Abstract
We have engineered the chemotaxis system of Escherichia coli to respond to molecules that are not attractants for wild-type cells. The system depends on an artificially introduced enzymatic activity that converts the target molecule into a ligand for an E. coli chemoreceptor, thereby enabling the cells to respond to the new attractant. Two systems were designed, and both showed robust chemotactic responses in semisolid and liquid media. The first incorporates an asparaginase enzyme and the native E. coli aspartate receptor to produce a response to asparagine; the second uses penicillin acylase and an engineered chemoreceptor for phenylacetic acid to produce a response to phenylacetyl glycine. In addition, by taking advantage of a ‘hitchhiker' effect in which cells producing the ligand can induce chemotaxis of neighboring cells lacking enzymatic activity, we were able to design a more complex system that functions as a simple microbial consortium. The result effectively introduces a logical ‘AND' into the system so that the population only swims towards the combined gradients of two attractants.
Keywords: signal transduction, synthetic biology, two-component systems
Introduction
Efforts to engineer complex biological systems are largely motivated by two goals: to understand biological processes by constructing systems that mimic their behavior or show new functions; and to build useful devices that can have practical applications in diverse fields ranging from medicine (Anderson et al, 2006) to synthetic chemistry (Keasling, 2008). Examples run from creating individual biological components with defined functions to a larger-scale engineering of pathways, cells, and multi-cellular systems (Drubin et al, 2007). Some recent studies have focused on engineering microbes to show population-level behavior, such as biofilm and pattern formation, cell-density regulation, consensus gene expression, and predator–prey dynamics (reviewed in (Brenner et al, 2008)). These systems share features with natural bacterial communities, known as consortia, which involve multiple interacting species (Wolfaardt et al, 2007). Here we describe an engineered bacterial-chemotaxis system that functions in a cooperative multi-cellular pathway.
Escherichia coli shows a chemotactic response to a set of small molecules, including amino acids, sugars, metals, and some small organic compounds (Adler, 1975). Signal detection is mediated by transmembrane receptor proteins, which sense external stimuli and transduce the signal into the cytoplasm. E. coli expresses five different receptors, including the Tar aspartate receptor, which feed into the same downstream pathway (Grebe and Stock, 1998). These receptors sense chemical signals either by direct binding of a small molecule to the periplasmic sensor domain of the protein, such as aspartate to Tar, or through interaction of the receptor with periplasmic-binding proteins.
We engineered E. coli to sense and respond to small molecules that are not chemoattractants for the wild-type bacteria. The system incorporates an enzyme from an unrelated pathway, whose end product is a native chemoattractant, into the chemotaxis system by expressing it in the periplasm, where the sensing domain of the chemoreceptor is located. As a result, the engineered bacteria respond to the molecule that is the substrate for this enzyme (Figure 1A). A second system was also designed, which further expands the range of attractants by incorporating a modified receptor that responds to a new ligand. The two systems yielded effective chemotaxis toward non-native molecules but also showed interactions between strains in mixed populations. By taking advantage of this population-level behavior, we constructed a more complex chemotaxis system, which involves two strains and shows characteristics of a simple microbial consortium.
Figure 1.
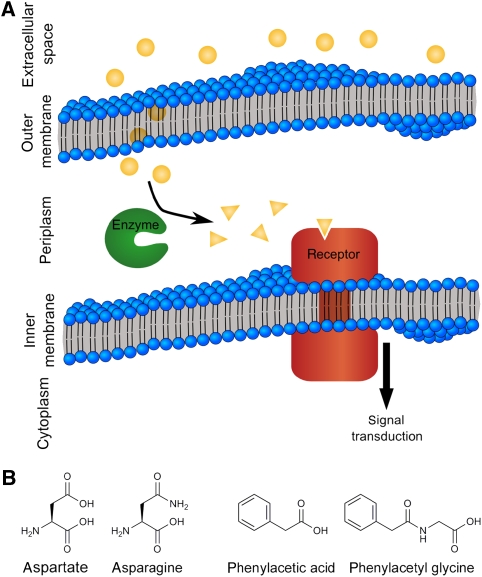
Engineered chemotaxis pathway. (A) The pathway incorporates an enzyme that converts the target molecule (gold spheres) into a product (gold triangles) that is a ligand for the chemotaxis receptor. (B) Two ligand–target molecule pairs used in this study. Asparagine is converted to aspartate, the ligand for the wild-type Tar chemoreceptor, by asparaginase. Phenylacetyl glycine is converted to phenylacetic acid, the ligand for the engineered chemoreceptor TarPA, by penicillin acylase.
