Abstract
Increased B-type natriuretic peptide (BNP) gene expression is regarded as one of the hallmarks of cardiac myocyte hypertrophy. Here we demonstrate that both basal and endothelin-1 (ET-1) -dependent stimulation of human (h) BNP gene transcription requires the presence of an intact Yin Yang 1 (YY1) binding site positioned at -62 bp relative to the transcription start site. Mutation of this site reduced both basal and stimulated hBNP promoter activity. This site was shown to bind YY1 both in vitro and within the context of the intact cell. The latter interaction increased following ET-1 treatment. Exposure to ET-1 also resulted in increased nuclear localization of YY1 and a reduction in acetylation of the YY1 protein. Overexpression of wild type YY1 increased both basal and endothelin-stimulated hBNP promoter activity, while a carboxy terminal deletion mutant of YY1 was devoid of activity. Treatment with the histone deacetylase inhibitor trichostatin A (TSA) resulted in decreased hBNP reporter activity. YY1 was shown to associate with histone deacetylase 2 (HDAC2), and HDAC2 was shown to associate directly with the hBNP promoter in the intact cell. Collectively these findings demonstrate that YY1 plays an important role in regulating the transcriptional activity of the hBNP gene promoter. These data suggest a model in which YY1 activates hBNP transcription through interaction with HDAC2.
Keywords: B-type Natriuretic Peptide, Cardiac Hypertrophy, Endothelin, Histone Deacetylase Ying Yang1
Cardiac hypertrophy is characterized by both positive and negative changes in the gene expression profile of the heart 1, 2. Increased expression of the cardiac natriuretic peptides, atrial natriuretic peptide (ANP) and brain or B-type natriuretic peptide (BNP), has been linked to the development of hypertrophy in both animal models and humans 3, 4. ANP and BNP both possess potent activity in the cardiovascular and renal systems. Through association with a shared receptor, they each reduce blood pressure, maintain or improve glomerular filtration rate, promote a natriuretic diuresis, inhibit secretion and/or action of vasopressin, renin, angiotensin and aldosterone and suppress hypertrophy, hyperplasia and/or fibrosis in the heart and vasculature 5. Thus, they function as endogenous antagonists of many of the factors that have been causally linked to the pathogenesis of human cardiovascular disease.
Under normal conditions, BNP is expressed at relatively low levels in the adult heart and without a pronounced chamber-specific pattern. Expression is increased in late fetal and early neonatal life and it increases dramatically in association with cardiac hypertrophy. Plasma BNP levels are used clinically in the diagnosis and management of heart failure 6, 7.
The transcription factor Ying Yang 1 (YY1), also known as NF-F1, UCRBP and CF1, is a ubiquitously expressed 68 kilodalton, zinc-finger transcription factor of the GLI-Kruppel family with the relatively unique ability to stimulate 8, suppress 9 or initiate 10 transcription of target genes in different genomic contexts. YY1 protein levels have been shown to be increased in both mouse models of hypertrophy as well as in human patients with heart failure 11. In addition, several genes involved in the hypertrophic program have been shown to be regulated by YY1 11–13.
A recent study from Bhalla et al. 14 investigated the role of YY1 in the regulation of rat BNP gene transcription with particular emphasis on the interaction of this protein with the transcription factor GATA-4 in heterologous cells (HeLa cells). In the present study, we have identified a YY1 site in the human BNP gene that appears to traffic both basal and agonist-stimulated promoter activation in the neonatal rat cardiac myocyte. YY1 occupies this site in an agonist-dependent manner within the context of the intact myocyte, and may acquire its activating properties through interaction with a histone deacetylase, HDAC2.
Methods
Plasmid Construction and Site-directed Mutagenesis
YY1 expression vector pCMV-YY1 and the mutant vector pCMV-YY1ΔC (deleted amino acids 334–414) were gifts from Bernhard Lüscher 15 of Medizinische Jochschule, Hanover, Germany. The -198 human (h) BNP luciferase has been described previously 16. Site-directed mutagenesis was carried out with the QuickChange kit (Stratagene, LaJolla, CA.). The sequence of the mutagenic primer (sense strand) was as follows (mutagenized bases are underlined): 5’-CAGAGATAGACCTGCCGTTCAGGCAGGCCCGACA-3’. Lentiviral YY1 and control siRNA plasmids construct were purchased from OpenBiosystems (Huntsville, AL).
Cell Culture
Ventricular myocytes were prepared from 1–2 day-old neonatal Sprague-Dawley rats by alternate cycles of 0.05% trypsin digestion and mechanical trituration and cultured as described 17.
