Abstract
Despite the knowledge of complex prokaryotic-transcription mechanisms, generalized rules, such as the simplified organization of genes into operons with well-defined promoters and terminators, have had a significant role in systems analysis of regulatory logic in both bacteria and archaea. Here, we have investigated the prevalence of alternate regulatory mechanisms through genome-wide characterization of transcript structures of ∼64% of all genes, including putative non-coding RNAs in Halobacterium salinarum NRC-1. Our integrative analysis of transcriptome dynamics and protein–DNA interaction data sets showed widespread environment-dependent modulation of operon architectures, transcription initiation and termination inside coding sequences, and extensive overlap in 3′ ends of transcripts for many convergently transcribed genes. A significant fraction of these alternate transcriptional events correlate to binding locations of 11 transcription factors and regulators (TFs) inside operons and annotated genes—events usually considered spurious or non-functional. Using experimental validation, we illustrate the prevalence of overlapping genomic signals in archaeal transcription, casting doubt on the general perception of rigid boundaries between coding sequences and regulatory elements.
Keywords: archaea, ChIP–chip, non-coding RNA, tiling array, transcription
Introduction
Systems-biology approaches have been successfully applied to construct quantitative and predictive models of biological networks (Bonneau et al, 2007; Faith et al, 2007). However, a significant amount of information is missing from these models because of incomplete parts lists (unannotated genes, non-coding RNAs (ncRNAs), poorly understood protein modifications and so on) as well as a lack of molecular detail associated with these processes. Incorporating such detail will make these models mechanistically accurate and useful for synthetic-biology approaches targeting large-scale biological-circuit re-engineering. Among the current systems-scale models most amenable for such large-scale redesign are those that describe gene-regulatory networks (GRNs).
GRN models are usually built upon transcriptome data, in which typically genes or gene modules (with similar expression patterns and shared regulatory motifs) are associated with their transcriptional regulators through linear or Bayesian models. However, although these models can be predictive (Bonneau et al, 2007), they often rely on approximations of the transcription process and lack finer details of dynamic environment-dependent assembly of transcription complexes at each of the numerous promoters in the genome. High-density tiling arrays can be used to define transcribed regions (David et al, 2006), start sites (McGrath et al, 2007), and protein–DNA interaction sites (Reiss et al, 2008), which can be used to identify some of these missing details associated with transcriptional regulation, and thereby enable us to construct systems-scale predictive models of GRNs that are also mechanistically accurate.
We recently constructed a model of an environment and gene-regulatory influence network (EGRIN) for the halophilic archaeon Halobacterium salinarum NRC-1. This model accurately predicts the transcriptional changes in 80% of all genes to new environmental and genetic perturbations (Bonneau et al, 2007). Using an integrated biclustering algorithm to identify regulons and their putative cis-regulatory motifs (Reiss et al, 2006), and a sparse regression procedure to statistically pair these regulons with their putative regulators (Bonneau et al, 2006), we were able to discover the combinatorial and conditional regulation of genes by multiple TFs and EFs (environmental factors) (Bonneau et al, 2007). Although several of the statistically inferred influences in this network were shown to be likely mediated through direct interactions with the promoters of regulated genes, a large number of influences are thought to be indirect. The logical next step is to make this quantitative and predictive network also mechanistically accurate on a systems scale.
Construction of a mechanistically accurate systems-scale model is a reasonable expectation for Halobacterium salinarum NRC-1, as its transcription is driven by a simplified version of a eukaryotic RNA polymerase (RNAP) II (Hirata et al, 2008) in a genome with prokaryotic organization. The archaeal RNAP requires only two general transcription factors – GTFs (TATA binding protein –TBP and transcription factor B –TFB) for promoter recruitment and basal transcription initiation. Furthermore, only ∼130 putative transcriptional regulators (TRs) are present among the ∼2400 genes encoded in the genome of H. salinarum NRC-1 (Ng et al, 2000). A relatively small number of genes and few TFs (GTFs and TRs) together make H. salinarum NRC-1 an attractive model system for characterizing gene-regulatory mechanisms at all promoters. Notably, the combinatorial action of multiple TFBs and TBPs (H. salinarum NRC-1 possesses 6 TBPs and 7 TFBs) in defining basal promoter architecture in most archaea (Baliga et al, 2000; Facciotti et al, 2007) provides a unique opportunity to characterize dynamic conditional regulation of a large fraction of genes during cellular responses to complex changes.
Here, we report a significant step toward a mechanistically accurate EGRIN model by characterizing the dynamic remodeling of the transcriptome structure of H. salinarum NRC-1 during a complex cellular response, and correlating these changes to genome-wide binding locations of 50% of all predicted GTFs as well as several specific TRs. By integrating diverse data types, we identified: (i) transcription start sites (TSSs) and termination sites (TTSs) for ∼64% of the genes, including new and revised protein-coding genes; (ii) 61 new ncRNA candidates; (iii) 5′ and 3′ untranslated regions (UTRs) of mRNAs; (iv) functional promoters upstream and internal to coding regions; (v) instances of transcription termination inside coding sequences; (vi) mRNA populations with variable 3′-end locations; (vii) transcripts with extensive overlaps in their 3′ termini; and (viii) operon-encoding transcripts of variable length. Significantly, these findings suggest that the incorporation of mechanistic accuracy into GRN models would require genes, operons, promoters, and terminators to be treated as dynamic entities.
Results
Genome-wide protein–DNA binding data show TF binding inside genes and operons
A detailed map of genomic locations where TFs bind DNA and modulate transcription is essential to model mechanisms of gene regulation on a systems scale. Chromatin immunoprecipitation of transcription complexes coupled to microarray (ChIP–chip Ren et al (2000)) or sequencing (ChIP–seq (Robertson et al (2007)) is a commonly used approach to construct such maps. In ChIP–chip, the resolution to which the protein–DNA binding sites (TFBSs) can be identified is often limited by the genomic spacing of the probes in the array. We utilized the MeDiChI algorithm (Reiss et al, 2008) to estimate precise TFBS locations and their corresponding local false discovery rates (LFDRs) from new and previously reported genome-wide ChIP–chip measurements for 11 TFs (with two or more biological replicates for each): all TFBs (TFBa, TFBb, TFBc, TFBd, TFBe, TFBf, and TFBg), one TBP (TBPb) and three TRs (Trh3, Trh4, and VNG1451C) in H. salinarum NRC-1 (see Materials and methods). On the basis of simulations similar to those of Reiss et al (2008), with a noise model customized to mimic the data used in this study, we estimated that the average positional uncertainty in TFBS locations identified by MeDiChI averaged ∼50 nucleotides (nt) (1SE) over all ChIP–chip data sets used in this study.
We found that the 3072 significant (LFDR<0.1) individual TFBSs for all data sets often fell within distinct loci where at least three different TFs were observed within a ±50 nt window (P<10−8). We therefore refined this TFBS list to a conservative set of 318 such distinct ‘multi-TF-binding loci', hereafter TFBS loci throughout the genome (Table I; see Supplementary Table 1 for each loci). As we applied to each individual data set an LFDR cutoff of 0.1, which by itself is rather stringent, the joint LFDR of these 318 TFBS loci is significantly smaller than that. Although each individual TF had a significant bias of binding in annotated intergenic regions (∼60%, on average, versus ∼16% expected), this fraction increased to ∼70% (276) when considering the 318 TFBS loci (P∼10−31). Monte Carlo simulations of TFBSs placed only in non-coding regions in the genome with a ∼50–75 nt positional uncertainty and an LFDR between 0.1–0.01 show that 80–85% of detected TFBSs should fall in intergenic regions (for more details, see Materials and methods). Thus, our assessment was that a small but significant fraction of these significant TFBS loci in our ChIP–chip data sets (as many as ∼10% of the multi-TFBS loci) fell within coding regions. Here onwards, we present detailed and systematic experimental validation that shows that many of these TF-binding events inside coding sequences have significant consequences on the transcriptional regulation of diverse aspects of cellular physiology.
Table 1.
Numbers of TFBS loci comprised of varying numbers of individual TFBS and their distribution in annotated coding sequences, predicted operons, and conditional predicted operons
| Number of loci | Total | In annotated coding sequence | In predicted operons | In conditional predicted operons (P-value) |
|---|---|---|---|---|
| With ⩾1 TFBS | 1249 | 368 | 82 | 58 (1.4 × 10−10) |
| With ⩾2 TFBS | 649 | 231 | 34 | 28 (4.3 × 10−8) |
| With ⩾3 TFBS | 318 | 96 | 13 | 13 (<1 × 10−30) |
| With >3 TFBS | 196 | 56 | 10 | 10 (<1 × 10−30) |
The reported results of this paper utilize the 318 very stringent ⩾3 TFBS loci but clearly the same conclusion holds (although the numbers increase) as this threshold is relaxed. The P-values were estimated for the probability of observing as many TFBS loci internal to conditional operons (column 4), given the number of TFBS loci observed internal to all operons (column 5), and the estimated fraction of conditional operons (∼43%; see Results and Discussion).
Analysis of transcriptome structure shows new expression features
The location of a TFBS in the vicinity of a TSS or a TTS could indicate whether a given binding event is functional, especially for the interactions localized within a gene or operon. We investigated this by systematically mapping transcript boundaries and their dynamic changes at the whole-genome level using genome-wide tiling array data and then integrating this information with the TF-binding information.