Results and discussion
Two different initial systems were designed to produce a chemotactic response to non-native molecules by incorporating an enzyme into the pathway. The first system was designed to respond to asparagine by introducing an asparaginase enzyme into cells expressing the wild-type aspartate receptor Tar. The second was designed to respond to phenylacetyl glycine (PAG) by introducing a penicillin-acylase enzyme into cells expressing a variant of Tar that responds to phenylacetic acid (PAA) (Figure 1B).
Despite the chemical similarity between asparagine and aspartate, wild-type E. coli is only weakly attracted to asparagine (Hedblom and Adler, 1983). To create an asparagine-sensing strain we used E. coli asparaginase II, a hydrolase that converts asparagine to aspartate (Jennings and Beacham, 1990). Asparaginase II is secreted into the periplasm, which is convenient as this is likely the necessary location for it to take part in the chemotaxis pathway, and is very efficient (Derst et al, 2000). The ansB gene, which encodes Asparaginase II, is ordinarily expressed only under conditions of low oxygen. We therefore constructed a strain, in which the chromosomal ansB was deleted and the gene was instead expressed from a weakened trc promoter (Weiss et al, 1999) to give low-level constitutive expression. The strain also lacked the chromosomal genes encoding all five chemoreceptors. The aspartate receptor Tar was expressed from a plasmid (Derr et al, 2006).
The second system was designed to give a chemotactic response to PAG. Compared with asparagine, PAG is much more chemically distinct from aspartate and thus provides a more stringent test of the design strategy. In addition, PAG is not involved in E. coli K-12 metabolism and does not show any chemoattractant activity with wild-type E. coli.
To create a PAG-sensing strain, we used penicillin acylase (Pac) from E. coli strain W, which efficiently hydrolyzes a range of phenylacetyl amides, including PAG, to produce PAA (Margolin et al, 1980). We had previously constructed a Tar variant by directed evolution, denoted TarPA, which responds to PAA (unpublished data), and we hypothesized that co-expression of TarPA and Pac would produce E. coli that would show chemotaxis toward PAG. As pac is absent in E. coli K-12 strains, which are used in this work, the pac gene with its native promoter was isolated from E. coli W and cloned into a plasmid. As above, the strain lacked the chromosomal copies of the native E. coli chemoreceptors. The PAA-responsive chemoreceptor TarPA was also expressed from a plasmid.
To evaluate the chemotactic behavior of the constructed strains, two methods were used: an assay on soft-agar plates containing a pre-established gradient of attractant (Derr et al, 2006), and a capillary assay in liquid media (Adler, 1973). For each designed strain, a corresponding strain was constructed that contained the same chemoreceptor plasmid (expressing Tar or TarPA) and a control plasmid lacking the gene for the enzyme.
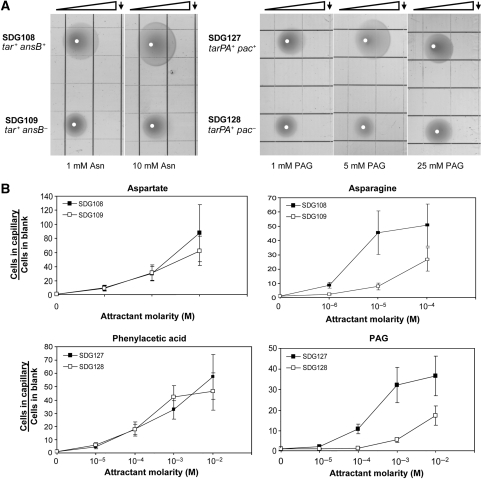
The ansB+ strain, which contains the plasmid expressing AnsB, showed a strong response to asparagine, whereas the corresponding ansB− strain did not. The two strains were tested on gradient plates formed with 1, 10, and 100 mM aspartate or asparagine. Chemotaxis was apparent for both strains on the gradient plates formed with 1 and 10 mM aspartate (Supplementary Figure S1a). For plates with 100 mM aspartate, both strains showed relatively little response, presumably because the high concentration of ligand saturates the chemoreceptor, effectively rendering the cells blind to the gradient. On the asparagine gradient plates, however, only the ansB+ strain showed a response to 1 and 10 mM asparagine (Figure 2A). Again, for a gradient formed with a very high concentration of attractant (100 mM asparagine), there was relatively little response (Supplementary Figure S1a).