3H Leucine Incorporation
After serum starvation for 12 hours, cells were treated with 10−7 mol/L ET-1 for 36 hours. During the final 12 hours they were incubated with 3H-leucine (5 µCi/mL). 3H-leucine incorporation was measured as previously described 17.
Lentiviral preparation and infection
Lentivirus was prepared as previously described 18. Virus was handled according to established bio-safety protocols. Following serum deprivation, lentivirus was directly applied to the media and cells were incubated an additional 24 hours prior to treatment with vehicle or ET-1 (10−7mol/L) for 48 hours.
RNA isolation and Quantitative PCR (Q-PCR)
Total RNA was isolated from cardiac myocytes with the RNeasy kit (Qiagen. Valencia, CA) and reverse transcribed into cDNA. Q- PCR was carried out with rat BNP (Rn00580641_m1) and GAPDH (Rn99999916_sl) Taqman primers (Applied Biosystems, Foster City, CA).
Transfection and Luciferase Assay
Ventricular myocytes were transiently transfected with the indicated reporters by electroporation (Gene Pulser; Bio Rad Laboratories, Hercules, CA) at 280 mV and 250 µF or by lipofectin, according to the manufacturer’s recommendation (Invitrogen, Carlsbad, CA). DNA content in individual cultures was normalized with pcDNA3. Cells were plated and cultured in DMEM H-21 containing 10% enriched calf serum for 24 hrs, and then changed to serum-free medium for 4 hours prior to treatment with ET-1, 10−7 mol/L. After 48 hrs, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega, Madison WI). Where indicated, firefly luciferase levels were normalized for Renilla luciferase.
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed with isolated myocyte nuclear extracts and 32P-labeled oligonucleotide harboring the candidate YY1 binding sequence as described previously 19. Double stranded oligonucleotides used for electrophoretic mobility shift assay are described above. Nuclear extracts were combined with 32P-end-labeled double-stranded wild-type or mutant oligonucleotide on ice. All samples were resolved on 4% nondenaturing polyacrylamide gels. Gels were dried and exposed to X-ray film.
DNA-Immunoprecipitation (IP) Assay
Cardiac myocytes were transfected with -198 hBNP-Luc using lipofectin (Invitrogen, Carlsbad, CA). After 24 hours, cell cultures were changed to serum free-media for an additional 4 hours, then treated with 10−7 mol/L ET-1 for different periods of time. The DNA-IP assays were performed using a modification of published methodology 20. See Supplemental Methods for additional details (please see http://hyper.ahajournals.org.)
Statistics
Data were analyzed using one-way ANOVA and the Newman-Keuls post hoc test or the student’s T-test to assess significance. Error bars represent standard deviation or standard error of the mean (SEM) as indicated
Results
YY1 knockdown inhibits ET -mediated total protein synthesis and BNP expression in cardiac myocytes
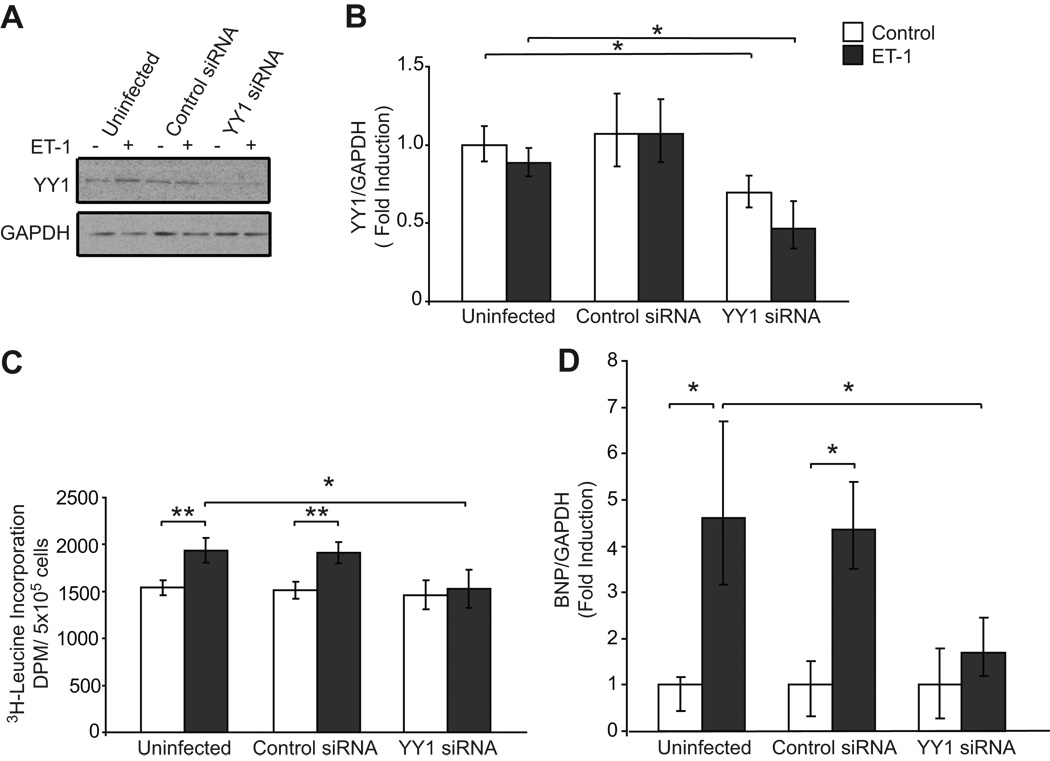