We define transcriptome structure as the collection of TSSs and TTSs that together characterize transcriptional units (mono- and polycistronic mRNAs, tRNAs, rRNAs, and other ncRNAs). Sequence signatures for these features are yet to be characterized in archaea, and computational predictions based on known signatures in bacteria and eukaryotes remain error prone due to incomplete understanding of transcription processes in all organisms (Jones, 2006). Therefore, we experimentally mapped the transcriptome structure of H. salinarum NRC-1 by hybridizing total RNA (including RNA species <200 nt) to genome-wide high-density tiling arrays (60mer probes with 40 nt overlap between contiguous probes).
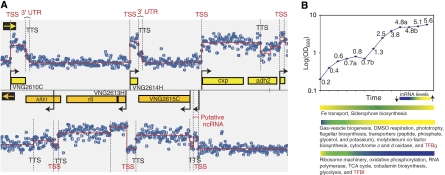
We first applied a segmentation algorithm based on regression trees (see Materials and methods) to map transcript boundaries in cells cultured under standard laboratory growth conditions (mid-logarithmic phase, 37°C, 225 r.p.m. shaking—hereafter ‘reference RNA') (Figure 1A). Although this approach effectively mapped TSSs for mRNAs, tRNAs, rRNAs and probable ncRNAs with significant expression levels, it was ambiguous for genes with low expression levels. Moreover, TTSs proved difficult to determine in general, even for highly expressed genes, because no sharp boundaries were observed for most transcripts at the 3′ termini (Figure 1A; Supplementary Figure 1B). We overcame these challenges and recovered further information by analyzing dynamic modulation of the transcriptome structure during typical growth of a batch culture under standard conditions (Figure 1B).
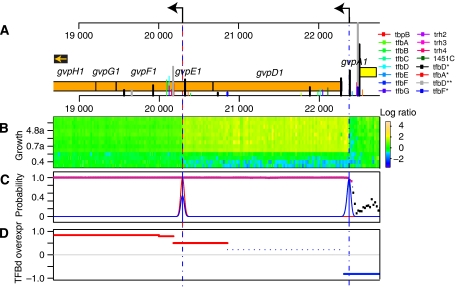
Figure 1.
Transcriptome structure and growth-phase-dependent changes in Halobacterium salinarum NRC-1. (A) Genome map of a segment of the main chromosome of H. salinarum NRC-1 (NC_002607) with corresponding signal intensity of total RNA from a mid-log phase culture (‘reference RNA') hybridized to 60mer overlapping probes in a high-density tiling array. Genes in the forward and reverse strands are shown in yellow and orange, respectively. Each blue dot represents probe intensity (in log2 scale) at the given genomic location in the forward (upper panel) or reverse (lower panel) strands. The overlaid red line is the result of a segmentation algorithm that was applied to determine transcription start sites (TSS and black arrows), transcription termination sites (TTS), untranslated regions in mRNAs (3′ UTR), and putative non-coding RNAs. (B) Dynamic changes in transcriptome structure were evaluated (Figure 2) at different phases of growth in a standard laboratory batch culture. Important physiological changes that are reflected in differential expression of corresponding mRNAs during the various phases of growth are indicated with a heat map (Facciotti et al, submitted).
H. salinarum NRC-1 presents a number of interesting switches in metabolism during growth (Facciotti et al, submitted) because of complex changes in EFs, including pH, oxygen, nutrition, and so on (Schmid et al, 2007). Although most single perturbations (radiation, oxygen, metals, and so on) affect the expression of only ∼10% of all genes (Baliga et al, 2004; Kaur et al, 2006; Whitehead et al, 2006), the changes during growth resulted in differential regulation of a significantly higher proportion of genes (∼63%, 1518 genes) (Figure 1B). These conditions thus enabled the investigation of a wider transcriptional landscape, which includes not only modulation of transcript levels (Figure 1B), but also extensive changes in transcriptome structure. We observed altered TSSs, TTSs, operon organizations, and differential regulation of putative ncRNAs (Supplementary Figure 1). By integrating hybridization signals (Figures 1A and 2B) with dynamic growth-related changes (Figure 2C and D), we estimated the probability that each tiling array probe was complementary to a transcribed region, mapped locations of putative transcript boundaries (Figure 2E; see Materials and methods) and identified 1574 TSSs and 1952 TTSs for most genes with some transcriptional variation. Subsequently, we manually assessed and curated gene assignment to each TSS and TTS. The error of these assignments is given by the resolution of probes on the tiling array (20 nt). In sum, TSSs were assigned to 64% (1156 singletons and 544 genes in 203 operons) of all annotated genes and TTSs were assigned to 1114 genes and 202 operons. A TSS and a TTS together define a unit of transcription (Supplementary Table 2). We describe below, how by correlating locations of these transcriptional units to predicted coding sequences in the genome, we were able to characterize and discover new features within the transcriptome structure.
Transcription of mono- and polycistronic mRNAs. In many organisms, especially prokaryotes, genes of related function are often co-transcribed as a single polycistronic mRNA (operons). Operon predictions based on genome-specific distance models, combined with comparative genomics and functional features identified 299 operons in H. salinarum NRC-1 (Price et al, 2005). According to our analysis of 1,698 genes with significant transcription signal, at least 544 (32%) genes were transcribed as polycistronic mRNAs in 203 operons. Comparative analysis with the predicted operon structures identified 123 new or truncated operons, which are dynamically regulated during growth.
Discovery of leaderless transcripts and 5′ and 3′ UTRs. UTRs in the proximal (5′) end of transcripts often contain signals, such as the Shine–Dalgarno (SD) sequence signature for ribosome loading (Sartorius-Neef and Pfeifer, 2004). Although some mRNAs spanned short distance beyond the coding-sequence boundaries, others were significantly longer (greater than the error in transcript-boundary assignment-20 nt), with 5′ (457 transcripts, 40% of genes assigned to an experimentally determined TSS) and/or 3′ (857 transcripts, 77% of the genes assigned to an experimentally determined TTS) UTRs (Supplementary Table 2 and Supplementary Figure 2). We validated the TSS and 5′ UTR lengths by comparing our observed UTR lengths with those experimentally measured in a closely related strain -H. salinarum R-1 (Brenneis et al, 2007). We found that, on average, the predicted 5′-UTR lengths correlate strongly with those determined by Brenneis et al (2007) (P<0.001); however the predicted NRC-1 3′ UTRs are usually longer (on average 1.8 ±1.3 times longer than those of R-1) (Supplementary Figure 2). Interestingly, 137 transcript pairs had overlapping 3′ ends (Supplementary Table 3) ranging from 25 to 788 nt in length, with a median length of 264 nt.
Distance between newly mapped TSSs and GTF-binding sites agrees with earlier knowledge of GTF binding. It is known that the archaeal pre-initiation complex lies between 25–30 nt upstream of the TSS (Bell et al, 1999). Although the relatively large uncertainty in the MeDiChI-mapped TFBSs precludes the quantification of this distance for individual TSSs, we found that the 318 TFBS loci (defined above) lie at an average of 24 nt (95% probability that the average falls between 35 and 16 nt) upstream of the nearest TSS. This may be compared with an average upstream distance of 59 nt (95% probability that the average lies between 69 and 49 nt) between the TFBS loci and the first (annotated) translation codon. This difference is further evidence of the significant number of genes with 5′ UTRs (see above).
Revisions of predicted translation start sites and discovery of new protein-coding genes. For 222 transcripts (178 genes and 31 operons), we observed that the TSS is downstream (by >20 nt) of the predicted start codon (Ng et al, 2000) and that the TSSs for 45% of these (52% of the genes and 26% of the operons) are corroborated by the locations of one or more TFBSs. We investigated this further by analyzing the distribution of peptides detected in 527 tandem mass spectrometry runs from 91 proteomics experiments representing 121 618 high-quality tandem mass spectra within the H. salinarum NRC-1 peptide atlas (PA) (Van et al, 2008). This study added 30 runs in four new experiments conducted specifically to catalog the proteome under conditions in which the reference RNA was prepared. Absence of peptides from these apparently untranscribed regions led to the revision of start codons for 61 genes and 12 operons. Conversely, we were also able to match previously unassigned tandem mass spectra to 10 new transcripts representing new protein-coding genes, and to detect longer proteins and transcripts for three genes (Supplementary Table 4). Comparison with strain H. salinarum R-1 (Pfeiffer et al, 2008) showed that most of these discrepancies with the original genome annotation of H. salinarum NRC-1 had been resolved in the newer annotation.
