unperturbed by isobutanol under fermentative growth, whereas quinol-linked PhoB target gene expressions remained activated. In addition, growth on glycerol was shown to be more significantly retarded by isobutanol than growth on glucose (glycerol requires quinones to enter glycolysis but glucose does not). Although quinones are found in the membrane and their malfunction conforms to a solvent-stress model centered on membrane disruption, they have not been previously implicated in the solvent-stress regulatory cascade. Repercussions of quinone malfunction include disproportionate growth defects for different carbon sources (glycerol versus glucose growth), and alterations in metabolism (TCA cycle, glyoxylate shunt), protein–protein interaction (ArcA–ArcB and PhoB–PhoR), and transcriptional regulation (ArcA, Fur, PhoB). The results presented here connect changes in respiratory, phosphate, and iron control to the malfunction of a single class of membrane components upon exposure to isobutanol stress, and therefore highlight the utility of a systems-biology approach for the study of isobutanol toxicity. Lastly, we identified the common and distinct toxicity features of the three alcohol-based biofuels, n-butanol, ethanol, and isobutanol, by comparing their initial response networks.
Results and discussion
Physiological response to isobutanol
As shown in Figure 1A, isobutanol caused E. coli growth arrest at concentrations ⩾1% vol/vol in minimal MOPS medium with 0.2% glucose. Accumulation of pyruvate was observed at 1% vol/vol isobutanol in that medium (Figure 1B), and was more pronounced at 2 and 3% vol/vol isobutanol. As pyruvate is a core metabolite, its accumulation suggests a significant degree of metabolic distress. In order to characterize the initial transcriptional changes induced by isobutanol, aerobic E. coli cultures were split; one half received a 1% isobutanol treatment whereas the other remained untreated, and transcriptome measurements were taken from both cultures at 10 min. Using the LOWESS normalization and rank-invariant selection method of lcDNA (Hyduke et al, 2003), which is an open-source Markov Chain Monte Carlo (MCMC) procedure that can identify statistically significant expression perturbations that do not meet the ad hoc two-fold change threshold (Materials and methods), 1452 genes were identified as significantly perturbed (Supplementary Table I). To identify whether these genes were enriched for any biological function, Gene Ontology (GO) classifications were obtained from EcoCyc (Karp et al, 2007) and analyzed using the BinGO plugin (Maere et al, 2005) for Cytoscape (Shannon et al, 2003). There were 53 diverse functional enrichments at a P-value ⩽0.05 (Supplementary Table II). This result highlights the fact that isobutanol has a widespread effect on cellular function. NCA was used to unravel this complex transcriptional response.
Figure 1.
Phenotypic changes of BW25113 in response to isobutanol. (A) Growth assay of E. coli BW25113 in the presence of 0, 0.5, 1, 1.5, and 2% (vol/vol) isobutanol. (B) Concentration of pyruvate in the presence of 0, 1, 2, and 3% isobutanol.
NCA identifies transcription factors perturbed by isobutanol
NCA uses transcription network connectivity to deduce transcription factor activities (TFAs) and control strengths (CSs, quantified TF–gene interaction) from gene-expression data. The following transcription regulation model is used:
where mRNAi is the mRNA transcript level of genei, TFAj is the activity level of TFj, CSij is the control strength of TFj on the expression of genei, and (t) and (0) designate condition t and reference condition 0. Equation (1) can be linearized by taking the log, and multiple experiments can be represented in matrix form using the equation
where E is an (N × M) matrix of expression ratios, A is an (N × L) matrix of CSs, P is an (L × M) matrix of TFAs, N is the number of genes, L is the numbers of TFs, M is the number of experiments, and Γ is the residual of the model. As the decomposition of E into component matrices is inherently non-unique, NCA uses topological constraints from the transcription network connectivity to guarantee a unique solution up to a scaling factor (Liao et al, 2003). The element aij in matrix A is set to 0 if there is no evidence to suggest regulation of genei by TFj. Others are estimated together with P using the expression data, E. If the 0s in A satisfy the NCA uniqueness criteria (Liao et al, 2003; Galbraith et al, 2006), the decomposition shown in equation (2) is unique up to a scaling factor for any given residual Γ. This criterion clearly links NCA results to the biological system and makes interpretation straightforward. A detailed derivation of NCA can be found in Liao et al (2003).
In this study, NCA was used to deduce TFA perturbations resulting from isobutanol stress. Gene-expression data were analyzed by NCA using transcription network connectivity obtained from Regulon DB (Gama-Castro et al, 2008) to quantify TFAs and CSs. For NCA, training data are not required and were not used in this study (Liao et al, 2003). The transcription network is presented in Supplementary Table III. As the number of experiments (M) was less than the number of TFs (L), a modified NCA criterion for use with limited microarray data was used (Galbraith et al, 2006). This criterion allows for the identification of a unique solution up to a scaling factor when M<L, by requiring each gene to be regulated by not more than M TFs. The statistical significance of TFA perturbations was evaluated by comparing each TFA with a null distribution generated from randomization of the data using a z-test (Materials and methods).
Owing to the imposition of NCA uniqueness criteria on the transcription network connectivity, we were able to quantify TFAs and CSs for 67 TFs from E. coli. Among the 67 TFs analyzed, 16 TFs had significantly perturbed activities in response to isobutanol (P-value ⩽0.01). These TFs are listed along with their significance level (low P-value=high likelihood TFA perturbed by isobutanol), biological function, and regulon members that were significantly perturbed in Table I. A complete list of TFs analyzed along with their significance level can be found in Supplementary Table IV. It is evident from the three most significantly perturbed TFs (ArcA, PdhR, and FNR) that isobutanol affects respiration. Under the assumption that isobutanol toxicity is mediated through membrane disruption, it is not surprising that respiration is affected. The purpose of respiration is to generate a PMF across the membrane, many critical respiratory processes occur at the membrane, and many respiratory components are membrane constituents. To explore this result, we looked at the known activation mechanisms for ArcA, PdhR, and FNR, and the identities of their regulon members that were perturbed by isobutanol.
Table 1.