To begin to address the function of YY1 in the ET-mediated hypertrophic response, we performed knockdown experiments using a lentiviral construct specific for YY1. As shown in Fig. 1A and 1B, YY1 protein and mRNA were reduced in whole cell extracts derived from cells infected with a lentiviral YY1 siRNA construct, but not in response to a control lentiviral siRNA. As a measure of cellular hypertrophy, protein synthesis was quantified by 3H leucine incorporation in the absence and presence of ET-1. Treatment with ET-1 failed to increase 3H-leucine incorporation in cells infected with the YY1 siRNA lentivirus, but not in uninfected or control siRNA infected cells (Fig. 1C). This suggests that YY1 plays a key role in the ET-1 mediated hypertrophic response. As YY1 has been implicated in the regulation of BNP expression, we next assessed the result of reduction of YY1 on ET-1-mediated BNP expression. We found that knockdown of YY1 resulted in a reduction of BNP expression as quantified by Q-PCR (Fig. 1D). This suggests that YY1 may function as a transcriptional activator to promote BNP expression in response to a hypertrophic stimulus.
Figure 1. Reduction of YY1 inhibits the ET-1 mediated hypertrophic response and BNP expression in cardiac myocytes.
YY1 expression is reduced by siRNA sequence specific for YY1. Cardiac myocytes were serum starved prior to treatment with vehicle or ET-1 (10−7 mol/L). Whole cell lysates were subjected to A. Western blot analysis with anti-YY1 and anti-GAPDH and B. Q-PCR analysis of YY1 mRNA normalized for GAPDH. N=3, results are representative of 4 independent experiments. C. Knockdown of YY1 reduces ET-1 mediated 3H-Leucine incorporation in cardiac myocytes. Pooled data, N= 12. D. ET-1 mediated BNP expression is reduced by YY1 knockdown as assessed by Q-PCR. N=3, results representative of 3 independent experiments reported as means ± standard deviation.*p < 0.05 ** p<.01.
YY1 binding element contributes to basal and ET-1 -dependent hBNP promoter activity
We identified a candidate YY1 binding site (consensus gCCAT) on the antisense strand between −62 bp and −58 bp relative to the transcription start site of the hBNP gene (Fig. 2A). Much like the site identified by Bhalla et al. 14 in the rat gene, this binding site lies in close proximity to two functional GATA binding elements in the BNP gene promoter. As shown in Fig. 2B, mutation of the YY1 binding site resulted in ~75% reduction in basal promoter activity and eliminated the ET-1 dependent stimulation of hBNP promoter activity. This suggests that the YY1 site plays a key role in promoting the increase in BNP gene transcription that accompanies transition to the hypertrophic phenotype.
Figure 2. Identification of a YY1 consensus binding site in hBNP promoter.
A. A candidate YY1 site (gCCAT; bold type) was identified on the antisense strand at position -62 relative to the transcription start site of the hBNP gene. B. Cardiac myocytes were transfected with wild type -198 BNP-LUC or a mutant harboring a modified YY1 binding element. Cells treated with 10−7 mol/L ET-1 as indicated. Firefly luciferase levels were normalized for Renilla luciferase activity. Pooled data from three independent experiments are presented as means ± SEM. * p<0.01 versus ET-1 control.