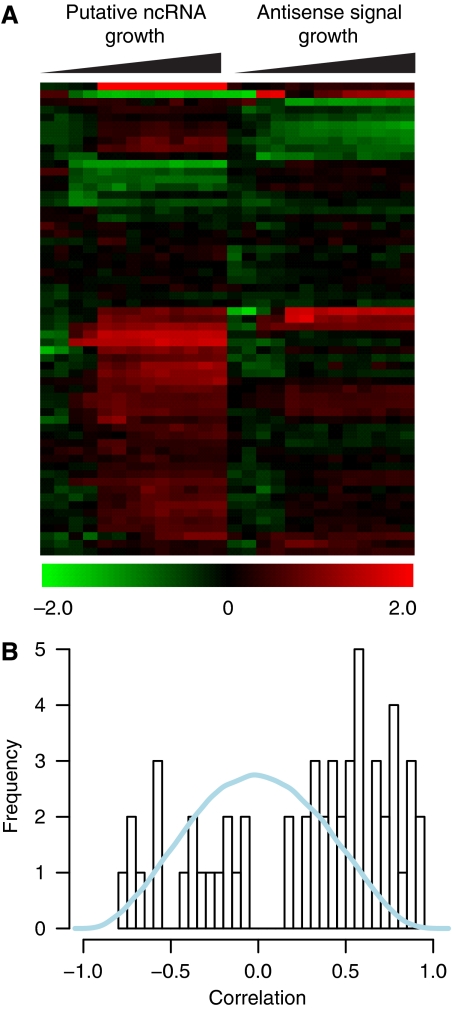
Discovery of putative ncRNAs. Non-coding RNAs are implicated in diverse regulatory processes from chromatin accessibility to mRNA translation and even modulation of protein activities (Storz et al, 2005). In archaea, 57 ncRNAs have been identified in Sulfulobus solfataricus (Tang et al, 2005) and 86 in Archaeoglobus fulgidus (Tang et al, 2002) by cDNA library cloning. Little is known about their functions except for the well-characterized snRNAs (Dennis and Omer, 2005). Although reliable bioinformatics approaches exist for identifying tRNAs and rRNAs, detection of other ncRNAs remains a challenge (Wang et al, 2006). We identified at least 61 transcripts experimentally with no bona fide coding sequence and/or matching tandem mass spectra from diverse proteomic analyses (Supplementary Table 5). These sequences were defined as putative ncRNAs. Fifty-two ncRNAs (85%) had increased expression levels at higher cell densities, whereas only 9 (15%) were downregulated. This is consistent with the predicted function of ncRNAs during stress (Shimoni et al, 2007), given the decrease in oxygen and nutrient availability at higher cell densities. Expression profiles for most ncRNAs were either positively or negatively correlated with corresponding changes in antisense transcripts (P<10−14) (Figure 3B).
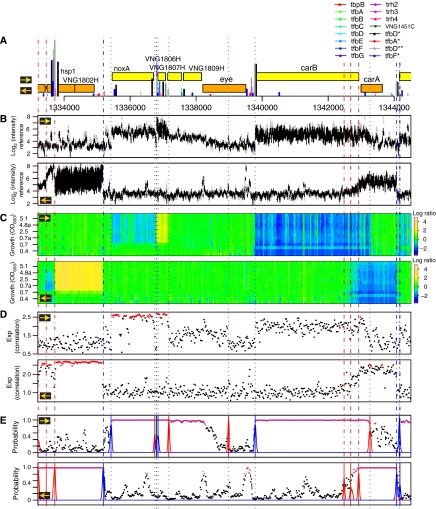
Figure 2.
A multitiered approach to characterize transcriptome structure. The transcriptome structure was determined through integration of RNA-hybridization signal (Figure 1), and analysis of relative changes in RNA levels corresponding to each probe. In each panel, the horizontal axis indicates genomic coordinates on the main chromosome and the two sub-panels show strand-specific signals (denoted by a yellow arrow for the forward and an orange arrow for the reverse strand). (A) Putative protein-coding genes on the forward and reverse strands are shown as yellow and orange rectangles, respectively, along with protein–DNA interaction sites (vertical bars, color coded per TF) determined by MeDiChI analysis of ChIP–chip data. Height of each vertical bar represents putative strength of binding event (proportional to chip signal intensity). Binding sites derived from high-resolution tiling array data are indicated by an asterisk (*) in the inset legend. (B) Mean reference-RNA hybridization signal (black dots) and associated error for each probe from 54 replicate experiments was normalized for sequence-content bias; the non-normalized data are shown with gray points (vertical axis: log2(signal intensity)). (C) Dynamic changes in the transcriptome are illustrated as a heat map along the genome (X-axis), with time along the growth curve increasing vertically from bottom (early log phase) to top (stationary phase); the color scale represents log2 ratio of transcript-level changes during growth relative to the reference RNA (blue is downregulated; yellow upregulated). (D) Correlation of growth-related transcriptional-change measurements for each probe with that of its neighboring (downstream) probe shown along the genome, exponentiated in this plot to enhance the visual contrast between correlated and uncorrelated probes. Probes with high correlation (r>0.9) are highlighted in red. (E) The probability of assigning each probe to a transcribed region was calculated by integrating data from panels A to D; probes with high probability (P>0.9) are highlighted in pink (Materials and methods). An integrated multivariate segmentation approach was used to identify and classify transcript boundaries as either TSSs (blue lines; dotted lines correspond to the forward strand and dashed lines to the reverse strand) or TTSs (red lines; dotted lines correspond to the forward strand, dashed lines to the reverse strand). Blue and red bootstrap density distributions indicate the relative likelihood of associating each position with a transcript boundary. This multivariate approach significantly improves the detection of transcript boundaries, in particular for TTSs. For example, the TTS for carB is difficult to determine from hybridization signals for reference RNA (B), but its differential expression during growth enables the identification of its TTS. The gradual decay in signal at the 3′ end of carA results in the assignment of multiple TTS.
Figure 3.
New non-coding RNAs in Halobacterium salinarum NRC-1. (A) Expression profiles of 61 putative ncRNAs and their respective antisense transcripts during growth. (B) The bimodal distribution of correlations between putative ncRNAs profiles and antisense transcripts suggests the ncRNAs might stabilize or destabilize transcripts on the opposite strand (the null distribution from randomly selected probes is shown in blue).
Integration of TFBSs with transcriptome structure shows conditional modulation of operon organization
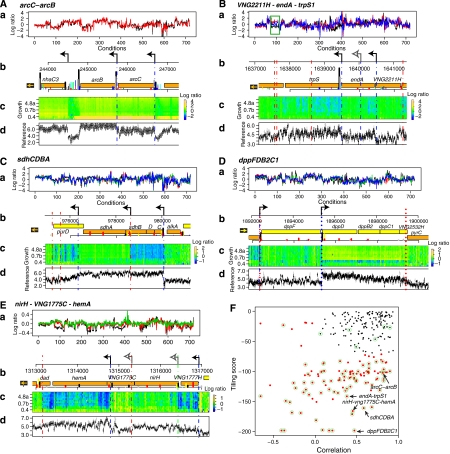
Changes in transcript levels and structure are ultimately the product of gene regulation mediated by dynamic assemblies of TF–DNA complexes. This mechanistic perspective of global gene-expression dynamics can be obtained by correlating transcript boundaries with genome-wide binding locations of TFs. We estimated that at least 13 of the 318 TFBS loci were internal to predicted operons (Price et al, 2005) (see Materials and methods). This is a conservative estimate (see Table I) as only TFBS with three or more binding loci were considered. Eight of these TFBS loci fall within 50 nt of an intergenic ‘gap' between predicted open-reading frames in the operon. An interesting example is the cluster of genes involved in arginine fermentation (arcRACB), at least two of which (arcC and arcB) constitute a predicted operon (Figure 4A) (Price et al, 2005). Notably, the strong co-expression of these genes across a wide range of environments (Bonneau et al, 2007) (Figure 4A(a)) does not necessarily result from a unique polycistronic message. The GTF-binding sites upstream to arcC and in the intergenic region between arcC and arcB (Figure 4A(b)) along with nearby TSSs (Figure 4A(d)) indicate two promoters that yield three possible transcripts: polycistronic arcCB and monocistronic messages for each arcC and arcB. The three possible transcripts suggested by our analysis (polycistron arcCB and monocistronic messages for each arcC and arcB) were consistent with previously reported northern-blot experiments that identified all three transcripts (Ruepp and Soppa, 1996), showing that our systems-level approach was able to independently recover known information.
Figure 4.
Conditional modulation of operon organization. Analysis of predicted operon structures identifies unexpected internal promoters that conditionally break the organization during cellular responses in differing environments. (A) The high degree of co-expression of arcC (red) and arcB (black) transcript-level changes in diverse environments (probed by ∼700 microarray experiments) (a) coupled to their genomic organization (b) strongly suggested co-transcription of these genes as an operon. Dynamic transcriptional changes of these genes during growth (c) also support this prediction. However, the integrated transcriptome-structure analysis identified a promoter (black arrow along genome coordinates of plasmid pNRC200 (NC_002608) in (b) and vertical blue line spanning panels b–d) in the 56-nt intergenic region between arcB and arcC. The location of the promoter is consistent with the different absolute levels of transcripts spanning the two genes (d) as well as with locations of TFBSs (vertical lines in the pNRC200 map in (b); for color code see Figure 2). (B) Although the predicted operon organization of VNG2211H (blue), endA (red), and trpS1 (black) is supported by their co-expression in most environments, their expression is not correlated during a few responses, including experiments investigating H. salinarum NRC-1 interaction with a unicellular alga (green box) (a). This differential regulation was also observed during growth (c) and could be explained by an alternate promoter within the coding sequence of endA (black arrow) whose location was corroborated by co-localized TFBSs (b) and a distinct TSS (c and d). A second weak TSS was also identified internal to endA (gray open arrow). (C) Genes in the predicted operon sdhCDBA (sdhC - blue, sdhD - green, sdhB – red, and sdhA - black) are co-expressed in most of the environmental perturbations, except for sdhA during a few responses. (b) TFBS (vertical lines, color coded as Figure 2) are found near the TSS for sdhC and in the coding region of sdhB (black arrows, blue dashed lines). (c) Dynamic changes during growth show that sdhCDB is downregulated and sdhA does not have the expression levels altered (d) and reference-RNA hybridization shows that sdhA is expressed. (D) Operon dppFDB2C1. (a) dppF (black) and dppD (red), dppB2 (green) and dppC1 (blue) are organized in a predicted operon and are co-expressed in most of the environmental perturbations. TSS identified for dppF and dppDB2C2 (black arrows and blue dotted lines) are localized near (b) TFBS (vertical lines, color coded according to Figure 2), which could explain the (c) differential expression of dppF and dppDB2C1 during growth. (E) Operon nirH-VNG1775C-hemA. (a) nirH (green), VNG1775C (red), and hemA (black) are organized in a predicted operon and co-expressed in most of the environmental perturbations. (b) TFBS localized internal to VNG1775C (vertical lines) are found near the TSS for hemA (black arrow), which could explain (c) the differential expression of this gene at higher cell densities. (F) Conditional operons were identified in a genome-wide manner by analyzing two parameters: minimum correlation score along all 719 environmental conditions between each gene in each predicted operon (horizontal axis) and minimum ‘tiling score', which quantifies the difference in the tiling probe levels for genes constituting the operon (vertical axis; see Results and Discussion for details). Green circles represent operons that were manually identified as condition dependent and were used as a training set for the conditional-operon classification. Red dots represent operons that were automatically classified as condition dependent (see Materials and methods for details). The conditional operons described above are highlighted.