Figure 2.
Assays for chemotaxis. (A) Panels show images of bacteria 19 h (for SDG108 and SDG109) or 24–27 h (for SDG127 and SDG128) after inoculation on soft-agar plates containing a gradient formed by spotting attractant onto the line indicated by the arrow. Images shown are representative of experiments that were carried out at least three times. White dots indicate the points where culture was applied to the plates. Grid squares are 13 mm × 13 mm. (B) Capillary assays of the designed strain SDG108 and the control strain SDG109 with a range of aspartate and asparagine concentrations, and of designed strain SDG127 and the control strain SDG128 with a range of PAA and PAG concentrations. For each strain and attractant concentration, the number of cells that accumulated in the capillary was normalized by dividing by the number of cells of the same strain that accumulated in a blank—that is, a capillary containing buffer but no attractant. Results are the averages of experiments on at least three capillaries. Error bars indicate standard errors.
E. coli cells have much greater mobility in liquid than in soft agar. Therefore, in liquid media it may be more difficult for cells to produce significant local concentrations of chemoreceptor ligand in their immediate environment. To test the behavior of the enzyme-mediated chemotaxis in liquid, capillary assays were carried out (Figure 2B). The two strains showed similar responses to aspartate, as would be expected as they both express the Tar chemoreceptor. For asparagine, however, the ansB+ strain did indeed show a robust chemotactic response that was five-fold greater than the ansB− strain.
The second engineered strain similarly showed robust chemotaxis to PAG. The pac+ strain (containing a plasmid expressing Pac) and the corresponding pac− strain were tested for their response to PAA and PAG on gradient plates and in capillary assays. On plates, both strains showed a pronounced response to PAA at 5 mM, and a weaker response at 1 and 25 mM (Supplementary Figure S1b). However, only the pac+ strain showed chemotaxis on PAG plates (Figure 2A). The response was strongest at 5 mM, and weaker at 1 and 25 mM. In capillary assays (Figure 2B), the pac+ and pac− strains showed similar abilities to swim toward PAA, whereas the pac+ strain showed a seven-fold stronger response towards PAG.
We next sought to determine for each system whether the engineered strain would enable a second strain, which expresses the chemoreceptor but lacks the enzyme that converts attractant to receptor ligand, to swim toward the attractant in a mixed population. We hypothesized that the catalytic activity of receptor+ enzyme+ cells would be sufficient to bring the receptor+ enzyme− cells along through a ‘hitchhiker effect'; that is, the receptor+ enzyme− cells would detect the gradient of chemoreceptor ligand produced by the receptor+ enzyme+ cells and effectively follow them up the gradient. To test this, we used a previously developed competition assay (Derr et al, 2006). Receptor+ enzyme+ and receptor+ enzyme− cells were combined in ratios ranging from 0.1 to 0.8 and applied to gradient plates. After ∼24 h, the leading edge of the growth up the gradient was collected and the ratio of receptor+ enzyme+ cells to receptor+ enzyme− cells was determined. The ratio was typically one to six-fold greater than in the input mixture. Thus, although there was an increase in the proportion of receptor+ enzyme+ cells, there was still a significant number of receptor+ enzyme− cells present in the leading edge. In contrast, when receptor+ enzyme+ cells were combined with cells lacking the appropriate receptor, the leading edge of the population was found to consist almost entirely of the receptor+ enzyme+ cells (Supplementary Table S1). This suggests that the receptor+ enzyme− cells are indeed able to follow the receptor+ enzyme+ cells up the gradient.