Transcription factors with significantly perturbed activities in response to isobutanol
| TF | P-valuea | Function (regulator of) | Regulon Members Significantly Perturbed by Isobutanolb |
|---|---|---|---|
| aTFs are listed in order of their significance. | |||
| bItalics font signifies a decrease in expression and bold italics signifies an increase in expression upon isobutanol exposure. | |||
| ArcA | 6.8E−19 | Respiration | aceBAK, aceEF, ackA, acnA, aldA, betBA, betT, caiA, cydAB, cyoABCDE, dctA, fadA, fadE, fadJ, fnr, fumA, fumC, gadX, gatYZBCD, gltA, hyaDE, hybOBF, icd, lldD, moeA, mdh, ndh, nuoABCEFGHIKL, oppABCDF, pflB, ptsG, rpoS, rpsJ–rplCDWB–rpsS–rplV–rpsC–rplP–rpmC–rpsQ, rutB, sdhCDAB, sodA, sucABCD, treB, uvrA, ydeA, yfiD |
| PdhR | 1.4E−15 | Electron transport | aceEF, cyoABCDE, hemL, hha, lldD, ndh, yfiD |
| FNR | 2.3E−09 | Respiration | aceEF, ackA, acnA, aldA, caiA, ccmB, cydAB, cyoABCDE, dcuB, fhlA, fnr, frdABC, fumA, gadXW, gcvTH, gltBDF, hmp, hyfA, hypE, moaB, moaC, moeA, ndh, nrdDG, nrfABF, nrfG, nuoABCEFGHIKL, pepT, pflB, pitA, pstSCAB–phoU, purMN, rpsJ–rplCDWB–rpsS–rplV–rpsC–rplP–rpmC–rpsQ, rpsP–rimM–trmD–rplS, sdhCDAB, sodA, ssuB, sucABCD, tdcB, upp, uraA, ycaC, yfiD, yhjA, ysgA |
| Fur | 6.3E−07 | Iron transport | cirA, cyoABCDE, entC, entS, exbBD, fecI, fepC, fhuAC, fhuF, fiu, flhDC, hmp, metH, nrdHIE, ompF, purR, rcnA, rcnR, sdhCDAB, sodA, sodB, sucABCD, tonB, ydfN, yhhY |
| OmpR | 7.4E−07 | Outer membrane | bolA, csgDF, flhDC, micF, nmpC, ompF, sra |
| Crp | 1.2E−06 | Catabolite repression | aceBAK, aceEF, acnA, agp, aldA, aldB, araE, araG, argG, bglG, caiA, cdd, cirA, crp, csgDF, cyoABCDE, cytR, dadX, dctA, dcuB, deoCB, dgsA–ynfK, dusB–fis, entC, epd–pgk, exuT, fiu, flhDC, fucO, fucU, fumA, gadBC, gadE–mdtF, gadX, gapA, gatYZBCD, gcd, gdhA, glgA, glgS, glnAL, glpEGR, glpFKX, gltA, gltBDF, gntK, gntT, gntY, grpE, guaBA, hupB, hyfA, ilvBN, lamB, malF, malT, malX, manXYZ, marA, mdh, melAB, melR, metK, mhpC, mhpE, mtlD, nmpC, nupC, ompA, ompF, ompR, osmY, oxyR, pflB, pncB, ppiA, ptsG, putP, rbsDK, rpoH, rpoS, sdhCDAB, serC–aroA, sodA, sodB, spf, srlAEB–gutQ, sucABCD, tdcB, treB, tsx, udp, ugpA, uidC, xseA, xylF, ychH, yfiD, yhbC–nusA–infB–rbfA–rpsO–pnp, yhcH, yiaJ, yiaL, yjcH, yjhT |
| GadE | 7.9E−06 | Acid resistance | cyoABCDE, gadE–mdtF, gadXW, gatBC, gltBDF, hdeBA, hlpA–lpxD |
| Nac | 1.4E−04 | Nitrogen metabolism | codBA, gdhA, gltBDF, mioC–mnmG, nac, nupC |
| LexA | 1.5E−04 | SOS response | ftsK, ftsL, lexA, recN, rpoD, rpsU–dnaG, umuC, uvrA |
| RpoH | 1.5E−04 | Heat shock | bssS, can, clpB, clpPX, creB, dgsA–ynfK, dnaKJ, fxsA, gapA, gntY, groSL, grpE, hslRO, hslUV, hspQ, htpG, htpX, ibpB, ldhA, lon, macB, metA, miaA–hfq–hflXKC, mutM, narP, nusB–thiL, osmF, phoPQ, pphA, prlC–yhiQ, raiA, rfaD, rpmE, rpoD, rrmJ–hflB, sdaA, valS, yafDE, ybbN, ybeD–lipB, ybeZX–lnt, yciH, yciSM, ycjFX–tyrR, ydhQ, yehW, ygaD, ygbFT, yhdN–zntR, yjhH |
| PurR | 3.8E−04 | Purine nucleotide biosynthesis | carAB, codBA, cvpA–purF, gcvTH, glnB, glyA, guaAB, hflD–purB, prs, purC, purE, purHD, purL, purMN, purR, pyrC, pyrD, speB |
| Fis | 7.8E−04 | Diverse processes | aldB, bglG, crp, deoCB, dusB–fis, glnAL, glnQ, guaAB, gyrB, hupB, marA, mtlD, ndh, nrfABF, nrfG, nuoABCEFGHIKL, pdxA–ksgA–apaGH, ptsG, queA, sra, tpr, xylF, yhbC–nusA–infB–rbfA–rpsO–pnp, ygjG, yjcH |
| IHF | 1.6E−03 | Diverse processes | aceBAK, caiA, carAB, cysJIH, dps, dusB–fis, fhlA, flhDC, gcd, glnHPQ, gltA, gltBDF, hycH, hypE, ibpB, ilvMD, micF, mtr, ndh, nmpC, nrfABF, nrfG, nuoABCEFGHIKL, ompF, ompR, osmY, pflB, pspBCE, pstSCAB–phoU, rpoH, rtcB, sodA, sodB, sra, ssuB, sucABCD, tdcB, uspA, uspB, ygjG, yiaJ, yiaL, yjcH |
| HNS | 4.2E−03 | Diverse processes | bolA, bglG, chiA, cspD, cydAB, cysWAM, degP, flhDC, fliY, gadXW, gspAB, gspJ, gspM, gspO, gutQ, hchA, hdeAB, hisJQP, micF, osmC, sodB, srlAEB, stpA, yciE |
| NtrC | 5.4E−03 | Nitrogen metabolism | argT, astB, cbl, ddpX, glnAL, glnHPQ, glnK, hisJQP, nac, potF, rutB, yeaGH, ygjG |
| PhoB | 6.6E−03 | Phosphate transport | argP, asr, phnCHN, phoA–psiF, phoBR, phoH, pstSCAB–phoU, ugpA |
The activities of AcrA, PdhR, and FNR are mediated by phosphorylation, pyruvate, and oxygen, respectively. All three are cytoplasmic proteins. However, ArcA forms a two-component system with ArcB, a membrane protein. Phosphorylation of ArcA occurs when quinone inhibition of ArcB is released. Quinones are metabolites embedded in the membrane that function as electron carriers for respiratory processes. PdhR shares 10 of its 12 perturbed regulon members with ArcA, whereas FNR shares 50 of its 96 perturbed regulon members with ArcA. It should be noted here that NCA might have difficulty in separating TFAs for TFs with highly overlapping regulons. Considering this information, it is likely that the ArcA–ArcB system is the major regulator of respiratory genes in response to isobutanol. PdhR is a minor transcriptional regulator not known to associate with the membrane, the majority of whose perturbed regulon members are also regulated by ArcA. FNR, which is inactive under aerobic growth due to Fe–S oxygen binding, is unlikely to be active under the growth conditions used for transcriptome measurements in this work (mid-log aerobic). In addition, more than half of the perturbed FNR regulon members are also regulated by ArcA, and 74 of its 96 perturbed regulon members are controlled by one or more of the other 14 TFs identified as significantly perturbed. Therefore, it is more likely that these genes were perturbed through the action of other regulators rather than by FNR.