This prompted us to search for operons interspersed with conditionally active promoters. For instance, we observed a TFBS internal to the operon VNG2211H-endA-trpS1 (Figure 4B). This putative operon encodes a hypothetical protein, a tryptophanyl tRNA synthetase and a tRNA intron nuclease, respectively (Figure 4B(b)). The internal TFBS correlates with both a nearby TSS and differential expression of the operon genes during growth: whereas trpS1 is downregulated, expression levels of endA and VNG2211H do not change (Figure 4B(c)). Although these genes are co-expressed in most of the ∼700 microarrays representing ∼20 different environmental perturbations, we could detect instances besides standard growth where they are not co-transcribed, such as during interaction of H. salinarum NRC-1 with Dunaliella salina, a unicellular alga (Figure 4B(a), green box). Notably, the conditionally activated promoter for trpS1 is located within the coding sequence of endA.
Using this approach, we manually identified 78 operons with conditionally altered gene-expression levels of constituent genes. To gain a global perspective on the prevalence of conditional operons, we classified all predicted operons in the H. salinarum NRC-1 genome based on two scores (Figure 4F): (1) correlations between expression profiles of genes in each operon among 719 microarrays and (2) the ‘tiling score', which is the log10(P-value) of the statistical significance of the difference in tiling array probe intensities or ratios between the genes in the operon (Materials and methods). A low ‘tiling score' indicates operons where member genes present significantly different transcript levels in the reference RNA or in any of the growth samples probed. We then classified the operons using the manually identified conditional operons as the training set (Figure 4F, green circles). Even for operons with genes that are co-expressed in most environmental conditions (high correlation), such as the arcC–arcB operon (Figure 4A(a)), the low tiling score (Figure 4F and 4A(d)) accurately identifies an internally located alternate promoter. The operon VNG2211H-endA-trpS1 has a low correlation score, as these genes are anti-correlated in some conditions (Figure 4B(a)), as well as a low tiling score because of significant difference in their absolute expression levels (Figure 4B(c)). Other examples of conditional operons are illustrated in Figure 4: sdhCDBA encodes the subunits of the succinate-dehydrogenase complex (Figure 4C), where sdhC, D, and B are downregulated at high cell densities, whereas the expression of sdhA remain unchanged (Figure 4C (c)); dppFDB2C2, which encodes the subunits of a dipeptide ABC transporter (Figure 4D), in which, with the exception of dppF, all other genes are upregulated at higher cell densities (Figure 4D(c)) and nirH-VNG1775C-hemA (Figure 4E), where hemA, encoding a glutamyl-tRNA reductase is downregulated at high cell densities.
We were able to compute tiling and correlation scores for the 269 operons in H. salinarum NRC-1 with significant expression in the tiling-array experiments, and classified 115 (∼43%) as condition dependent (Figure 4F, red dots; Supplementary Table 6). Interestingly, conditional operons were highly enriched for internal TFBS loci relative to their non-conditional-classified peers (P<10−9, Figure 4F, black dots); indeed all 13 operon-internal TFBS loci fell within conditional operons (P<10−30, see Table I).
Interaction of TFs within coding regions is associated with transcript boundaries
The operons in H. salinarum NRC-1 usually have very short (∼50 nt) or no intergenic regions between constituent annotated coding regions (‘gaps'). We find that although only 27% of the gaps between coding regions in all 299 predicted operons are longer than 20 nt, this fraction increases to 37% in the conditional operons (Supplementary Figure 3). Although this might partly explain some of the internal promoter activity, the lack of a significant intergenic region (⩽20 nt) between at least 53 conditionally co-transcribed gene pairs within operons suggest the presence of alternate internal promoters within coding sequences, as illustrated with operons VNG2211H-endA-trpS1, sdhCDBA, and nirH-VNG1775C-hemA. Notably, absence of a significant intergenic gap and high degree of correlation in transcription profiles for these genes would have precluded discovery of their conditional co-transcription based on generally accepted rules for operon organization (Supplementary Figure 3E).
TF binding in the middle of coding sequences can also result in transcription initiation or termination internal to a single annotated protein-coding gene. We highlight this with an example that focuses on gas-vesicle biogenesis—a hallmark response of H. salinarum NRC-1 to low toxic conditions under high cell density (Yang and DasSarma, 1990). Several TFs, including TFBd, bind internally to two distinct loci within gvpE1, a transcriptional regulator of gas-vesicle biogenesis (Scheuch et al, 2008). The binding locations of one set of TFs correlates with the termination of a transcript initiated upstream to gvpD. Moreover, when we observe the relative transcript levels in a strain overexpressing TFBd, it results in upregulation of a transcript downstream of its binding location in the second locus (Figure 5d). Together these results show both a regulated TFBd-dependent promoter and a growth-phase dependent terminator located inside the gvpE coding sequence. It is noteworthy that despite extensive prior analysis of the transcriptional regulation of gas-vesicle biogenesis, this aspect of growth-dependent regulation was never discovered.
Figure 5.
TF binding internal to coding regions results in transcriptome-structure changes. A putative promoter and a terminator internal to the coding sequence of gvpE1, a gas-vesicle biogenesis regulator, is corroborated by co-localized TFBSs for several TFs, including TFBd (A). Although the activity of the terminator was verified by growth-phase-dependent termination of transcription originating upstream to gvpD1 (B), this region also presented high probability of being transcribed (P>0.9 are highlighted in pink) and a putative transcription start site from an internal promoter (blue line) (C); the internal promoter could be validated by analyzing the transcriptome structure in a strain overexpressing TFBd (D). The red line indicates a break in the transcription levels of the strain overexpressing TFBd relative to the reference RNA. This evidence associated with mapped TFBd-binding site and TSS suggests the presence of an internal promoter.
We note that our estimate of the prevalence of internal TFBSs as described above is conservative given the stringent nature of our automated analysis with the inclusion of only significant multi-TFBS loci, as well as the limited range of conditions under which both transcriptome and ChIP–chip data were collected. Indeed, although 69% of the 318 TFBS loci lie in intergenic regions, a fraction of the remaining 31% (∼100 sites) which fall within annotated coding regions (in particular, 42 of these, which fall >50 nt from any annotated start or stop site) are likely functional (Table I). After revising the predicted translation start sites based on the transcriptome structure as described previously, we observed 610 TFBSs over all 11 TFs (LFDR<0.1) that fell within coding regions (by >50 nt) and 47 (7.7%) were nearby (within 100 nt) a putative internal transcription break point (P∼0.015 relative to randomly placed internal break points), suggesting that they might constitute functional promoters and/or terminators (Supplementary Table 7).
However, using the automated procedure, about half of all detected TSSs for annotated genes were not associated with any detectable TFBSs and about half of all detected TFBSs were not associated with any identifiable TSS. Although a significant fraction of transcript boundaries might also result from alternate regulatory mechanisms, such as transcript cleavage, our inability to correlate these features to TF-binding locations might also reflect the dynamics and complexity of combinatorial TF binding and TSS selection.
Discovery of conditional promoter binding of GTFs
All ChIP–chip data described above were collected at mid to late phase of growth in batch cultures (OD600>1.0), and therefore are not specific to the conditions over the entire growth curve, which were investigated in the mRNA-expression experiments. We investigated the effect of this condition dependence on the ChIP–chip data by comparing genome-wide binding locations of TFBd during three different phases of growth (OD600=0.3, 0.8, and 1.4). Surprisingly, even though TFBd was strongly overexpressed in all three ODs, only the ChIP–chip data obtained at OD600=0.8 (mid phase) showed highly enriched over-representation of TFBSs within intergenic regions (P∼10−19 for OD600=0.8, versus P∼10−3 for OD600=0.3 and P∼10−2 for OD600=1.4), a stringent criterion which we have used throughout this and other studies (e.g. Reiss et al, 2008) to enrich ChIP–chip TFBS selections for likely functional binding. Moreover, the locations of the strongest TFBSs were more in agreement (within 50 nt) between the early- and mid-phase (OD600=0.3 and 0.8, respectively) data (P∼10−3) and between the mid- and late-phase (OD600=0.8 and 1.4, respectively) (P∼10−3) than between the early- and late-phase (OD600=0.3 and 1.4, respectively) data (P∼0.35).