YY1 binds the human BNP promoter
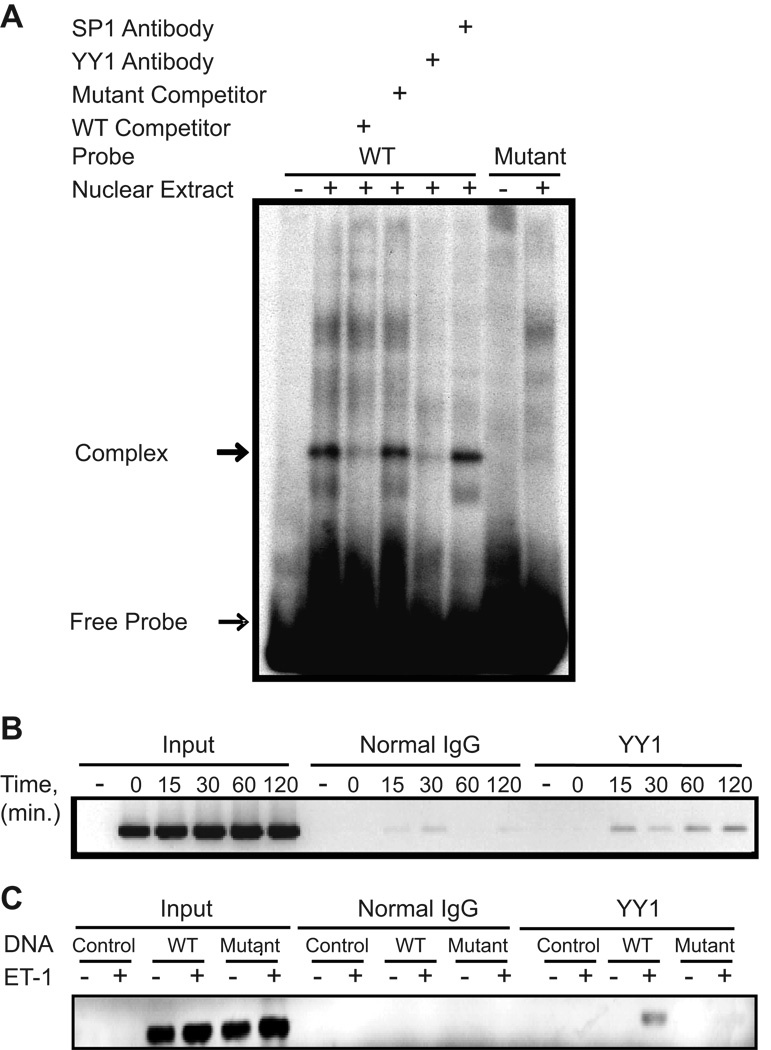
Binding of YY1 protein to this candidate site was confirmed by electrophoretic mobility shift assay (EMSA) {Fig. 3A and Supplemental Fig. S1 (please see http://hyper.ahajournals.org.)} using an oligonucleotide probe spanning the candidate site. A protein(s) in the nuclear extract of cardiac myocytes formed a complex with the radiolabeled oligonucleotide harboring the YY1 binding site but not to the oligonucleotide containing the mutant sequence. Binding to the wild type sequence was appropriately competed by unlabeled excess (100X) wild type oligonucleotide but not mutant oligonucleotide. Importantly, an antibody directed against YY1, but not SP1, disrupted the putative YY1-DNA binding complex, suggesting that the complex contained the YY1 transcription factor. Disruption of the binding complex by the YY1 antibody has been reported previously 21–23and similar findings were obtained using 35S labeled recombinant YY1 (Supplemental Fig. S1, please see http://hyper.ahajournals.org.).
Figure 3. YY1 binds the hBNP promoter.
A. 32P-labeled double stranded oligonucleotide harboring the wild type 5’-GATAAGACCTGCATGGCAGGCAGGCCCG-3’or mutant sequence, 5’-GATAAGACCTGCcgttCAGGCAGGCCCG-3’ were used for EMSA. Where indicated antibody directed against YY1 or Sp1 was included in the binding reaction. B. Myocytes were transfected with wild type -198 hBNP LUC or the mutant -198 hBNP LUC harboring a modified YY1 binding element. Cells were treated with 10−7 mol/L ET-1 for the times indicated (B) or 60 min (C) and the DNA-IP assay performed as described in Methods. The band (214 bp product) in B. and C. represents the PCR product obtained using primers specific for the -198-BNP luciferase plasmid. Results are representative of three independent experiments.