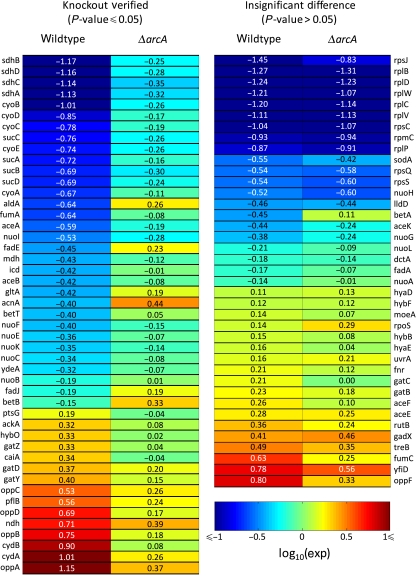
To verify that ArcA modulates gene expression in response to isobutanol, transcriptome measurements were obtained from a ΔarcA strain treated with and without 1% isobutanol (Materials and methods). We reasoned that if ArcA regulates geneX in response to isobutanol, then the response of geneX to isobutanol in ΔarcA would be different from the response of geneX to isobutanol in wild type. Figure 2 shows the log10 expression ratios of ArcA-regulon members from wild-type and ΔarcA experiments, along with an indication of whether the difference was significant at a P-value ⩽0.05 (for exact P-values, see Supplementary Table V). The majority of ArcA-regulon members perturbed by isobutanol in wild type (47 out of 86) had significantly different expression ratios in ΔarcA (P-value ⩽0.05). This provided additional evidence that ArcA participates in the E. coli response to isobutanol. The remaining 39 regulon members showed indistinguishable activation/repression (P-value >0.05) in response to isobutanol in ΔarcA compared with wild type. This could have resulted from the inherent noise of DNA microarrays, compensatory action by other regulators, or expression of that gene being under the control of a different TF under isobutanol stress.
Figure 2.
Log10 expression ratios for ArcA-regulon members from wild-type and ΔarcA experiments. Left: regulon members with significant expression differences (P-value ⩽0.05); right: regulon members with non-significant expression differences (P-value >0.05). Expression ratios are 1% isobutanol/0% isobutanol.
Isobutanol stress causes a loss of quinone function and ArcA activation
With ArcA verified by knockout to be involved in the isobutanol response, we sought to identify the upstream isobutanol target responsible for ArcA activation. ArcA is activated through phosphorylation by ArcB, a membrane protein. This phosphorylation occurs after quinone (primarily ubiquinone) inhibition of ArcB autophosphorylation is released. Therefore, given this two-component mechanism, the likely target for isobutanol is quinone function. Quinones are electron carriers with an isoprenoid side chain that anchors them to the membrane. They function as the primary electron carriers for respiration, and are thought to regulate the ArcA–ArcB two-component system in response to cellular redox state (reduced Qs/QH2s cannot inhibit ArcB autophosphorylation) (Georgellis et al, 2001; Malpica et al, 2004, 2006). A mechanism for the isobutanol activation of ArcA through quinone malfunction conforms to a mode of toxicity through membrane disruption. We hypothesize that isobutanol disrupts the membrane, leading to quinone malfunction (dissociation from or disruption of interaction with the membrane), which results in a release of quinone inhibition on ArcB, and subsequent autophosphorylation of ArcB and activation of ArcA.
To support this hypothesis, we tested the effect of isobutanol on metabolic processes that require quinones. One such example is glycerol degradation, which requires quinones in the second step (glycerol 3-phosphate dehydrogenase reaction). Figure 3A shows the glycolytic entry pathways for glucose and glycerol under normal aerobic growth conditions. Quinones are the only components of the respiratory chain found along the glycolytic separation of glycerol and glucose. As glycerol directly requires functional quinones to enter glycolysis and glucose does not, growth on glycerol should suffer from more significant retardation than growth on glucose if the presence of isobutanol causes quinone malfunction. Thus, growth rates were measured for E. coli in glycerol and glucose minimal media in the presence of a spectrum of isobutanol concentrations (Materials and methods). Figure 3B shows the relative time to four doublings for E. coli grown in glycerol and glucose medium over a spectrum of isobutanol concentrations (see Supplementary Figure 1 for growth curves). These results show that isobutanol hinders growth on glycerol more significantly than growth on glucose, and that this effect becomes more pronounced at higher concentrations of isobutanol. This result supports the hypothesis that isobutanol causes the loss of quinone function either by membrane damage or by quinone depletion, and shows possible repercussions of these findings for isobutanol production in terms of carbon-source selection, which will be discussed further in the Conclusion section.
Figure 3.
(A) Metabolic pathways for glycolytic entry of glycerol and glucose. (B) Relative time to four doublings for BW25113 grown on glycerol or glucose as the sole carbon source in a spectrum of isobutanol concentrations (growth curves presented in Supplementary Figure 1). Red/blue bars represent average; error bars represent maximum and minimum of two biological replicates. DHAP, dihydroxyacetone phosphate; FBP, fructose-1,6-bisphosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; G6P, glucose-6-phosphate, glycerol-3-P, glycerol-3-phosphate; Pi, phosphate; UQ, ubiquinone; UQH2, ubiquinol.
To support the hypothesis that ArcA activation is caused by the loss of quinone function, we measured ArcA-regulated genes under fermentative conditions using quantitative real-time PCR. As ubiquinone is a component of the respiratory chain, it is not needed in fermentative conditions. Thus, if the hypothesis is true, ArcA activity will not change in response to isobutanol under fermentative conditions. Fermentative expression ratios are presented alongside their aerobic equivalents in Figure 4. The genes sdhC and oppA were selected as ArcA target genes because they were the lead genes in the most significantly repressed and induced operons, respectively, shown to be regulated by ArcA in ΔarcA experiments. The expression changes of sdhC and oppA in response to isobutanol under fermentative conditions (red bars) are negligible compared with their expression changes in an aerobic environment (blue bars). The observed fermentative oppA repression (average approximately two-fold) under fermentative conditions is in contrast to the strong aerobic activation of oppA (average approximately 14-fold) and may have resulted from the action of other known regulators of oppABCDF expression, GcvB, Lrp, or ModE. These results support a mechanism of ArcA activation through malfunction of the respiratory chain, as in the absence of respiration (fermentation) isobutanol fails to perturb the expression of ArcA-regulon members to the same degree or direction observed under aerobic conditions.
Figure 4.
Log10 expression ratios for ArcA, Fur, and PhoB targets under aerobic and fermentative culture conditions. Expression ratios are for (isobutanol/untreated), red/blue bars represent average, error bars for aerobic treatment are 95% confidence intervals as calculated by lcDNA, and those for fermentation are the maximum and minimum of two biological replicates.