Validation of regulated transcriptional initiation from promoters in coding sequences
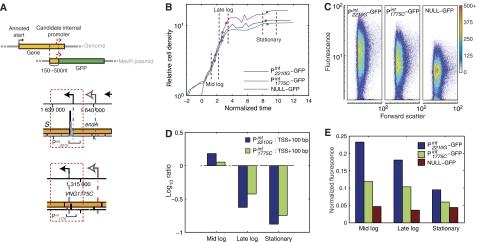
The proximity of chromosomal loci with multiple TFBSs and experimentally mapped TSSs within coding regions strongly indicates that the observed conditional modulation of operon organization is achieved through the activation of promoters in coding sequences. However, it is imperative to rule out alternate hypotheses, such as conditional transcript cleavage followed by differential degradation of the mRNA fragments as such a process could also result in truncated transcript(s) with a 5′ or a 3′ boundary within a coding sequence. Verifying our precision of mapping a transcript boundary and TFBSs with other technologies (EMSA, northern blot, 5′ RACE and so on) cannot definitively refute this alternate hypothesis. Instead, the ultimate proof for functional promoters within a coding sequence is the in vivo demonstration of regulated transcription initiation at these loci. To provide such evidence we developed a fluorescence-based assay using a fast-degrading variant of GFP (Reuter and Maupin-Furlow, 2004) and showed that the synthesis and degradation of this reporter accurately reflects dynamics observed at the level of transcription (Supplementary Figure 4). Next, we constructed GFP transcriptional fusions to assess the activity of promoters inside coding sequences of two genes encoding a tRNA endonuclease (VNG2210G) and a siroheme biosynthesis enzyme (VNG1775C) (Figure 6A; Materials and methods). The 100-bp long internal promoter regions were selected to include the TSS and the adjacent experimentally determined TFBS locus. Using flow cytometry, we then monitored the change in fluorescence along the growth curve (Figure 6B). Fluorescence of H. salinarum NRC-1 transformants with internal promoter–GFP (Pint2210G–GFP and Pint1775C–GFP) fusions and a no-promoter construct (NULL–GFP) was measured along the growth curve using flow cytometry and calibrated against a fluorescent bead internal standard (Figure 6C; Materials and methods). This validated functional transcription initiation from these promoters. Moreover, it also showed that the activity of the promoter was conditional on the growth phase (Figure 6E). Although it is difficult to assess every instance of internal promoters using this approach, it nonetheless validates that integrated analysis of globally determined TSSs and TFBSs coupled to the analysis of gene-expression changes can lead to the discovery of conditionally activated promoters—even when they are located inside genes and operons.
Figure 6.
GFP expression validates promoters localized in the coding regions. (A) Internal promoter–GFP fusion construction strategy. The 150–500 nt regions upstream of internal transcription start sites (within tiling probe error) were fused to a GFP coding sequence on a mevinolin resistance (MevR) selectable expression plasmid. Selected internal promoters located within coding sequences of VNG2210G and VNG1775C used for GFP-expression validation are shown. (B) Sampling points along transformants growth curves. Batch cultures of strains carrying these internal promoter–GFP transcriptional fusions were sampled at mid-log, late-log, and stationary phase. For purpose of comparison, the growth curves were normalized in time relative to the characteristic growth rate observed during exponential growth. (C) The 2D densitometric scattergram of fluorescence versus forward scatter for cells with active internal promoters. Population density increases from blue to red. (D) Growth-phase-dependent transcriptional activity of the internal promoters was measured as the mean signal intensity for probes covering a region of 100 nt downstream of the internal TSS. (E) GFP production was used as proxy to validate that the growth-phase-dependent change in transcription observed in panel D was due to regulated transcription initiation by the internal promoters. The plots indicate normalized mean population fluorescence values during various growth phases (calculated from distributions shown in panel C for samples noted in panel B).
Discussion
Our analysis of genome-wide protein–DNA binding sites suggested that ∼10% of the multi-TFBS loci fell within coding regions. To show that these TFBS have significant functional consequences on transcriptional regulation and cellular physiology, we carried out a series of systematic experimental validations. First, we analyzed the transcriptome structure of H. salinarum NRC-1 under dynamic conditions. By correlating locations of the transcriptional units to predicted coding sequences in the genome, we were able to discover and characterize new features within the transcriptome. Next, by integrating TFBS locations with the transcriptome structure, we were able to show that some of these internal binding sites were indeed functional in the conditional modulation of operon organization, including promoters in coding regions. Finally, using transcriptional fusions to GFP reporters we provided in vivo validation of growth-phase-regulated transcription initiation from two of these promoters localized in coding sequences. We will discuss in detail each of these findings and their impact on our understanding and modeling of GRNs.
New features in H. salinarum transcriptome: increasing and detailing the parts lists
By analyzing the transcriptome of H. salinarum NRC-1 using high resolution and under dynamically changing growth conditions, we were able to assign TSSs to 64% of all annotated genes, TTSs to 46% of the genes and verify the expression of 203 operons. Notably, the average distance of ∼24 nt between these TSSs and experimentally mapped locations of TFBSs for one or more of 11 TFs is consistent with earlier knowledge of the relative position of the pre-initiation complex from the TSS. One important outcome of mapping TSSs and TTSs was the discovery of 5′and 3′ UTRs for genes and operons. The observed absence of a 5′ UTR in most of the genes with an identified TSS suggests either internal SD translation signals or more likely, an alternate (eukaryotic-like) mechanism of translation initiation. In contrast, most transcripts with an experimentally detected TTS had a 3′ UTR, which was on average longer than those determined by Brenneis et al (2007). This difference can be explained by the smooth decay observed in the signal at the 3′ end of the transcripts (Figures 1A and 2), which suggests that for a given gene, there are mRNA populations with the same 5′ end but different 3′-end locations. The shorter messages are usually more abundant and have higher probability of being selected for 3′-end determination by sequencing approaches (Brenneis et al, 2007). We hypothesize that this smooth signal decay at the 3′ termini of most transcripts is a product of imprecise termination, degradation, and/or paused or incomplete elongation complexes. Overall, ∼5% of the H. salinarum NRC-1 genome seems to be transcribed simultaneously in both strands. We also observed an overlap of 3′ ends of 137 transcript pairs, a phenomenon that has also been observed in yeast at a similar scale (Nagalakshmi et al, 2008), but the functional implication of this overlap remains to be investigated.
By correlating the transcriptional units contained within pairs of TSS and TTS with chromosomal coordinates of predicted genes (Ng et al, 2000) and experimentally mapped peptides from large-scale proteomics studies (Van et al, 2008), we were able to revise the translation start site for 61 genes and detect 10 new protein-coding genes (Supplementary Table 4). This highlights the importance of constant genome re-annotation on the basis of evidence presented by new high-throughput experimental data. Another important feature was the identification of 61 new putative ncRNAs in H. salinarum genome. Although the physiological roles and mechanism of action of specific ncRNAs remain to be uncovered, the significant correlation (positive or negative) between the profiles of the ncRNAs and the antisense strand (Figure 3) are consistent with the characterized roles of ncRNAs in the regulation of their cognate antisense transcripts. The discovery of these ncRNAs represents new information in the parts lists of regulatory elements encoded by the H. salinarum NRC-1 genome.
Mapping of all these new features of the H. salinarum NRC-1 transcriptome is expected to pave the way toward a detailed functional and mechanistic analysis of GRNs, thereby improving global models of cellular behavior.
Dynamics of operon organization
By observing the dynamics of the transcriptome structure, we noted that the organization of some operons seemed to be conditionally modulated. To assess the global prevalence of such operons, we devised a quantitative measure for classifying any operon as conditional, by integrating both the data from the transcriptome structure (‘tiling score') with correlations derived from expression profiles of H. salinarum NRC-1 genes in 719 microarray experiments (see Materials and methods). Using these measures, we classified 43% of the measured operons as condition dependent (Figure 4F, red dots; Supplementary Table 6). There was a strong functional link between transcription factor binding inside operons and their classification as ‘conditional' (P<10−9).
The conditional modulation of the operon arcBCD (Figure 4A) involved in arginine fermentation could be validated independently. Northern-blot experiments from (Ruepp and Soppa, 1996) showed the existence of the three possible transcripts suggested by our analysis. The failure to predict an alternate promoter for arcB from co-expression and genome organization analysis (Price et al, 2005) (Figure 4A (c)) emphasizes the importance of an integrated approach that incorporates TSS and protein–DNA interactions to identify detailed mechanistic information for modeling GRNs. We have provided further evidence that two newly discovered internal promoters inside the coding sequences of VNG2210G and VNG1775C (Figure 6) can also drive regulated transcriptional initiation during complex changes associated with growth.
It is arguable whether, in some cases, our data simply refute the initial prediction of operon organization (Price et al, 2005). However, operons with low correlation and high tiling score have high probability of being conditionally co-transcribed, as there is no difference in their absolute transcript levels, suggesting that these genes are transcribed as a polycistron in some, if not in most, conditions. Many operons with low overall correlation and low tiling score still present meaningful co-expression of genes in specific conditions (see Supplementary information 1 at http://baliga.systemsbiology.net/regulatory_logic/). Likewise, many operons have high correlation but low tiling score, suggesting that they have identical relative transcript levels (and are probably co-regulated) even if their absolute transcript levels differ. On the basis of this evidence, we posit that considering operons as dynamic entities is more appropriate than refuting the initial prediction, given that the currently available data sets do not exhaust the universe of possible environmental perturbations. This raises interesting questions on how the annotation databases will evolve to represent the complicated dynamics of biological features.