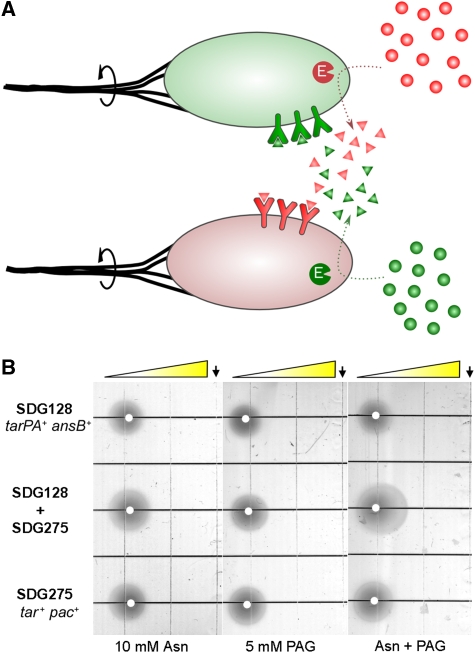
By using this population-level behavior of the engineered bacteria, we developed a new system consisting of two strains that function cooperatively. One strain is tar+ pac+ and the other is tarPA+ ansB+. Each strain contains a subset of the components necessary for chemotaxis to asparagine or PAG, and is thus unable to respond to either attractant alone. The two strains in combination, however, contain all of the components for a chemotactic response (Figure 3A). This system comprises a simple microbial consortium, as both of the strains are necessary to produce the behavior that neither can generate alone. Note that the mixture of strains should not be able to respond to a gradient consisting of only one of the two compounds. Although the target molecule would be converted by one strain into a molecule that the second strain can swim toward, the first strain and its associated enzymatic activity would not receive a chemoattractant signal and therefore would not move up the chemical gradient. As the production of chemoreceptor ligand for the second strain should remain constrained to the location of the first strain, no movement up the gradient should be observed for either strain.
Figure 3.
A designed microbial consortium. (A) The system consists of two interacting strains that produce a mutually interdependent chemotactic response. Red and green spheres represent target molecules; red and green triangles represent ligands for the chemoreceptors (Y-shaped objects). Dotted arrows represent conversion by enzymes (marked ‘E'). Solid arrows indicate rotation of the flagella (thick black lines). (B) Comparison of chemotaxis of strains SDG128, SDG275, and the SDG128+SDG275 mixture on soft-agar gradient plates, as in Figure 2A. The plates show 22 h of growth at 30°C. The leading edge of growth of the middle spot (SDG128+SDG275) on the Asn+PAG plate was collected and plated on LB agar. By counting fluorescent and non-fluorescent colonies (corresponding to SDG275 and SDG128, respectively), the mixture was found to consist of 55±5% of strain SDG275.
To test the designed consortium, the two strains were spotted separately and as a mixed population on soft-agar plates containing gradients of asparagine, PAG, or both compounds (Figure 3B). The mixed population showed robust chemotaxis on the combined asparagine+PAG gradient, but not on plates containing gradients of asparagine or PAG alone. The individual strains, on the other hand, did not show chemotaxis on any of the gradients.
The mixed population was also tested in the capillary assay. It was unclear whether chemotaxis of the consortium would be evident in these conditions if, as proposed, chemotaxis of the consortium depends on gradients of chemoreceptor ligands that are established by enzymatic conversion within, and diffusion out of, neighboring cells. In particular, as cells move relatively rapidly over long distances in liquids, the local concentrations of converted ligands may not accurately reflect the gradient of attractants. We measured the accumulation of the mixed population and the accumulation of each strain alone in capillaries containing both attractants (Supplementary Figure S2). If the movement of each cell in the gradient were independent of other cells in the environment, we would expect that the net rate of accumulation of the mixed population in the capillary would be the sum of the accumulations seen with each individual strain; instead, the effect was roughly 1.5-fold greater than this, which may indicate an effect of the consortium. In contrast with the multi-cellular pathway of the consortium, the single-cell pathways in tar+ ansB+ and tarPA+ pac+ strains do not necessarily depend on ligand produced by other cells, which may enable more efficient chemotaxis in a liquid environment. Using techniques, such as video microscopy (Gestwicki et al, 2000) and microfluidics (Mao et al, 2003; Ahmed and Stocker, 2008), it may be possible to study the time scale and local structure of the chemotactic response in these engineered systems.