Isobutanol activations of PhoB and Fur are likely mediated by quinone function
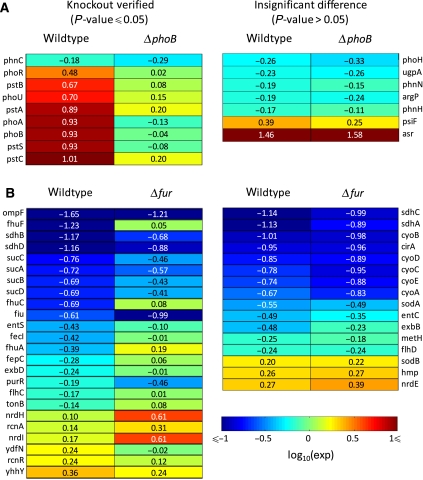
Owing to the profound effect isobutanol had on the respiratory system through quinone malfunction, we sought to identify additional TFs whose activity perturbations can be linked to quinone malfunction. We started by identifying additional two-component systems known to be regulated by quinones. Three other two-component systems are known to have quinone-related activation mechanisms. These include EvgAS of E. coli (ubiquinone mediated), BvgAS of Bordetella pertussis (ubiquinone mediated), and PhoPR of Bacillus subtillis (menaquinol mediated) (Bock and Gross, 2002; Schau et al, 2004; Eldakak and Hulett, 2007). EvgAS is homologous to BvgAS in structure, and the PhoBR system of E. coli is homologous to the PhoPR system of B. subtillis in structure and function (BLAST results presented in Supplementary Table VI). Of these systems, PhoBR was identified as significantly perturbed by NCA. Although the EvgAS system is known to be regulated by quinones, EvgA was not identified as a significantly perturbed regulator by NCA (P-value=0.47), and therefore was not investigated further in this study. To verify that PhoB was involved in the isobutanol response, transcriptome measurements were obtained from ΔphoB treated with and without 1% isobutanol. Figure 5A lists the log10 expression ratios of PhoB-regulon members from wild-type and ΔphoB experiments, along with an indication of whether the difference was significant at a P-value ⩽0.05 (for exact P-values, see Supplementary Table VII). The majority of PhoB-regulon members perturbed by isobutanol in wild type (nine out of 16) had significantly different expression ratios in ΔphoB (P-value ⩽0.05). This provided additional evidence that PhoB is activated by isobutanol. Taken together with the knowledge that the PhoPR system of B. subtillis is homologous to the PhoBR system of E. coli and they regulate similar operons in response to the same stimulus (phosphate starvation), we postulate that isobutanol disrupts the membrane and releases quinol inhibition of PhoR, which allows PhoR to autophosphorylate and activate PhoB. To support this hypothesis, expression levels for PhoB target genes were measured under fermentative growth conditions using quantitative real-time PCR (Materials and methods). As reduced Qs/QH2s are the primary form of quinone under fermentative conditions (Shestopalov et al, 1997; Georgellis et al, 2001; Bekker et al, 2007), our hypothesis would predict that unlike ArcA, isobutanol treatment should activate PhoB under fermentative conditions to a similar, if not stronger, extent than it does in an aerobic environment. Fermentative expression ratios are presented alongside their aerobic equivalents in Figure 4. The genes phoB and pstS were selected as PhoB target genes because they were the lead genes in the most significantly induced operons shown to be regulated by PhoB in ΔphoB experiments. These results clearly show isobutanol activation of PhoB under fermentative and aerobic conditions is similar, and support a mechanism of PhoR inhibition by quinols and isobutanol-mediated PhoB activation through quinol malfunction.
Figure 5.
(A) Log10 expression ratios for PhoB-regulon members from wild-type and ΔphoB experiments. (B) Log10 expression ratios for Fur regulon members from wild-type and Δfur experiments. Left: regulon members with significant expression differences (P-value ⩽0.05); right: regulon members with non-significant expression differences (P-value >0.05). Expression ratios are 1% isobutanol/0% isobutanol.
In addition to their regulatory role, quinones serve as important electron carriers for many respiratory processes. We reasoned that if isobutanol caused general Q/QH2 malfunction the activity of all membrane-bound enzymes that rely on Q/QH2 for their electron-carrier capabilities, including all cytochromes and both NADH dehydrogenases, would be disrupted. With this in mind, there may be TFs whose isobutanol response can be linked to quinone electron-transport malfunction. To identify these TFs, we identified all aerobic respiratory machineries known to utilize quinones when glucose is the sole carbon source. These include succinate dehydrogenase (sdhCDAB), NADH dehydrogenase I (nuo operon), NADH dehydrogenase II (ndh), and cytochrome bo3 (cyoABCD). Malfunction of all of these complexes in E. coli would diminish the ability to convert NADH to NAD+ and generate PMF. It is also well documented that aerobic respiration generates superoxide ions (O2−), with NDH-II as the main generator of endogenous superoxide and NDH-I and SDH as smaller contributors (Gennis and Stewart, 1996). Therefore, malfunction of these enzymes would also decrease the O2− level. Combined with an inability to convert NADH to NAD+, a decrease in endogenous O2− would cause reductive stress. Interestingly, it has been suggested that a reducing environment may activate Fur (Jovanovic et al, 2006), which is the most significantly perturbed regulator not associated with respiration in this study.
Fur requires binding of Fe2+ to become active and repress genes of its regulon. It has been shown previously that O2− deactivates Fur after its conversion to H2O2 by superoxide dismutases, through the Fenton reaction (Blanchard et al, 2007):
Therefore, a decrease in endogenous O2− generation would increase the availability of Fe2+, through a decrease in H2O2 levels, and in effect activate Fur relative to the control. Indeed, on isobutanol stress, NCA deduced an increase in Fur activity. To verify that Fur was involved in the isobutanol response, transcriptome measurements were obtained from Δfur treated with and without 1% isobutanol. Figure 5B lists the log10 expression ratios of Fur-regulon members from wild-type and Δfur experiments, along with an indication of whether the difference was significant at a P-value ⩽0.05 (for exact P-values, see Supplementary Table VII). The majority of Fur-regulon members perturbed by isobutanol in wild type (24 out of 40) had significantly different expression ratios in Δfur (P-value ⩽0.05). This provided additional evidence that Fur is activated by isobutanol. We postulate that membrane disruption interferes with the ability of quinones to act as electron carriers, and thereby inhibits the enzymatic activity of NDH-I, NDH-II, and SDH. Inhibition of these complexes results in reductive stress through a decrease in endogenous O2− production and an increase in the NADH/NAD+. This results in Fur activation through a diminished Fenton reaction and stabilization of Fe2+. Once active, Fur represses the expression of genes related to iron homeostasis. To support the involvement of quinones in this hypothesis, expression levels for Fur target genes were measured under fermentative growth conditions using quantitative real-time PCR (Materials and methods). Fermentative expression ratios are presented alongside their aerobic equivalents in Figure 4. The genes fhuF and fecI were selected as Fur target genes because fhuF was the most strongly repressed gene singly regulated by Fur, fecI is an iron-associated sigma factor, and they were both shown to be regulated by Fur in Δfur experiments. The average expression changes of fhuF, an fecI, in response to isobutanol under fermentative conditions (red bars) are negligible compared with their expression changes in an aerobic environment (blue bars). The observed fhuF activation under fermentative conditions (average ∼2.5-fold) is in contrast to the strong aerobic repression of fhuF (average ∼17-fold). These results show that in the absence of respiration isobutanol fails to perturb the expression of Fur target genes to the same degree or direction observed under aerobic conditions, and supports a mechanism of isobutanol-mediated Fur activation through quinones malfunction.
Isobutanol disrupts general quinone function
Disruption of quinone function could result from a shift in the Q/QH2 ratio or disruption of quinone–membrane interactions. Considering the activations of ArcA, Fur, and PhoB, it is unlikely that isobutanol produces a shift in the Q/QH2 ratio. It has been shown that shifts in the Q/QH2 ratio can activate ArcA-ArcB in E. coli and PhoPR in B. subtillis. Under normal growth, ArcA becomes active when the Q/QH2 ratio decreases, which corresponds to NADH-dehydrogenase activity exceeding cytochrome activity. This decrease along with ArcA activation is present during a shift from aerobiosis to anaerobiosis when oxygen becomes limiting for the cytochromes, and for chemicals, such as KCN or NO, which target the cytochromes (Hyduke et al, 2007). However, PhoPR activation is inhibited by menaquinols and activated by an increase in the Q/QH2 ratio (Eldakak and Hulett, 2007). Consistent with this phenomenon is the observation that members of the PhoBR regulon are unperturbed by NO-mediated cytochrome-bo malfunction shown to activate the ArcA–ArcB regulon (Hyduke et al, 2007). This would suggest that the Q/QH2 ratio were not responsible for isobutanol activation of ArcA and PhoB. However, the major quinone inhibitors of the ArcA–ArcB and PhoPR systems are different (ubiquinone for ArcB and menaquinol for PhoR), and it is possible that isobutanol affects the UQ/UQH2 and MQ/MQH2 ratios differently. To investigate whether this was the case, we obtained a ΔubiE strain from the Keio collection (Baba et al, 2006) and took transcriptome measurements under isobutanol stress conditions. UbiE is an enzyme that catalyzes the final step of MQ synthesis and an intermediary step of UQ synthesis. In ΔubiE, both UQ and MQ are absent and the only functional quinone is demethylmenaquinone (DMQ). As DMQ is the only acting quinone, both ArcB and PhoR inhibition would be mediated by DMQ/DMQH2 ratio. Therefore, if a change in UQ/UQH2 were responsible for ArcA activation, in ΔubiE, ArcA should be activated and PhoB should be non-responsive. In contrast, if a change in MQ/MQH2 were responsible for PhoB activation, in ΔubiE, PhoB should be activated and ArcA should be non-responsive. However, in ΔubiE, both ArcA and PhoB were activated similar to wild type (Supplementary Table I), which suggests that a shift in the Q/QH2 is not responsible for ArcA and PhoB activation. It is unclear how a shift in the Q/QH2 would affect Fur activity, as a breakdown of cytochrome function under aerobic conditions does not necessarily mean a decrease in NDH-II or SDH activity. However, expression data from cultures with NO-perturbed cytochromes clearly show an unperturbed Fur regulon (Hyduke et al, 2007). All together, this information suggests that a shift in the Q/QH2 ratio is unlikely to cause ArcA, Fur, and PhoB activations.