Physiological implications for conditional modulation of operon structures
Modulation of transcript levels within certain operons suggests a change in stoichiometry or composition of subunits within protein complexes. Alternatively, it can also be a mechanism for maintaining stoichiometry of a complex with differential turnover or translation rates of specific subunits (Hayter et al, 2005). We illustrate this with two examples:
In the sdhCDBA operon (Figure 4C), which encodes four subunits of succinate-dehydrogenase (SDH) enzyme complex, the Fe–S, cytochrome b, and membrane anchor subunits (SdhC, D, and B, respectively) are downregulated at high cell densities, whereas the expression of the FAD-binding flavoprotein component (SdhA) remain unchanged (Figure 4C-c). Downregulation of SDH at later phases of growth correlates with a drop in oxidative phosphorylation and concomitant reduction in cellular ATP levels (Schmid et al, 2007). Interestingly, differential regulation of the SdhA subunit relative to the three other subunits has also been observed in mitochondria and has been linked to O2 sensing (Piantadosi and Suliman, 2008). Although the implications of this differential regulation are bound to be different in H. salinarum NRC-1, it nonetheless suggests potential physiological consequences of changing stoichiometry of SDH subunits.
In the dppFDB2C2 operon for a dipeptide ABC transporter (Figure 4D), all subunits, including the periplasmic and permease components (DppD, B2, and C2) with the exception of the ATPase subunit (DppF) were upregulated at higher cell densities (Figure 4D(c)). Among other speculations, including possible subunit exchange with other ABC transport systems, it is possible that this complex may function without an ATPase subunit (Hebbeln et al, 2007) to conserve a limited ATP pool at higher cell densities (Schmid et al, 2007) by taking advantage of a nutrient rich environment.
The association of a TFBS with a TSS internal to an operon strongly suggests that the operon's genes are conditionally transcribed through alternate promoters. Alternative mechanisms that could result in such behavior include post-transcriptional cleavage followed by differential turnover of mRNAs, intra-cistronic transcription termination (Adhya, 2003; Lee et al, 2008), or even the condensation state of the genome by chromatin proteins in regulating gene expression (Reeve and Sandman, 2007). Although specific consequences of this phenomenon on the function of SDH and the ABC transporter will require further investigation, these observations nevertheless challenge our assumption that the operon organization of genes encoding these complexes reflects static subunit composition and fixed stoichiometry in diverse environments. Our findings reinforce the fact that even simple prokaryotes possess complex mechanisms to fine tune the expression of genes in an operon through prevalent use of alternate promoters and/or terminators that are combinatorially induced or repressed in response to dynamically changing environments (Adhya, 2003).
The significance of conditional activity of transcriptional promoters
Although ∼57% of the operons were classified as not condition dependent by our analysis (Figure 4F, black dots), we cannot rule out their conditional modulation in environments yet to be investigated. This is because transcription is a regulated and dynamic process with activities of any given TF dependent not only on its own abundance in the cell but also on the simultaneous presence and availability of relevant cofactors, accessibility of binding sites and/or absence or diminished activity of other competing factors. Intragenic binding sites could also indicate re-association of TFs with the transcription complex, aiding the polymerase in the recognition of pausing or termination signals (Reppas et al, 2006; Lee et al, 2008) or may even indicate previously unknown activity of GTFs (Wade et al, 2007). Our ChIP–chip experimental data rely on the overexpression of a given TF from a heterologous promoter on a plasmid, which might alter the relative concentrations of the TFs in the cell. In order to overcome this experimental constraint, we took a highly conservative approach, where only multi-TFBS loci were considered for further analysis.
Notably, there were numerous examples where bona fide associations of TFBSs and TSSs or TTSs were not made by our conservative automated procedure (see Materials and methods). For instance, although some TFBSs presented weak binding-signal intensity, they could be manually associated with TSSs or TTSs, suggesting they too might be functional; and vice versa, weak TSSs or TTSs could be assigned to nearby TFBSs. Furthermore, by charting growth-phase-dependent changes in genome-wide binding patterns of TFBd we also provided further evidence for the conditional nature of TF binding. Although this shows some limitations in the coverage afforded by our automated approach and the single data set covering only growth-related transcriptional changes, it also gives a perspective on how to incorporate such valuable information with less stringent modeling approaches that integrate orthogonal evidences to mechanistically define GRNs.
In other words, protein–DNA binding is probabilistic and combinatorial, depending on the relative abundance of TFBs, TBPs, and TRs under specific environmental conditions, thus, when we observe a TFBS that is weak or not associated with a transcript boundary or a strong promoter consensus (Muller et al, 2007), it does not necessarily mean that it is not functional. In fact, some weak-affinity TFBSs in yeast have been shown to be functional (Tanay, 2006). Only given sufficient experiments for localizing most TFBSs in a wide array of environments, one could map these types of conditional interactions to construct a comprehensive map of dynamic transcriptional regulatory mechanisms at all promoters.
Conclusions
Historically, studies of transcriptional regulation have often focused on TFBSs upstream to coding sequences. Furthermore, TFBSs inside coding regions are usually viewed as spurious or non-functional and used as a quality control metric for genome-wide ChIP–chip experiments. Computational (Ward and Bussemaker, 2008) and experimental (Lee et al, 2002) analyses of promoters restricted to intergenic regions add a significant bias in the distribution of documented TFBSs in the genome, reinforcing the overall assumption that transcriptional regulation occurs exclusively in intergenic regions.
Interestingly, many studies that focus on a single gene or operon do find internal promoters (Tsui et al, 1994; Guillot and Moran, 2007). We have significantly extended these findings, by showing that the widespread and conditional affinity for several TFs does not discriminate between coding and intergenic regions. It also shows how a simple prokaryote can use the same set of genes in different combinations to elicit different responses according to the environmental challenge. Given recent reports of extensive TF binding within coding regions in many organisms (Tanay, 2006; Zhu et al, 2006; Yochum et al, 2007; Shimada et al, 2008), evidence is mounting to suggest that this is a general phenomenon. This is not surprising given that the genome is known to encode multiple levels of information within the same sequence (Itzkovitz and Alon, 2007)—here TF binding and gene coding.
Irrespective of the specific underlying mechanism(s), our observations of widespread modulation of operon architecture, as well as transcription initiation and termination inside genes, etc. all constitute evidence that archaea can intersperse regulatory logic within their coding sequence and blur the boundaries between coding and non-coding elements. We have shown that it is possible to use new high-throughput technologies to find these biologically important instances in which transcriptional regulation does occur within coding sequences and, furthermore, that it is possible to globally characterize specific regulatory mechanisms responsible for these phenomena. Combined with new high-throughput sequencing technologies, our results will expand the view of genetic information processing that can be investigated at high resolution (Nagalakshmi et al, 2008; Wilhelm et al, 2008). These data will enable construction of mechanistically accurate models for reliable systems re-engineering of biological circuits.
Materials and methods
Strains, culturing, and growth conditions
H. salinarum NRC-1 growth-curve experiments were conducted in CM media, in a water bath incubator at 37°C with agitation of 125 r.p.m. Reference samples were cultured under standard growth conditions (Baliga and DasSarma, 1999), at mid-log phase (OD600=∼0.6), as well as all the strains used for ChIP–chip experiments (Facciotti et al, 2007).
High-resolution tiling array construction, RNA hybridization, and reference normalization
Whole-genome high-resolution tiling arrays for H. salinarum NRC-1 were designed with e-Array (Agilent Technologies), using strand-specific 60mer probes tiled every 20 nt for the main chromosome (NC_002607) and every 21 nt for the plasmids pNRC200 (NC_002608) and pNRC100 (NC_001869), consisting a total of 244K probes, including manufacturers' controls. The microarrays were printed by Agilent technologies and hybridized to total RNA, which was isolated using mirVana miRNA Isolation kit (Ambion) and direct labeled with Alexa547 and Alexa647 dyes (Kreatech) (Baliga et al, 2004). We used direct chemical labeling of RNA (Baliga et al, 2004) to avoid enzymatic labeling artifacts (Perocchi et al, 2007) and enable strand-specific signals for transcribed segments. Hybridization and washing were carried out according to array manufacturer's instructions. Arrays were scanned in ScanArray (Perkin Elmer) and spot finding was carried out using Feature Extraction (Agilent Technologies). Two biological replicates were sampled and dye-flip experiments were conducted for each sample. Resulting intensities were quantile normalized across all experiments. Log ratios were calculated for each probe (growth-curve sample/reference). The reference-RNA signals were quantile normalized and then jointly normalized by sequence content using a linear model similar to that of Johnson et al (2006). This model attempts to capture the effect on hybridization signal or efficiency from duplicate probes (cross-hybridization), G-C content, and sequence-specific factors. The ‘sequence-based' correction was subtracted from the probe intensities and resulted in a reduction by ∼10% in residual sum-of-squares between the intensities of neighboring probes. Interactive visualization of the data was carried out in the Gaggle Genome Browser (Bare et al., in preparation), available at http://baliga.systemsbiology.net/regulatory_logic/.