The combination of the tar+ pac+ and tarPA+ ansB+ strains results in a simple microbial consortium that senses the coincidence of two chemical gradients; the cells will only swim up the combined gradients of asparagine and PAG. Thus, the population effectively functions as a chemotactic AND gate, showing a level of signal integration in chemotaxis that cannot be easily achieved with a single strain. The system involves cooperative behavior between the two strains' enzymatic reactions and their responses to small molecules, much like the synergistic interactions found in many natural contexts (Budrene and Berg, 1995; Schink, 2002). This mutualist system, which modulates the bacterial behavior but does not depend on changes in gene expression, is complementary to recently designed E. coli consortia that use quorum sensing to regulate gene expression (Brenner et al, 2007, 2008; Balagadde et al, 2008). The system that we have constructed could serve as a model for exploring aspects of microbial consortia, such as co-evolution, and mathematical modeling of growth dynamics and metabolite cycling. It may also have applications in bioremediation and various industrial processes, where synthetic pathways that are restricted to a single cell line can often have deleterious growth effects.
We have also shown that the repertoire of bacterial chemotaxis can be expanded by incorporating ligand-producing enzymes. In principle, this strategy could be used to produce a chemotactic response to any molecule that can be converted enzymatically to a ligand for a native or engineered chemoreceptor. It is likely that this diversity can be expanded even further by combining the approach described here with strategies for re-engineering other components of the chemotaxis circuit (Topp and Gallivan, 2007, 2008).
Materials and methods
Media
Cells were grown in LB medium or minimal A medium (60 mM K2HPO4/33 mM KH2PO4/8 mM (NH4)2SO4/2 mM sodium citrate) supplemented with 0.2% (v/v) glycerol, 1 μg/ml of thiamine HCl and 20 μg/ml each of methionine, leucine, and histidine (40 μg/ml each in agar plates). Semisolid agar was made from the above minimal medium with 0.25% (w/v) agar. Media were supplemented with 100 μg/ml ampicillin (50 μg/ml in minimal medium) and 34 μg/ml chloramphenciol (17 μg/ml in minimal medium) as needed. Note that no IPTG was added to the growth media. The basal expression from the IPTG-inducible promoter was sufficient. All chemicals were obtained from Sigma-Aldrich unless otherwise noted. PAG was synthesized from PAA and glycine methyl ester (Advanced Chemtech) by standard peptide-coupling chemistry.
Strains and plasmids
The strains and plasmids used in this work are summarized in Table I. The ansB deletion in strains SDG104 and SDG105 was constructed by lambda red-mediated recombination (Datsenko and Wanner, 2000). E. coli W was obtained from ATCC (strain #9637). The TG1 strain of E. coli was from Stratagene.
Table 1.