The remaining hypothesis focuses on disruption of quinone–membrane interaction. For example, quinones, which under normal physiological conditions diffuse freely in the membrane, might aggregate at membrane lesions created by isobutanol, or changes in membrane fluidity might alter quinone anchoring and result in sub-optimal distances between quinone headgroups and membrane-bound proteins, or membrane disruption could result in leakage of quinones into the cytoplasm. It is unclear at this point what type of malfunction would cause ArcA, Fur, and PhoB activation. As our aim was to identify the isobutanol response network, identification of the exact abnormality that leads to quinone malfunction is beyond the scope of this study.
Isobutanol response network
Figure 6 illustrates the postulated isobutanol response network associated with quinone malfunction. Quinone has an important role in respiration, and its isobutanol-associated malfunction alters the respiratory machinery (TCA cycle, glyoxolate shunt, cytochromes, and NADH dehydrogenases), as well as iron and phosphate homeostasis. On isobutanol treatment, ArcB senses a decrease in respiratory performance and activates its two-component-system partner ArcA, which adjusts the cellular metabolism for growth with decreased respiratory efficiency (e.g. low oxygen). This decrease in respiratory performance results in reductive stress due to an increase in the NADH/NAD+ and a decrease in endogenous O2−. Fur senses this reductive stress and represses genes associated with iron uptake in preparation for conditions in which Fe–S clusters are safe from damage by reactive oxygen species. PhoR senses a disruption in quinol inhibition by isobutanol and activates its two-component-system partner PhoB, which signals a shift to an environment with higher phosphate demand.
Figure 6.
Isobutanol response network related to quinone malfunction. Isobutanol disrupts quinone/quinol function, which releases inhibition of ArcB and PhoR autophosphorylation. Once phosphorylated, ArcB activates ArcA and PhoR activates PhoB, each of which goes on to control its regulon. Disruption of enzymatic activity of cytochromes, NADH dehydrogenases, and succinate dehydrogenase by quinone malfunction results in a decrease in endogenous O2− leading to a reduction in H2O2 and increase in Fe2+ through a diminished Fenton reaction. Fur then binds free Fe2+ to become active and control its regulon.
Comparison of ethanol, n-butanol, and isobutanol response networks
Transcriptome measurements were taken from wild-type E. coli exposed to 1% n-butanol and 3% ethanol to identify similarities and differences with the isobutanol response. These concentrations were selected because of similar growth inhibition compared with 1% isobutanol. The expressions of 342 and 644 genes were significantly perturbed (P-value ⩽0.01) between n-butanol and ethanol stress when compared with isobutanol, respectively. The GO annotations of these genes were analyzed for functional enrichment by BinGO (Shannon et al, 2003; Maere et al, 2005), and the results are presented in Supplementary Table VIII. These results suggest that the butanol and isobutanol responses regulate amino-acid metabolism differently (P-value=1.9E−9), whereas the list of genes whose expression differed between isobutanol and ethanol stress contain only nominal enrichment (P-value ∼0.03) for any GO category, which represents broad differences not specific to any category.
NCA was carried out on the expression data to determine which TFAs were significantly different under isobutanol, n-butanol, and ethanol stress (Materials and methods). In all, six TFAs differed significantly (P-value ⩽0.01) between isobutanol and n-butanol stress conditions (CpxR, GadX, LeuO, MalT, QseB, and TyrR), and 19 TFAs differed significantly between isobutanol and ethanol stress conditions (ArcA, BolA, Crp, CysB, ExuR, FNR, Fur, GadE, IHF, LeuO, Lrp, MetR, PdhR, PurR, RpoH, RpoN, SoxS, TyrR, and Zur). Consistent with the GO-term enrichment analysis, regulators of amino-acid metabolism (LeuO and TyrR) had significantly different TFAs under butanol stress compared with isobutanol stress. Indeed, 15 of the 21 enzymes required for branched-chain amino-acid synthesis from pyruvate and oxaloacetate (aspC, leuABCD, thrABC, tyrB, ilvN, ilvB, ilvG_1, ilvM, ilvD and ilvH) are more strongly repressed by n-butanol than by isobutanol. The large number of regulators with significant TFA differences between isobutanol and ethanol suggests widespread differences in the response network.
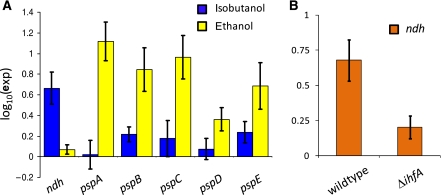
An interesting result not captured by NCA involves pspABCDE and ndh, which encode the phage shock protein operon and NDH-II, respectively. This result was excluded from NCA because the psp operon is regulated by PspF, which violated NCA uniqueness criteria, necessitating the removal of its target genes for the analysis. On isobutanol treatment, ndh was strongly induced and pspABCDE expression did not change appreciably, whereas on ethanol treatment ndh expression did not change significantly and pspABCDE was strongly induced. These results are depicted in Figure 7A. Expression of the psp operon is induced by a variety of environmental stresses, including ethanol treatment, psp induction of which requires IHF (Weiner et al, 1995; Darwin, 2005; Jovanovic et al, 2006). It is thought that the psp gene products act to restore PMF lost due to environmental stress (Darwin, 2005). Ethanol is known to reduce PMF by disruption of the membrane (Sikkema et al, 1995; Jovanovic et al, 2006; Kobayashi et al, 2007). NDH-II is the NADH dehydrogenase that does not generate any PMF, and therefore is thought to manage redox balance when the capacity to generate energy is greater than demand (Calhoun et al, 1993; Jackson et al, 2004). These results suggest that ethanol decreases PMF and isobutanol treatment creates an environment in which PMF exceeds the demand. This implies that one of the most important membrane-associated processes, generation and regulation of PMF, is handled differently by E. coli under ethanol stress compared with n-butanol and isobutanol.
Figure 7.
(A) Log10 expression ratios of ndh and psp operon in the presence of isobutanol and ethanol in wild type. (B) Log10 expression ratios of ndh in wild type and ΔihfA (error bars are 1 s.d.).