ChIP–chip experiments and analysis
ChIP–chip experiments were carried out for all TFBs (TFBa, TFBb, TFBc, TFBd, TFBe, TFBf, and TFBg), one TBP (TBPb) and three TRs (Trh3, Trh4, and VNG1451C) in H. salinarum NRC-1 using the HaloSpan array (Facciotti et al, 2007), which consists of 500 nt PCR products of successive regions of H. salinarum NRC-1 genome. The data for all TFBs were retrieved from Facciotti et al (2007). For TFBa, TFBd, and TFBf, data for additional biological replicates were also acquired using 13-nt resolution tiling arrays on the Nimblegen platform (for TFBd, two such distinct biological replicates were acquired). The TRs encoding genes were cloned in pMTFcmyc vector; chromatin immunoprecipitation and identification were carried out as described by Facciotti et al (2007). Binding locations were defined by applying the MeDiChI algorithm (Reiss et al, 2008) to each data set. This regression-based method deconvolves the ChIP–chip enrichment ratios along the genome by fitting them with a ‘peak profile' model of binding events, assuming a distribution in enriched DNA fragment lengths. It was shown that MeDiChI can increase the effective resolution of TFBS locations by a factor of five relative to the probe spacing of the tiling array, even for overlapping peaks (Reiss et al, 2008). P-values reported by MeDiChI (based upon peaks detected in bootstrap-resampled data that statistically seem to contain only noise; see (Reiss et al, 2008)) for each data set were converted to LFDR estimates through a semi-parametric two-component mixture model (Robin et al, 2007). A comparison of the peak intensities derived from MeDiChI for all three TFs (TFBd, TFBf, and TFBa) for which there were biological replicate measurements using both microarray platforms (500 nt resolution spotted arrays versus 13 nt resolution Nimblegen arrays) (Supplementary Figure 5) provided strong validation (with R2 of 0.66, 0.52, and 0.81, respectively) of most (∼500) TFBSs included in the analysis, and even for TFBSs with LFDR >0.1. The 318 multi-TFBS loci described in Results and Discussion were computed by locating peaks in a kernel density estimate (bandwidth=50 nt) of all TFBSs with LFDR<0.1 across the genome. Only density peaks generated by >2 individual TFBSs were counted. Monte Carlo simulations were used to estimate the expected fraction of TFBSs, which would be detected in intergenic regions, as a function of detection positional uncertainty σ, and FDR f, given that the true TFBS locations fall only in intergenic regions across the genome. In these simulations, n TFBSs were simulated by placing n (1–f) TFBSs in intergenic regions and n f in annotated coding regions, and Gaussian-distributed random offsets (±σ in nucleotides) were added to each simulated TFBS location. We used n=20 000 for our simulations. A binding event was considered internal to a transcribed or annotated coding region only if it was internally localized at a distance of >50 nt from the respective region's boundaries.
The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (GEO accession no. GSE13150).
Identification of probes hybridizing with transcribed regions of the genome
Probes in the tiling arrays were assessed as to whether they were complementary to a region that is transcribed in one or more of the observed conditions. We integrated the following probe measurements: (a) their log intensities in the 54 reference-RNA tiling arrays, (b) their relative changes across the growth curve in the 12 growth-curve tiling arrays, and (c) their Pearson correlations with the changes of their two neighboring probes across the growth curve (McGrath et al, 2007) into an iteratively reweighted logistic regression model that used annotated coding regions as the ‘training' set. The resulting model was used to estimate a probability that each probe was complementary to a transcribed region.
Integrated, multivariate segmentation defines transcript boundaries
Regression trees (CART; Breiman et al (1984)) were used to partition the tiling array data (log probe intensity values) into regions of constant intensity, separated by abrupt ‘break points', by fitting a constant value to a large, contiguous region, and recursively dividing the regions to significantly improve the residual sum-of-squares. The number of splits (and hence the complexity of the model) was determined using 100-fold cross-validation, to choose the most parsimonious model within 1σ from the optimal one. The relative likelihood of each break was estimated using 100 bootstraps with symmetric wild resampling (as in Reiss et al (2008)). Each segment was constrained to contain no fewer than five probes, restricting the procedure to detect only larger (⩾100 nt) segments (putative transcripts). Using the multivariate implementation of this procedure in the mvpart R library, we could apply the procedure simultaneously to all tiling arrays described above, enabling us to constrain the segments in an integrated manner using the reference-RNA and growth-curve tiling arrays, as well as the growth-curve correlations and the probe transcription probabilities. The maximum resolution of the derived transcription break points is no better than the tiling array resolution (here, 20 nt). The resulting breaks were subsequently classified into transcription ‘starts' and ‘stops' based upon whether the signal increased or decreased across the break in both the RNA references and the probe transcription probabilities.
Detection of putative non-coding RNAs
The procedure described above for segmenting the data and identifying transcriptional start/stop sites was constrained to omit smaller (⩽100 nt) transcripts. To identify putative non-coding RNAs (ncRNAs), including smaller ncRNA candidates, we individually partitioned each growth-curve sample data set (log ratios of the growth-curve samples relative to the reference RNA) using recursive partitioning trees as previously described. For each sample, P-values for each segment were computed relative to the log ratios of reference-RNA samples localized in the segment's coordinates. A segment was classified as a putative non-coding RNA if it presented a high probability of being expressed (P-value<0.05), its neighboring segments were not differentially expressed (P-value >0.05, in order to filter out possible UTR regions of genes), and no annotated gene or repeat overlapped the segment's coordinates. Further filtering was carried out through the model used to estimate a probability that each probe was complementary to a transcribed region (see section ‘Identification of probes hybridizing with transcribed regions of the genome'), and segments with P>0.05 were discarded. Many ncRNAs were complementary to repeat regions of the genome, so all duplicate ncRNA candidates were removed if they contained a 12-nt contiguous sequence similarity with any other ncRNA candidate.
Peptide atlas update
H. salinarum NRC-1 Peptide Atlas was updated with the addition of three new experiments corresponding to cultures grown under standard conditions. Sample preparation and mass spectrometry analyses were carried out as described by Van et al (2008), resulting in an additional 30 mass spectrometry runs and 33 986 tandem mass spectra. A new search database was constructed, including sequences from newly identified transcribed regions in the tiling array experiments. The updated version of H. salinarum NRC-1 peptide atlas is available at https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/buildDetails?atlas_build_id=130, including a total of 527 mass spectrometry runs and 121 618 tandem mass spectra.
Identification of conditional operons
For each of the predicted operons obtained from Price et al (2005), three different statistics were computed in a pairwise manner over all genes in that operon: (1) the log10(P-value) for the two-sample Student's t-test of the mean levels of the probes complementary to each pair of genes; (2) the Spearman rank correlation of the gene-expression profiles across 719 microarray experiments covering several diverse environmental perturbations (oxygen (Schmid et al, 2007), transition metals—Mn, Fe, Co, Ni, Cu, and Zn (Kaur et al, 2006), UV (Baliga et al, 2004) and gamma (Whitehead et al, 2006) radiation, interaction with Dunaliella salina, growth curve in CM media and in defined media, light–dark cycle, and oxidative stress—H2O2 and paraquat (unpublished; see http://gaggle.systemsbiology.net/projects/halo/2007-04), and (3) the Spearman rank correlation of the genes' tiling array probes over the growth curve. Thus, conditional operons (as opposed to classical operons) were identified on the basis of (1) the similarity in expression levels of the probes for each gene in the operon in the tiling array data, and (2) co-expression of the operon's genes across 719 microarrays. To obtain a probability that each operon is conditional, we computed the minimum values of each of these statistics for each operon (resulting in three ‘scores' per operon), and applied a quadratic discriminant classifier to these scores, using a set of 73 manually identified conditional operons as the training set. A probability cutoff of P=0.64 was chosen to minimize the false classification rate for the manually classified training set. This protocol resulted in a total of 123 classified conditional operons. Finally, the cumulative hypergeometric distribution was used to assess the P-value for the over-representation of TFBSs internal to the 123 conditional operons (from the ChIP–chip data for all GTRs and TFs described above) versus the number of TFBSs internal to the 176 non-conditional (classical) operons.
Construction of promoter–GFP fusions and evaluation of promoter activity
A 150–500 bp region surrounding the TSS localized in coding sequences of VNG2210G and VNG1775C was PCR amplified (primers VNG2210G-F: 5′-CGAAAACCGGATTCAAGTTC-3′, VNG2210G-R: 5′- ATCGTGTCGTCTGTGTCGTC-3′, resulting in a 208-bp PCR product corresponding to region 1 639 638–1 639 431 in H. salinarum main chromosome and VNG1775C-F: 5′-CTTCGGTCGACAGGGTTATC-3′ and VNG1775C-R: 5′-TGTCGACCAATCTACGTCGC-3′, resulting in a 162-bp PCR product corresponding to region 1 314 877–1 314 716 in H. salinarum main chromosome) and fused to a GFP coding sequence on a MevR selectable expression plasmid. H. salinarum NRC-1 cells were transformed and selected on CM agar containing 20 μg/ml mevinolin. Cells were sampled at mid-log, late-log, and stationary phases. On sampling, the cells were simultaneously washed, diluted to a nominal density of OD600 0.2, and fixed in a basal salt solution with 0.25% (w/v) formaldehyde. Cells were incubated in the fixative for 10 min at 4°C, followed by a second wash in basal salt solution. The fixative concentration was previously determined to adequately arrest cell function while also preserving GFP fluorescence and the combined wash steps served to remove as much of the peptone-based growth medium as possible to reduce fluorescence background. The dynamic range of the flow cytometer (InFlux, Cytopia/BD) was calibrated using fixed, non-fluorescent H. salinarum NRC-1 cells and 1-μm Y/G fluorescent beads (Polysciences, Inc). Before running on the flow cytometer, 1-μm beads were spiked into each sample to a final density of 1 × 107 beads/ml (approximately ten-fold less than the nominal cell density) to serve as an internal calibration standard. The mean fluorescence from 100 000 cells in each sample was normalized relative to the bead fluorescence level.