To assess binding of YY1 to the hBNP gene promoter within the context of the intact cardiac myocyte, we performed DNA-immunoprecipitation (DNA-IP) analysis 20 using a transfected -198 hBNP-LUC reporter or CMV-pcDNA3 plasmid as a control. As shown in Fig. 3B, YY1 bound the proximal hBNP promoter following 15 min of ET-1 (10−7 M) treatment. It remained bound for at least 120 min following stimulus application. Amplifications of DNA in the input samples were not significantly different. Similar to the EMSA findings, YY1 could not be shown to associate with the -198 hBNP-LUC construct harboring the mutant YY1 binding element (Fig. 3C)
YY1 enhances basal and ET-mediated BNP transcriptional activity
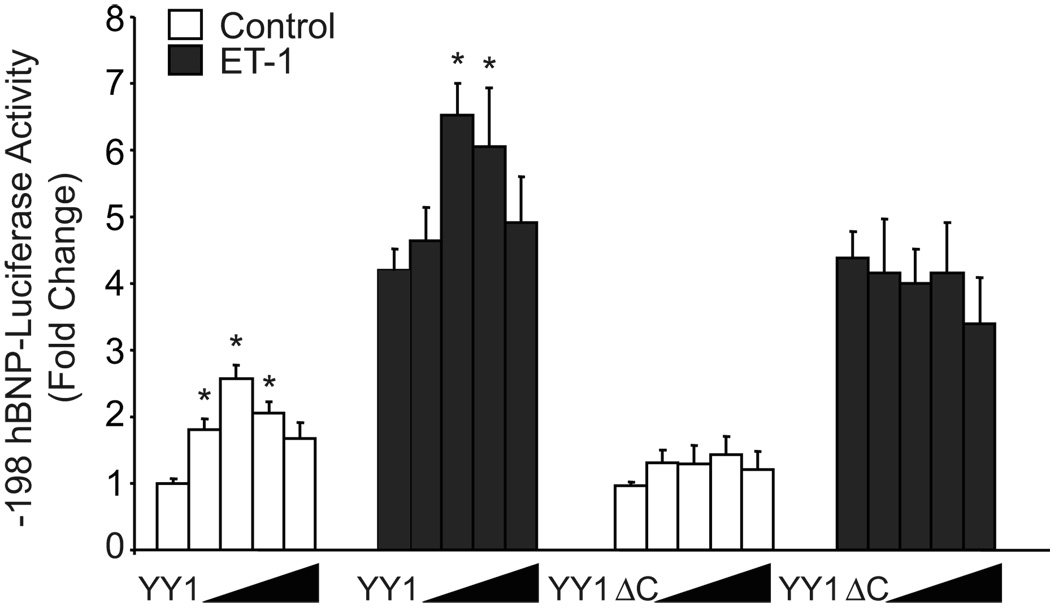
Co-transfection of the wild type YY1 expression vector with -198 hBNP-LUC resulted in a dose dependent increase in both basal and ET-stimulated hBNP promoter activity (p<0.05) that peaked at 0.125 µg of YY1 expression vector and fell off at higher concentrations of transfected plasmid (presumably reflecting a non-specific “squelching” effect of the transcription factor) (Fig. 4). It has been shown previously that the DNA binding activity of YY1 is located in the C-terminal zinc finger domain of the protein 24. Transfection of an expression vector encoding a YY1 mutant that lacks the C-terminal DNA binding domain failed to stimulate BNP promoter activity (Fig. 4). This contrasts with the previous report of Bhalla et al. 14 in which the YY1 DNA binding domain was not necessary for GATA4-dependent synergistic activation of the rat BNP promoter by YY1. These results support our hypothesis that YY1 plays a key role in regulating hBNP promoter activity and suggest that YY1 levels are, to some degree, limiting (i.e. non-saturating) in the neonatal myocyte.
Figure 4. Effect of co-transfection of wild type or mutant YY1 on basal and ET-dependent BNP promoter activity in neonatal rat ventricular myocytes.
Cells were co-transfected with expression plasmid encoding wild-type YY1 or a YY1 mutant lacking the C-terminal DNA binding domain, together with the –198 hBNP- LUC reporter. Cells were treated with vehicle alone or 10−7 mol/L ET-1. After 48 hrs, cells were collected and lysates were prepared for measurement of firefly or Renilla luciferase activity. Pooled data from three independent experiments are presented as means ± SEM. * p<0.01 versus control-transfected cells in each group.