It has been shown previously that IHF regulates psp expression upon ethanol treatment (Weiner et al, 1995), and IHF participates in the regulation of ndh expression (Karp et al, 2007; Gama-Castro et al, 2008). In addition, IHF was identified by NCA to be significantly perturbed by isobutanol stress. To identify whether IHF regulates ndh induction upon isobutanol treatment, transcriptome measurements were obtained from ΔihfA with and without 1% isobutanol treatment. The results for ndh are presented in Figure 7B, whereas the results for the entire IHF regulon are presented in Supplementary Table IX. These data suggest that IHF is necessary for full induction of ndh expression in response to isobutanol. Therefore, it is possible that the differences in psp and ndh expression between isobutanol- and ethanol-stressed E. coli could result from a difference in IHF activity, as IHF is required for ethanol induction of psp and isobutanol induction of ndh.
Conclusion
In this study, we identified the initial transcriptional response network of E. coli towards isobutanol. NCA was used to quantify TFAs for 67 TFs in response to isobutanol, of which 16 were significantly perturbed. Regulators beyond these 67 could not be analyzed due to insufficient regulatory information (ill-defined regulon) or non-compliance with NCA uniqueness criteria. On the basis of the annotated TF function, the 16 significantly perturbed TFs fall into three categories: those associated with stress mitigation (GadE, LexA, OmpR, and RpoH), those that regulate metabolism (ArcA, Crp, FNR, Fur, Nac, NtrC, PdhR, PhoB and PuR), and nucleoproteins that regulate diverse function (Fis, HNS and IHF). Evidence presented suggests that isobutanol interrupts the function of membrane-bound Q/QH2, which leads to respiratory distress and the activations of ArcA (the most significantly perturbed TF), Fur, and PhoB. This malfunction most likely results from disruption of quinone–membrane interaction, such as decreased diffusivity, sub-optimal anchoring, or leakage into the cytoplasm. The implications of this finding include decreased activity for enzymes that utilize quinones for their electron-carrier capability. To demonstrate the significance of this finding, E. coli was grown in a spectrum of isobutanol concentrations in MOPS media with glycerol or glucose as the sole carbon source. As the degradation of glycerol requires quinones before entry into glycolysis but glucose does not, we predicted that growth of E. coli on glycerol would be more severely affected by isobutanol than growth on glucose. As measured by the time it took the culture to double four times, growth on glycerol was more severely retarded by isobutanol than growth on glucose. This result suggests that the quinone dependence of a carbon source should be considered before selection of a feedstock or strain for production. This would be especially important for aerobic production. (Supplementary Figure 2 shows aerobic isobutanol production.) Perhaps, metabolic engineering efforts could be used to change the quinone dependence of a carbon source and enhance its conversion to isobutanol. In addition to repercussions from quinone malfunction, the activations of ArcA, Fur, and PhoB could be utilized to create synthetic switches that turn off or on particular enzymes when the concentration of isobutanol reaches a desired level. For instance, we have shown that branched-chain amino-acid synthesis is downregulated in response to isobutanol. As these pathways supply the precursors of isobutanol, it may be desirable to use the endogenous signaling provided by ArcA or PhoB to overexpress branched-chain amino-acid synthesis enzymes as the concentration of isobutanol reaches inhibitory levels.
In addition, the response networks of ethanol, n-butanol, and isobutanol were compared. The response networks of n-butanol and isobutanol were qualitatively the same, with the most significant difference being the stronger repression of amino-acid synthesis by n-butanol. The response network of ethanol varied more significantly, with a major difference in regulation of genes responsible for PMF management. Data presented here and in earlier studies (Weiner et al, 1995) suggest that this difference may be dependent on IHF. This result is of particular interest because it implies that one of the most important membrane-associated processes, generation and regulation of PMF, is handled differently by E. coli under ethanol stress compared with n-butanol and isobutanol, which are treatments commonly thought to disrupt the membrane in a similar manner.
The isobutanol response network presented here is consistent with previous work on ethanol and n-butanol that attributed the toxic effect of these alcohols to membrane disruption (Ingram, 1976; Borden and Papoutsakis, 2007). The central role of ArcA in response to isobutanol under aerobic conditions mirrors the finding of Gonzalez et al (2003) of a non-functional fnr gene contributing to ethanol tolerance under fermentative conditions. This suggests that broad rewiring of metabolism is a mechanism employed by E. coli to mitigate alcohol stress. In addition, earlier studies on C. acetobutylicum have observed that n-butanol stress elicits a response similar to heat shock and that overexpression of heat-shock proteins confers increased tolerance (Tomas et al, 2003). Consistent with this finding was the fact that NCA deduced a strong activation of RpoH, the heat-shock sigma factor, in response to isobutanol. This suggests that, similar to C. acetobutylicum, E. coli could benefit from priming for stress before accumulation of isobutanol to toxic levels. Lastly, our finding that ΔarcA, Δfur, and ΔphoB did not significantly increase tolerance (Supplementary Table X) highlights the complex nature of tolerance mechanisms and agrees with conclusions that solvent tolerance is unlikely to result from single-gene mutations, but results from multiple successive mutations (Gonzalez et al, 2003).
Conceivably, both the rational design of solvent tolerance and comprehension of super-tolerant mutants can be aided by this network, and the integrated systems-biology approach presented could be applied to any stimulus in E. coli or other organisms.
Materials and methods
Phenotypic assays
MOPS medium (using 0.2% glucose as the sole carbon source) was prepared according to Wanner (1994). For growth measurements, five 250-ml baffled shake flasks with 1/10th the volume of MOPS medium with varying concentrations of isobutanol (0, 0.5, 1, 1.5, 2% vol/vol) were inoculated with 1% of a BW25113 overnight and grown aerobically in a 37°C water bath shaken at 300 r.p.m. OD600 measurements were taken at time 0 and every hour for 6 h on a Beckman-Coulter DU800 spectrophotometer. For metabolite measurements, BW25113 was grown aerobically from a 1% inoculation to mid-log (OD600=0.3–0.4) in 500-ml baffled flasks filled to 1/10th total volume in a 37°C water bath shaken at 300 r.p.m. Cultures were centrifuged (Beckman J2-21 centrifuge) at 8000 r.p.m. for 10 min, the supernatant was removed, and the cell pellet was resuspended in 1-ml MOPS media without a carbon source. The resuspension was centrifuged (Eppendorf 5415D microcentrifuge) at 13 000 r.p.m. for 5 min, the supernatant was aspirated, and cell pellet was resuspended in MOPS media without a carbon source. Isobutanol was added to the resuspensions for final concentrations of 0, 1, 2, and 3% vol/vol. After 10 min, glucose was added to a final concentration of 0.2%, and samples were taken and filtered through a 0.22-μm filter at 0, 10, 30, and 60 min. Metabolites were separated on an Aminex HPX-87H exclusion HPLC column using a 5-mM H2SO4 mobile phase, and concentrations were measured by UV–Vis and RID (Agilent 1100 series HPLC).
Cell harvesting, RNA purification, and microarray
BW25113 was grown aerobically from a 1% inoculation to mid-log (OD600=0.3–0.4) in 500-ml baffled flasks at 1/10th the total volume in a 37°C water bath shaken at 300 r.p.m. The culture was split into half, with one half receiving a 1% isobutanol treatment. Growth was allowed to continue in both cultures for 10 min, and then both were transferred to chilled centrifuge tubes and swirled for 2 min in an ice bath. Cultures were then harvested by centrifugation at 4°C and 8000 r.p.m. for 10 min, the supernatant was decanted, and cell pellet immediately stored at −80°C. Thawed cells were disrupted by lysozyme and total RNA was extracted using the RNAeasy kit (Qiagen). Indirect labeling with amino-allyl dUTP, array design, hybridization, and analysis were carried out as described previously (Braatsch et al, 2006; Hyduke et al, 2007). For each condition/strain, four biological replicate transcriptome measurements were taken, with two technical replicates for each biological replicate, totaling eight microarray slides, unless otherwise noted (wild-type isobutanol treatment → 10 arrays, ΔubiE isobutanol treatment → 4 arrays).