Supplementary Material
(A) Significant Transcription factor binding sites (TFBS, LFDR < 0.1) and (B) Multi transcription factor binding loci
Transcription start sites, termination sites and untranslated 5' and 3' regions for annotated genes and operons that were expressed in H. salinarum NRC-1 during growth and/or reference conditions.
Overlapping transcripts in H. salinarum NRC-1. Overlaps greater than 20 nt were considered as significant, given the error of transcript boundary detection using tiling arrays.
Revision of gene start codons based on detected TSS, Peptide Atlas information and H. salinarum R-1 annotation.
Newly transcribed elements in H. salinarum NRC-1 genome. Genome location, estimated length of the transcript and transcript levels (log ratio) during growth.
Conditional operons in H. salinarum NRC-1. Minimum correlation, tiling score values and combined score are reported. Manually verified conditional operons are also indicated.
Transcription factor binding sites (TFBS) internal to genes that are associated with a transcript boundary.
This file contains Supplementary figures S1-5, Supplementary table legends SI-VII
Acknowledgments
Thanks to Kenia Whitehead and Sacha Coesel for helpful discussions, Dan Tenenbaum for the construction of the webpage and Kenichi Masumura for help in the growth-curve experiments. This work was supported by grants from NIH (P50GM076547 and 1R01GM077398-01A2), DoE (MAGGIE: DE-FG02-07ER64327 and DE-FG02-07ER64327), NSF (EF-0313754, EIA-0220153, MCB-0425825, DBI-0640950) and NASA (NNG05GN58G) to NSB.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adhya S (2003) Suboperonic regulatory signals. Sci STKE 2003: pe22. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J (2004) Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res 14: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, DasSarma S (1999) Saturation mutagenesis of the TATA box and upstream activator sequence in the haloarchaeal bop gene promoter. J Bacteriol 181: 2513–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S (2000) Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol 36: 1184–1185 [DOI] [PubMed] [Google Scholar]
- Bell SD, Kosa PL, Sigler PB, Jackson SP (1999) Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci USA 96: 13662–13667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, Longabaugh W, Vuthoori M, Whitehead K, Madar A, Suzuki L, Mori T, Chang DE, Diruggiero J, Johnson CH, Hood L et al. (2007) A predictive model for transcriptional control of physiology in a free living cell. Cell 131: 1354–1365 [DOI] [PubMed] [Google Scholar]
- Bonneau R, Reiss DJ, Shannon P, Facciotti M, Hood L, Baliga NS, Thorsson V (2006) The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol 7: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and Regression Trees. Pacific Grove, CA: Wadsworth & Brooks/Cole Advanced Books & Software [Google Scholar]
- Brenneis M, Hering O, Lange C, Soppa J (2007) Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet 3: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Omer A (2005) Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694 [DOI] [PubMed] [Google Scholar]
- Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE, Baliga NS (2007) General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci USA 104: 4630–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS (2007) Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot C, Moran CP Jr (2007) Essential internal promoter in the spoIIIA locus of Bacillus subtilis. J Bacteriol 189: 7181–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayter JR, Doherty MK, Whitehead C, McCormack H, Gaskell SJ, Beynon RJ (2005) The subunit structure and dynamics of the 20S proteasome in chicken skeletal muscle. Mol Cell Proteomics 4: 1370–1381 [DOI] [PubMed] [Google Scholar]
- Hebbeln P, Rodionov DA, Alfandega A, Eitinger T (2007) Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA 104: 2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Klein BJ, Murakami KS (2008) The X-ray crystal structure of RNA polymerase from Archaea. Nature 451: 851–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Alon U (2007) The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res 17: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS (2006) Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SJ (2006) Prediction of genomic functional elements. Annu Rev Genomics Hum Genet 7: 315–338 [DOI] [PubMed] [Google Scholar]
- Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS (2006) A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res 16: 841–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jeon HJ, Ji SC, Yun SH, Lim HM (2008) Establishment of an mRNA gradient depends on the promoter: an investigation of polarity in gene expression. J Mol Biol 378: 318–327 [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH (2007) High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol 25: 584–592 [DOI] [PubMed] [Google Scholar]
- Muller F, Demeny MA, Tora L (2007) New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem 282: 14685–14689 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA 97: 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM (2007) Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res 35: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D (2008) Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 91: 335–346 [DOI] [PubMed] [Google Scholar]
- Piantadosi CA, Suliman HB (2008) Transcriptional regulation of SDHa flavoprotein by nuclear respiratory factor-1 prevents pseudo-hypoxia in aerobic cardiac cells. J Biol Chem 283: 10967–10977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Huang KH, Alm EJ, Arkin AP (2005) A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res 33: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve JN, Sandman K (2007) Chromatin and regulation. In Archaea: evolution, physiology and molecular biology, Garret RA, Klenk H (eds) 147–158. Wiley-Blackwell [Google Scholar]
- Reiss DJ, Baliga NS, Bonneau R (2006) Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics 7: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss DJ, Facciotti MT, Baliga NS (2008) Model-based deconvolution of genome-wide DNA binding. Bioinformatics 24: 396–403 [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA (2000) Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K (2006) The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell 24: 747–757 [DOI] [PubMed] [Google Scholar]
- Reuter CJ, Maupin-Furlow JA (2004) Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl Environ Microbiol 70: 7530–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Robin S, Bar-Hen A, Daudin J, Pierre L (2007) A semi-parametric approach for mixture models: Application to local false discovery rate estimation. 51: 5483–5493
- Ruepp A, Soppa J (1996) Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J Bacteriol 178: 4942–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorius-Neef S, Pfeifer F (2004) In vivo studies on putative Shine-Dalgarno sequences of the halophilic archaeon Halobacterium salinarum. Mol Microbiol 51: 579–588 [DOI] [PubMed] [Google Scholar]
- Scheuch S, Marschaus L, Sartorius-Neef S, Pfeifer F (2008) Regulation of gvp genes encoding gas vesicle proteins in halophilic Archaea. Arch Microbiol 190: 333–339 [DOI] [PubMed] [Google Scholar]
- Schmid AK, Reiss DJ, Kaur A, Pan M, King N, Van PT, Hohmann L, Martin DB, Baliga NS (2007) The anatomy of microbial cell state transitions in response to oxygen. Genome Res 17: 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Ishihama A, Busby SJ, Grainger DC (2008) The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res 36: 3950–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H (2007) Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Altuvia S, Wassarman KM (2005) An abundance of RNA regulators. Annu Rev Biochem 74: 199–217 [DOI] [PubMed] [Google Scholar]
- Tanay A (2006) Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res 16: 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A (2002) Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A (2005) Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 55: 469–481 [DOI] [PubMed] [Google Scholar]
- Tsui HC, Zhao G, Feng G, Leung HC, Winkler ME (1994) The mutL repair gene of Escherichia coli K-12 forms a superoperon with a gene encoding a new cell-wall amidase. Mol Microbiol 11: 189–202 [DOI] [PubMed] [Google Scholar]
- Van PT, Schmid AK, King NL, Kaur A, Pan M, Whitehead K, Koide T, Facciotti MT, Goo YA, Deutsch EW, Reiss DJ, Mallick P, Baliga NS (2008) Halobacterium salinarum NRC-1 PeptideAtlas: toward strategies for targeted proteomics and improved proteome coverage. J Proteome Res 7: 3755–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Struhl K, Busby SJ, Grainger DC (2007) Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol Microbiol 65: 21–26 [DOI] [PubMed] [Google Scholar]
- Wang C, Ding C, Meraz RF, Holbrook SR (2006) PSoL: a positive sample only learning algorithm for finding non-coding RNA genes. Bioinformatics 22: 2590–2596 [DOI] [PubMed] [Google Scholar]
- Ward LD, Bussemaker HJ (2008) Predicting functional transcription factor binding through alignment-free and affinity-based analysis of orthologous promoter sequences. Bioinformatics 24: i165–i171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K, Kish A, Pan M, Kaur A, Reiss DJ, King N, Hohmann L, DiRuggiero J, Baliga NS (2006) An integrated systems approach for understanding cellular responses to gamma radiation. Mol Syst Biol 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Yang CF, DasSarma S (1990) Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J Bacteriol 172: 4118–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochum GS, Rajaraman V, Cleland R, McWeeney S (2007) Localization of TFIIB binding regions using serial analysis of chromatin occupancy. BMC Mol Biol 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM (2006) Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Significant Transcription factor binding sites (TFBS, LFDR < 0.1) and (B) Multi transcription factor binding loci
Transcription start sites, termination sites and untranslated 5' and 3' regions for annotated genes and operons that were expressed in H. salinarum NRC-1 during growth and/or reference conditions.
Overlapping transcripts in H. salinarum NRC-1. Overlaps greater than 20 nt were considered as significant, given the error of transcript boundary detection using tiling arrays.
Revision of gene start codons based on detected TSS, Peptide Atlas information and H. salinarum R-1 annotation.
Newly transcribed elements in H. salinarum NRC-1 genome. Genome location, estimated length of the transcript and transcript levels (log ratio) during growth.
Conditional operons in H. salinarum NRC-1. Minimum correlation, tiling score values and combined score are reported. Manually verified conditional operons are also indicated.
Transcription factor binding sites (TFBS) internal to genes that are associated with a transcript boundary.
This file contains Supplementary figures S1-5, Supplementary table legends SI-VII