Histone deacetylase activity and hBNP promoter activity
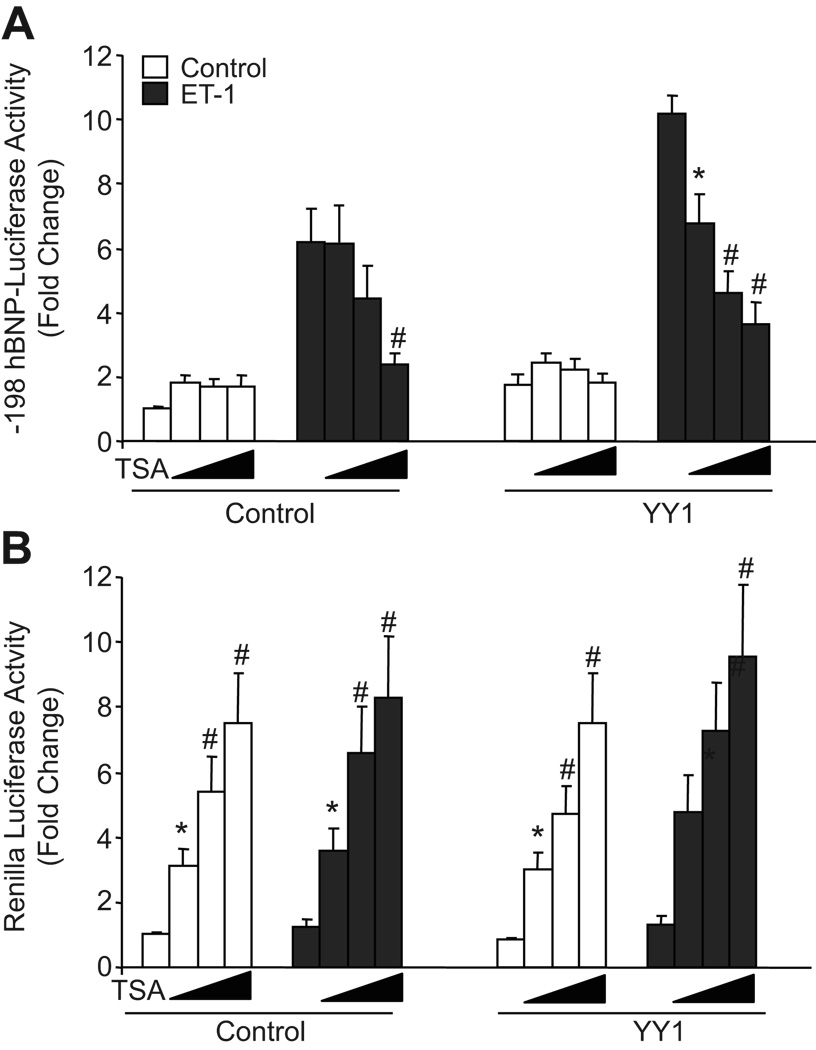
Recently, the histone deacetylase inhibitor TSA has been shown to reverse hypertrophy, including activation of hypertrophy-dependent gene expression, in both mouse and rat models 11, 25–27. As BNP gene transcription is known to be a marker for hypertrophy in cultured ventricular myocytes, we examined the effects of TSA on hBNP promoter activity. As shown in Fig. 5A, TSA inhibited hBNP promoter activity under both basal (~61% inhibition) and ET-stimulated (~65% inhibition) conditions. Interestingly, co-transfection of additional YY1 expression plasmid failed to enhance hBNP reporter activity in the presence of TSA. This did not reflect non-specific inhibition of myocyte gene transcription, as activity of a co-transfected cytomegalovirus (CMV) promoter (pRL-CMV; assessed as Renilla luciferase activity) was increased ~8 fold by 100 nmol/L TSA (Fig. 5B). These results suggest that suppression of histone deacetylase activity results in selective inhibition of the hBNP promoter.
Figure 5. Effect of TSA on ET-1 stimulation of -198 hBNP promoter activity.
Ventricular myocytes were transfected with -198 hBNP-LUC and CMV-Renilla. After 24 hours, cells were pre-incubated with or without TSA (0, 25, 50 or 100 nmol/L) for 30 minutes, then cultured for an additional 48 hours in the presence or absence of 10−7 mol/L ET-1. Firefly luciferase (A) and Renilla luciferase (B) values are presented here as relative light units/microgram of soluble protein. Pooled data from four independent experiments are presented as means ± SEM. * p<0.05, # p7#x0003C;0.01 versus vehicle (without TSA)-treated cells in each group.