Transcriptome data processing and analysis
Normalized expression ratios were obtained from lcDNA, which is an open-source MCMC procedure used to normalize and assess the statistical significance of gene expression perturbations from DNA microarray (Hyduke et al, 2003). Using the Gene Expression Profile Analysis Suite version 4, spot replicate ratios were averaged, genes that were absent in >30% of the slides in an experiment set (e.g., wild-type isobutanol, 10 arrays) were removed, and the remaining missing values were imputed using KNN imputation (k=15) by an experiment set (8–10 arrays) (Montaner et al, 2006). The entire gene-expression dataset in MIAME-compliant format has been deposited in the GEO database (www.ncbi.nlm.nih.bo/geo) under accession number GSE13444. Expression ratios were log10 transformed before NCA, which is a publicly available program that has been developed previously (Liao et al, 2003; Tran et al, 2005; Galbraith et al, 2006). Connectivity data were obtained from RegulonDB and are presented in Supplementary Table III (Gama-Castro et al, 2008). A confidence level of 95% was used to identify significant expression perturbations using lcDNA (Supplementary Table I). NCA was used to decompose the significant expression perturbations into TFA and CS (Liao et al, 2003; Tran et al, 2005; Yang et al, 2005; Galbraith et al, 2006; Hyduke et al, 2007). The method used to determine whether a TF was significantly perturbed in a given experiment was that described previously (Hyduke et al, 2007) with the following modifications: (1) resultant TFAs were averaged over a dataset (8–10 microarrays), whereas previously the gene expression ratios were averaged over a dataset before NCA; and (2) 50 randomizations of the data were used. In brief, NCA was used to deduce TFAtrue from the expression data, a null distribution was built by randomizing/shuffling gene expression, NCA was used to deduce TFArandom(i) from each randomization i, and a z-test was then used to identify the probability that TFAtrue could have been drawn from the null distribution, TFArandom. The reported P-value is the probability that TFAtrue could have been drawn from the null distribution, and thus a low P-value for TFAj equates to a high likelihood that the activity of TFj was perturbed by the specified treatment. Randomization of data amounted to the shuffling of gene expression data. For example, if in the true network gene1 was controlled by TF1, in the random data gene1-expression data would be replaced by expression data for a gene chosen at random from the genome. In this way, the network used to construct the null distributions is identical to the network used for NCA of the real data. The only difference is in the gene-expression data being decomposed. This way, each TF controls the same number of genes in the network and co-regulates with the same TFs.
Knockout strains
All knockouts were obtained from the Keio collection (Baba et al, 2006) and cultured identically to wild type, except for the presence of kanamycin in the medium (Keio collection has kanR cassette in place of the knocked-out gene). All downstream processes including harvesting and RNA purification were performed in the same manner as for wild type, except for ΔubiE, which suffered from significant growth lag and was harvested at OD600 ∼0.15.
Comparison of glycerol and glucose growth
MOPS medium (0.2% glucose or 0.2% glycerol as the sole carbon source) was prepared using the MOPS minimal media kit distributed by Teknova. Six 14-ml test tubes with 2.5 ml of MOPS media with varying concentrations of isobutanol (0, 0.5, 0.75, 1, 1.25, 1.5% vol/vol) were inoculated with 1% of BW25113 and grown overnight aerobically in a 37°C shaker at 300 r.p.m. OD600 measurements were taken at time 0, every 2 h for 12 h, and every 6 h from 12 to 24 h on a Tecan SPECTRAFluor Plus platereader. Time to four doublings was calculated by fitting an exponential curve over three time points, within which the culture doubled four times. For 1% isobutanol treatment in glucose and 0.75% isobutanol treatment in glycerol two points were used, and for 1% isobutanol treatment in glycerol an exponential curve was extrapolated from the final two time points.
Quantitative real-time PCR
Fermentative MOPS minimal media with 0.2% glucose as the sole carbon source was prepared by a procedure adapted from Holdeman and Moore (1977). Hungate anaerobic culture tubes (Belco Glass #; 2047-16125) were filled with 15 ml of sterile media, needles were placed in the septum to allow venting, and tubes were heated for 90 min in a boiling water bath. Rezuserin was used as an oxygen indicator for this preparation step, but not added to culture media used in experiments. Once removed from the water bath, needles were removed immediately. Culture tubes were then transferred to an anaerobic chamber equipped with palladium catalyst boxes to react with O2 in the presence of H2 (atmosphere maintained at 98/2 N2/H2). The medium was transferred in the anaerobic chamber to 250-ml baffled flasks and shaken at 200 r.p.m. for a minimum of 16 h at 37°C. Cultures were inoculated with an anaerobic overnight culture of BW25113 to obtain an initial OD600 of 0.01–0.02, and then shaken at 200 r.p.m. in 250-ml baffled flasks filled to 1/10th the volume of the flask at 37°C. Once cultures reached an OD600 of 0.15–0.3, they were split into half, with one half receiving a 1% isobutanol treatment. Growth was allowed to continue in both cultures for 10 min, and then they were harvested with RNAprotect (Qiagen) according to the manufacturer's protocol. RNA was purified with RNeasy mini columns, with a DNase digestion carried out before RNA cleanup according to the manufacturer's protocol (Qiagen). Any residual genomic DNA was then digested using Turbo DNA-Free according to the manufacturer's protocol (Ambion). Reverse transcription PCR was carried out using a Superscript III First Strand Synthesis kit (Invitrogen) and 50–100 ng of total RNA as template according to the manufacturer's protocol. An RNase-H treatment was carried out after the RT reaction according to the manufacturer's protocol. Real-time PCR was carried out using the Roche Lightcycler 480 SYBR Green Master kit on a Roche Lightcycler 480 II according to the manufacturer's protocol. Absolute quantification was measured using the 2nd-derivative maximum of the Lightcycler software. RNA was checked for genomic-DNA contamination by running reverse transcription in the absence of Superscript III, followed by real-time PCR. Transcript abundance was normalized to ypfI and hybE, which were both not perturbed in any of the expression data. Details of real-time PCR primers can be found in Supplementary Table XI.
Solvent comparison
A t-test was used to determine whether there was a statistically significant difference in expression (P-value ⩽0.01) between cultures treated with 1% isobutanol and 1% n-butanol or 3% ethanol. Genes whose expression differed from one solvent to another are presented in Supplementary Table XII. Genes with significant expression differences between the two solvent stresses were analyzed using NCA, and statistically significant TFA differences were identified (P-value ⩽0.01, as indicated by a t-test).