Strains and plasmids used in this study
| Strain | Description | Reference |
|---|---|---|
| UU1250 | RP437 Δtsr-7028 Δ(tar-tap)5201 Δtrg-100 Δaer-1 ygjG∷Gm zbd∷Tn5 | Bibikov et al (2004) |
| MDG162 | UU1250 Φ(ompC+–gfp+) | Derr et al (2006) |
| W | Wild-type E. coli strain | ATCC |
| TG1 | Cloning strain | Stratagene |
| SDG104 | MDG162 ΔansB | This study |
| SDG105 | UU1250 ΔansB | This study |
| SDG108 | SDG104+pPD12, pSG49 | This study |
| SDG109 | SDG105+pPD12, pSG50 | This study |
| SDG127 | MDG162+pPD12-TarPA4, pSG115 | This study |
| SDG128 | UU1250+pPD12-TarPA4, pSG49 | This study |
| SDG275 | MDG162+pPD12, pSG115 | This study |
| Plasmid | ||
| pACYC184 | Cloning vector | Chang and Cohen (1978) |
| pDSW206 | Promoter down mutations in −35 and −10 of pTrc99A | Weiss et al (1999) |
| pPD12 | Tar expression plasmid | Derr et al (2006) |
| pPD12- TarPA4 | Tar variant, phenylacetic-acid responsive | This study |
| pSG49 | pACYC184 lacIq, attenuated Ptrc-ansB+ | This study |
| pSG50 | pACYC184 lacIq, attenuated Ptrc | This study |
| pSG115 | pACYC184 pac+ | This study |
The plasmid pPD12-TarPA4, encodes a Tar variant, TarPA, which responds to PAA. The tarPA allele was isolated by directed evolution as part of a larger project to evolve Tar to respond to a variety of compounds. The details of this work will be published elsewhere, Here we provide a concise description of the construction of tarPA. The experimental procedure essentially followed the methods described in (Derr et al, 2006). The starting point for mutagenesis was the tar allele tarPhe2, which encodes a receptor that responds to phenylalanine (Derr et al, 2006). The tarPhe2 gene was subjected to mutagenesis, through error-prone PCR and DNA shuffling, followed by selection on semisolid agar plates containing gradients of PAA. After a total of four rounds of mutagenesis, each of which was followed by multiple cycles of selection, mutants were isolated that showed a strong response to PAA. One of these isolates was tarPA. TarPA has the following substitutions relative to wild-type Tar: L21I, S27C, F29I, S32P, S39N, V41E, S43C, R73W, M76V, A101S, T135S, E136G, Y143N, Y149F, F150L, Q152H, T154A, N159D, F165L, I179F, T181I, D182V, V196A, D249V, and S255T. We have not determined which of these substitutions are necessary for the receptor to respond to PAA.
The ansB gene was amplified from E. coli TG1 genomic DNA using the primers 5′-GACATCGCTCATATGGAGTTTTTCAAAAAGAC-3′ and 5′-CAGCGTGACCTCGAGTTAGTACTGATTGAAGATCTG-3′. The lacIq gene and the attenuated trc promoter were amplified from pDSW206 using the primers 5′-GATATCGTCTAGACGCGCGAAGGCGAAG-3′ and 5′-GTCACTCTCATATGCTGTTTCCTGTGTGAAATT-3′. Both fragments were inserted into a modified pACYC184 plasmid, which had the multiple cloning site from pET26b (Novagen) previously placed between the BamHI/SalI sites, into the NdeI/XhoI site and XbaI/NdeI site respectively, ultimately producing the plasmid pSG49. The pSG50 plasmid is identical to pSG49 except that the ansB gene is replaced by an inactive gene.
The pac gene (open reading frame plus promoter) was amplified from E. coli W (ATCC #9637) genomic DNA using the primers 5′-GACATCGTCTAGACTCTGCAAATAGATAACCG-3′ and 5′-GACGCTAGTCGACCGTTACAAAGGGATGCTG-3′ and inserted into the XbaI/SalI site of pACYC184 to generate the plasmid pSG115.
Plate assays
The gradient plates were made essentially as described earlier (Derr et al, 2006). Briefly, attractant was spotted onto semisolid agar plates containing minimal A medium with 0.25% (w/v) agar, and allowing it to diffuse at 4°C for 12–20 h. Bacteria were grown in LB medium supplemented with ampicillin and chloramphenicol to saturation and then inoculated at 1:1000 into minimal A medium. The cultures were grown to mid-exponential phase at 30°C, diluted to an OD600 of 0.05, and spotted onto the gradient plates. A total 5 μl of a culture was spotted approximately 2–2.5 cm from the attractant spots, and incubated at 30°C for 16–30 h.
Images of plates were acquired using an HP Scanjet 4470c flatbed scanner (color setting, 300 dpi resolution). All images are shown as negatives; the contrast has been heightened slightly and uniformly to improve visibility.
Capillary assays
The assays were carried out essentially as described earlier (Derr et al, 2006). Briefly, bacteria were grown to an OD600 of 0.3–0.4 in minimal A medium at 30°C. The culture was washed twice with chemotaxis buffer (10 mM potassium phosphate buffer pH 7.0, 0.1 mM Na–EDTA, 1 μM L-methionine, and 10 mM glycerol), which was pre-warmed to 30°C. The cells were then diluted in chemotaxis buffer to an OD600 of 0.05. Sealed calibrated pipettes (ID=0.27 mm, Drummond Scientific 2-000-001-90, Broomall, PA) were filled to a volume of 3±0.3 μl by heating and then inserting the open end into attractant. Filled capillaries were inserted into the chemotaxis buffer containing bacteria and left at 30°C for 1 h. Capillaries were then emptied into LB medium, serial dilutions were plated, and colonies were counted.