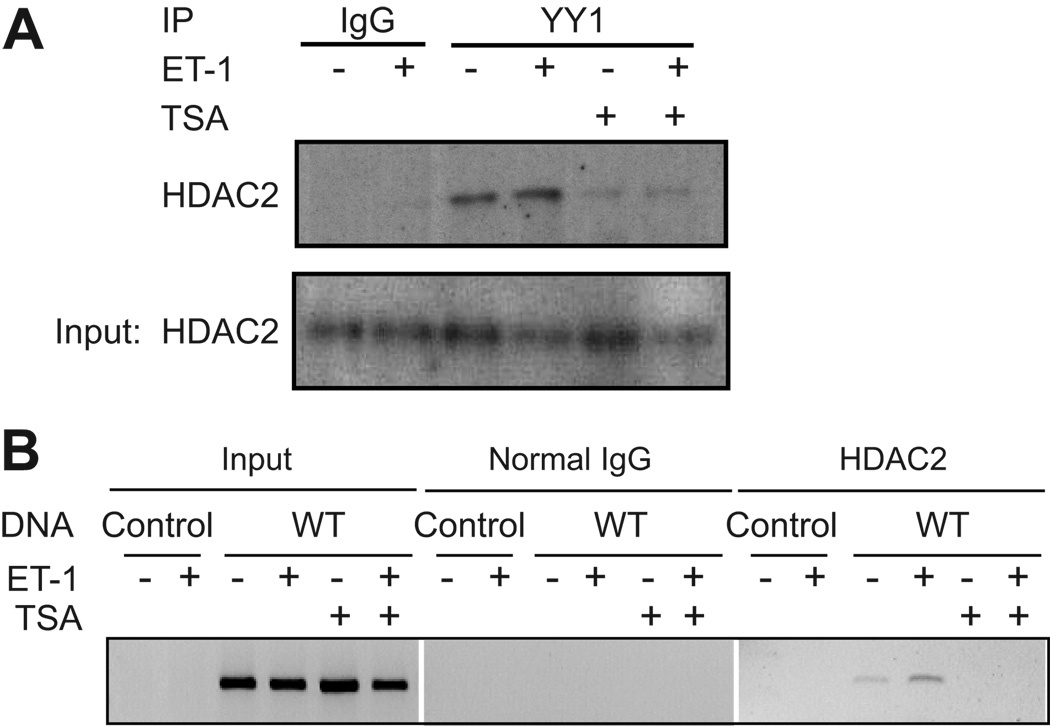
YY1 has been shown to interact with both class I and II histone deacetylases (HDACs). The role of HDACs in regulating cardiac gene expression is complex 28. However, it appears that class I HDACs, specifically HDAC2, serve to promote the hypertrophic program in cardiac myocytes 29. We examined the ability of HDAC2 to interact with YY1 within the context of the cardiac myocyte using a co-immunoprecipitation approach (Fig. 6A). Endogenous YY1 associates with HDAC2 in cultured myocytes. This interaction was not enhanced by ET-1 treatment, but it was completely inhibited following treatment with TSA.
Figure 6. HDAC2 binds YY1 and is associated with the hBNP promoter.
A Cultured cardiac myocytes were treated with 10−7 mol/L ET-1 in the presence or absence of 400 nmol/L TSA, as indicated. Nuclear extracts were incubated with antibody to YY1, followed by immunoprecipitation and Western blot analysis with an antibody to HDAC2. Results are representative of three independent experiments. B. Cardiac myocytes were transfected with -198 hBNP LUC or control pcDNA3 plasmid. Cells were pretreated with 400 nmol/L TSA for one hour and then treated with 10−7mol/L ET-1 for an additional hour, as indicated. Cells were collected and the DNA-IP assay was performed as described in Methods. Results are representative of three independent experiments.
To determine if HDAC2 directly associated with the hBNP promoter, DNA-IP was carried out using the transfected -198 hBNP-LUC reporter or CMV-pcDNA3 as a control. As shown in Fig. 6B, HDAC2 associated with the -198 hBNP promoter and the association was enhanced following treatment with ET-1. Pretreatment with TSA, however, prevented HDAC2 interaction with the promoter.
Discussion
The present studies provide direct evidence for the importance of the transcription factor YY1 in mediating both basal and ET-dependent human BNP promoter activity. We have demonstrated that YY1 binds to a specific element in the proximal promoter of the hBNP gene, both in vitro and in the context of the intact cell. YY1 has been shown to function as a stimulator, inhibitor or initiator of transcription in different contexts. In cardiac myocytes, YY1 seems to function as an activator of BNP gene transcription. This activation may, in part, be mediated through direct association with the histone deacetylase HDAC2.
YY1 has been shown to be increased in models of heart disease and in patients with heart failure 11, 30, suggesting a role for YY1 in mediating the transcriptional changes associated with cardiac dysfunction. Indeed, YY1 has been shown to both positively and negatively regulate several genes important in the hypertrophic program. In the cardiac myocyte, YY1 functions as a negative regulator of alpha myosin heavy chain (αMHC) 12, a key component of sarcomeric structure. In rodent models of cardiac hypertrophy, αMHC is down regulated in favor of the β isoform, which is thought to result in more energy-efficient sarcomeric activation and decreased contractility 1. YY1 has also been shown to reduce expression of the skeletal α-actin gene 31, which is increased in hypertrophic states 32. A mutation which selectively blocks YY1 binding to that promoter resulted in increases in both basal and TGFβ-dependent skeletal α-actin gene transcription implying that YY1 is a constitutive inhibitor of this gene. These changes in cardiac gene expression, taken together with the results in the present study and those of Bhalla et al. 14 with the rat BNP promoter, suggest that YY1 may serve as a point of molecular integration to coordinate expression of the fetal gene program.