Supplementary Material
Expression ratios for genes with significantly perturbed expression in the presence of isobutanol over all conditions and knockouts tested
Gene Ontology term enrichment of genes perturbed by isobutanol
Transcription network connectivity used to perform NCA
List of transcription factors (TF) analyzed by NCA
Expression ratios for ArcA regulon members
Protein BLAST results for B. subtillis PhoPR components
Expression ratios for PhoB regulon members and Fur regulon members
Gene Ontology term enrichment of genes with significant expression differences between ethanol and isobutanol, and n-butanol, and isobutanol stress
Expression ratios for IHF regulon members
Growth data for ÄarcA, Äfur, and ÄphoB in the presence of isobutanol
Quantitative real time PCR primers
Genes with significant expression differences between ethanol, n-butanol, and isobutanol stress
This file contains supplementary figures S1 and S2
Acknowledgments
We thank Jeffrey Bernstein for his technical assistance with microarray, Jonathan Lo for growth curve measurements of ΔarcA, Δfur, and ΔphoB strains, and Kevin Smith for his aerobic isobutanol production data. This work was supported by the National Institutes of Health Grant 1R01GM076143 and Department of Energy Grant DE-FG02-07ER64490. MPB was supported in part by a UCLA Dissertation Year Fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alsaker KV, Spitzer TR, Papoutsakis ET (2004) Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell′s response to butanol stress. J Bacteriol 186: 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, de Mattos MJ (2007) Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153: 1974–1980 [DOI] [PubMed] [Google Scholar]
- Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ (2007) Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS ONE 2: e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock A, Gross R (2002) The unorthodox histidine kinases BvgS and EvgS are responsive to the oxidation status of a quinone electron carrier. Eur J Biochem 269: 3479–3484 [DOI] [PubMed] [Google Scholar]
- Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol 73: 3061–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles LK, Ellefson WL (1985) Effects of butanol on Clostridium acetobutylicum. Appl Environ Microbiol 50: 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braatsch S, Bernstein JR, Lessner F, Morgan J, Liao JC, Harwood CS, Beatty JT (2006) Rhodopseudomonas palustris CGA009 has two functional ppsR genes, each of which encodes a repressor of photosynthesis gene expression. Biochemistry 45: 14441–14451 [DOI] [PubMed] [Google Scholar]
- Calhoun MW, Oden KL, Gennis RB, de Mattos MJ, Neijssel OM (1993) Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J Bacteriol 175: 3020–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin AJ (2005) The phage-shock-protein response. Mol Microbiol 57: 621–628 [DOI] [PubMed] [Google Scholar]
- Dombek KM, Ingram LO (1984) Effects of ethanol on the Escherichia coli plasma membrane. J Bacteriol 157: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldakak A, Hulett FM (2007) Cys303 in the histidine kinase PhoR is crucial for the phosphotransfer reaction in the PhoPR two-component system in Bacillus subtilis. J Bacteriol 189: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith SJ, Tran LM, Liao JC (2006) Transcriptome network component analysis with limited microarray data. Bioinformatics 22: 1886–1894 [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, Bonavides-Martinez C, Abreu-Goodger C, Rodriguez-Penagos C, Miranda-Rios J, Morett E, Merino E, Huerta AM, Trevino-Quintanilla L, Collado-Vides J (2008) RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res 36: D120–D124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis RB, Stewart V (1996) Respiration. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Böck A, Curtiss III R, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, Squires CL (eds). Washington, DC: ASM Press [Google Scholar]
- Georgellis D, Kwon O, Lin EC (2001) Quinones as the redox signal for the arc two-component system of bacteria. Science 292: 2314–2316 [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Tao H, Purvis JE, York SW, Shanmugam KT, Ingram LO (2003) Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: Comparison of KO11 (Parent) to LY01 (resistant mutant). Biotechnol Prog 19: 612–623 [DOI] [PubMed] [Google Scholar]
- Holdeman L, Moore W (eds). (1977) Preparation of reduced media for growing anaerobes. In Anaerobe Laboratory Manual, 4th edn. Blacksburg, VA: Virginia Polytechnic Institute and State University [Google Scholar]
- Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC (2007) Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci USA 104: 8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduke DR, Rohlin L, Kao KC, Liao JC (2003) A software package for cDNA microarray data normalization and assessing confidence intervals. OMICS 7: 227–234 [DOI] [PubMed] [Google Scholar]
- Ingram LO (1976) Adaptation of membrane lipids to alcohols. J Bacteriol 125: 670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Blake T, Green J (2004) Regulation of ndh expression in Escherichia coli by Fis. Microbiology 150: 407–413 [DOI] [PubMed] [Google Scholar]
- Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M (2006) Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem 281: 21147–21161 [DOI] [PubMed] [Google Scholar]
- Karp PD, Keseler IM, Shearer A, Latendresse M, Krummenacker M, Paley SM, Paulsen I, Collado-Vides J, Gama-Castro S, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Bonavides-Martinez C, Ingraham J (2007) Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res 35: 7577–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R, Suzuki T, Yoshida M (2007) Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol 66: 100–109 [DOI] [PubMed] [Google Scholar]
- Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP (2003) Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci USA 100: 15522–15527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D (2004) Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA 101: 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Sandoval GR, Rodriguez C, Franco B, Georgellis D (2006) Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal 8: 781–795 [DOI] [PubMed] [Google Scholar]
- Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, Valls J, Pujana MA, Alloza E, Herrero J, Al-Shahrour F, Dopazo J (2006) Next station in microarray data analysis: GEPAS. Nucleic Acids Res 34: W486–W491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schau M, Eldakak A, Hulett FM (2004) Terminal oxidases are essential to bypass the requirement for ResD for full Pho induction in Bacillus subtilis. J Bacteriol 186: 8424–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov AI, Bogachev AV, Murtazina RA, Viryasov MB, Skulachev VP (1997) Aeration-dependent changes in composition of the quinone pool in Escherichia coli. Evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett 404: 272–274 [DOI] [PubMed] [Google Scholar]
- Sikkema J, de Bont JA, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59: 201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Beamish J, Papoutsakis ET (2004) Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol 186: 2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Welker NE, Papoutsakis ET (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell′s transcriptional program. Appl Environ Microbiol 69: 4951–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LM, Brynildsen MP, Kao KC, Suen JK, Liao JC (2005) gNCA: a framework for determining transcription factor activity based on transcriptome: identifiability and numerical implementation. Metab Eng 7: 128–141 [DOI] [PubMed] [Google Scholar]
- Wanner B (1994) Gene expression in bacteria using TnphoA and TNphoA′ elements to make and switch phoA gene, lacZ(op), and lacZ(pr) fusions. In Methods in Molecular Genetics, Adolph K (ed), Vol. 3, pp 291–310. Orlando, FL: Academic Press [Google Scholar]
- Weiner L, Brissette JL, Ramani N, Model P (1995) Analysis of the proteins and cis-acting elements regulating the stress-induced phage shock protein operon. Nucleic Acids Res 23: 2030–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Suen J, Brynildsen MP, Galbraith SJ, Liao JC (2005) Inferring yeast cell cycle regulators and interactions using transcription factor activities. BMC Genomics 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomano LP, York SW, Ingram LO (1998) Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J Ind Microbiol Biotechnol 20: 132–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression ratios for genes with significantly perturbed expression in the presence of isobutanol over all conditions and knockouts tested
Gene Ontology term enrichment of genes perturbed by isobutanol
Transcription network connectivity used to perform NCA
List of transcription factors (TF) analyzed by NCA
Expression ratios for ArcA regulon members
Protein BLAST results for B. subtillis PhoPR components
Expression ratios for PhoB regulon members and Fur regulon members
Gene Ontology term enrichment of genes with significant expression differences between ethanol and isobutanol, and n-butanol, and isobutanol stress
Expression ratios for IHF regulon members
Growth data for ÄarcA, Äfur, and ÄphoB in the presence of isobutanol
Quantitative real time PCR primers
Genes with significant expression differences between ethanol, n-butanol, and isobutanol stress
This file contains supplementary figures S1 and S2