Supplementary Material
2 supplementary figures are included providing additional data from chemotaxis plate assays and capillary assays. 1 supplementary table is included containing data from the competition assays.
Acknowledgments
We thank Dena Pichette for synthesis of PAG. This work was supported in part by the NSF MRSEC program under award number DMR05-20020 and grants from the NIH (GM54616 to WFD, GM080279 to MG, AI74866 to WFD and MG, and GM073499 to SDG).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adler J (1973) A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol 74: 77–91 [DOI] [PubMed] [Google Scholar]
- Adler J (1975) Chemotaxis in bacteria. Annu Rev Biochem 44: 341–356 [DOI] [PubMed] [Google Scholar]
- Ahmed T, Stocker R (2008) Experimental verification of the behavioral foundation of bacterial transport parameters using microfluidics. Biophys J 95: 4481–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 [DOI] [PubMed] [Google Scholar]
- Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L (2008) A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol 4: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikov SI, Miller AC, Gosink KK, Parkinson JS (2004) Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J Bacteriol 186: 3730–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, Karig DK, Weiss R, Arnold FH (2007) Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA 104: 17300–17304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, You L, Arnold FH (2008) Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol 26: 483–489 [DOI] [PubMed] [Google Scholar]
- Budrene EO, Berg HC (1995) Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature 376: 49–53 [DOI] [PubMed] [Google Scholar]
- Chang AC, Cohen SN (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derr P, Boder E, Goulian M (2006) Changing the specificity of a bacterial chemoreceptor. J Mol Biol 355: 923–932 [DOI] [PubMed] [Google Scholar]
- Derst C, Henseling J, Rohm KH (2000) Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci 9: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DA, Way JC, Silver PA (2007) Designing biological systems. Genes Dev 21: 242–254 [DOI] [PubMed] [Google Scholar]
- Gestwicki JE, Strong LE, Kiessling LL (2000) Tuning chemotactic responses with synthetic multivalent ligands. Chem Biol 7: 583–591 [DOI] [PubMed] [Google Scholar]
- Grebe TW, Stock J (1998) Bacterial chemotaxis: the five sensors of a bacterium. Curr Biol 8: R154–R157 [DOI] [PubMed] [Google Scholar]
- Hedblom ML, Adler J (1983) Chemotactic response of Escherichia coli to chemically synthesized amino acids. J Bacteriol 155: 1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MP, Beacham IR (1990) Analysis of the Escherichia coli gene encoding L-asparaginase II, ansB, and its regulation by cyclic AMP receptor and FNR proteins. J Bacteriol 172: 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling JD (2008) Synthetic biology for synthetic chemistry. ACS Chem Biol 3: 64–76 [DOI] [PubMed] [Google Scholar]
- Mao H, Cremer PS, Manson MD (2003) A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc Natl Acad Sci USA 100: 5449–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AL, Svedas VK, Berezin IV (1980) Substrate specificity of penicillin amidase from E. coli. Biochim Biophys Acta 616: 283–289 [DOI] [PubMed] [Google Scholar]
- Schink B (2002) Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81: 257–261 [DOI] [PubMed] [Google Scholar]
- Topp S, Gallivan JP (2007) Guiding bacteria with small molecules and RNA. J Am Chem Soc 129: 6807–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP (2008) Random walks to synthetic riboswitches—a high-throughput selection based on cell motility. Chembiochem 9: 210–213 [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J (1999) Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfaardt GM, Korber DR, Lawrence JR (2007) Cultivation of microbial consortia and communities. In Manual of Environmental Microbiology, Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mils AL, Stetzenbach LD (eds), 3rd edn. Washington, DC: ASM Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2 supplementary figures are included providing additional data from chemotaxis plate assays and capillary assays. 1 supplementary table is included containing data from the competition assays.