YY1 activity can be modified through changes in YY1 expression or YY1 activation, through increased translocation of YY1 from the cytoplasmic to the nuclear compartment 33 or through post-translational acetylation of the YY1 protein 34. YY1 has been shown to be acetylated by pCAF and p300 and deacetylated by members of the HDAC gene family. In this study, we have shown that ET-1 treatment results in a reduction in acetylated YY1 (Data Supplement, please see http://hyper.ahajournals.org.) which correlates with transcriptional activation of the BNP promoter. Whether it is this deacetylation (vs. others involving proteins associated with the BNP promoter) that leads the increase in transcriptional activity remains to be defined.
Recently, several members of the HDAC family have been shown to be expressed in the heart and associated with cardiac hypertrophy. In mammals, HDACs are subdivided into three classes termed class I, II and III. Classically, HDACs are thought to associate with and deacetylate histones and, thereby, promote transcriptional repression through condensation of chromatin. Several transcription factors have also been shown to be targets of HDACs, including the myocyte enhancer factor 2 (MEF2) which associates with class II HDACs 35 and YY1 which has been shown to associate with both class I and II HDACs 34, 36.
Both class I and II HDAC enzymatic activity is inhibited pharmacologically by the inhibitor, TSA. TSA has been shown to block the development of cardiac hypertrophy induced by thoracic aortic banding (a model of pressure induced hypertrophy), angiotensin II 26, 27 and isoproterenol 25 suggesting a key role for at least some HDACs in the regulation of hypertrophy. In our study, TSA treatment of cardiac myocytes resulted in a dose dependent inhibition of BNP reporter activity, a recognized marker for activation of the hypertrophic gene transcription program. Gene deletion studies of class II HDACs suggest that HDAC5 and HDAC9 act as antagonists of cardiac hypertrophy induced by either aortic banding or constitutive calcineurin signaling 37, 38. Interestingly, HDAC5 and HDAC9 null mice did not display an enhanced hypertrophic phenotype in response to isoproterenol mediated hypertrophy, raising the possibility of distinct signaling mechanisms associated with the hypertrophic response to β adrenergic stimulation. However, TSA treatment does block isoproterenol mediated hypertrophy. This taken together with the apparent anti-hypertrophic activity of HDAC5 and HDAC9, suggests that class I HDACs may be both pro-hypertrophic and likely targets of TSA treatment in the heart. In support of this, recent studies of HDAC2 null mice have revealed the importance of this class I HDAC in both normal heart development and in hypertrophy 39, 40. HDAC2 null mice are resistant to isoproterenol mediated hypertrophy and seem to be protected from hypertrophy associated with overexpression of the pro-hypertrophic transcription factor Hod (also known as Hop) 39. In addition, mice expressing a transgene for HDAC2 in the heart demonstrate increased hypertrophy 39.
We have shown that HDAC2 physically associates with the BNP promoter and that the association is inhibited by TSA. This result suggests that HDAC2 participates in the activation of the BNP promoter directly. Of note, ANP gene expression, which closely tracks with BNP expression, is blunted in HDAC2 null mice 29. In addition, HDAC5 and HDAC9 gene deletion result in enhanced expression of BNP, especially in the setting of enhanced calcineurin signaling. Thus, although YY1 is capable of binding class I and II HDACs, it seems likely that interaction with class I HDACs, specifically HDAC2, is the more probable link to hypertrophy-dependent BNP gene transcription.
Perspective
We have shown that the transcription factor YY1 plays a pivotal role in the regulation of hBNP gene promoter activity. Treatment with the pro-hypertrophic agonist (ET-1) results in translocation of YY1 to the nuclear compartment, deacetylation of the YY1 protein and increased YY1 binding to the hBNP promoter. Activation of the hBNP gene promoter is accompanied by increased association with the class I HDAC, HDAC2. Taken in context with other studies of this regulatory transcription factor, the present study suggests an important integrative role for YY1 in controlling BNP gene transcription.
Acknowledgements
We would like to thank Allison Brincat for her assistance in preparation of the lentivirus used in this study.
Source of Funding
Grant Support: Supported by HL45637 to D.G.G. and F32 HL086158 to D.J.G. from NIH.
Footnotes
Disclosures
none
REFERENCES
- 1.Gupta MP. Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure. Journal of Molecular and Cellular Cardiology. 2007;43:388–403. doi: 10.1016/j.yjmcc.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durocher D, Grepin C, Nemer M. Regulation of gene expression in the endocrine heart. Recent Prog Horm Res. 1998;53:7–23. discussion 22–23. [PubMed] [Google Scholar]
- 3.Chien KR, Knowlton KU, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. Faseb J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 4.Burnett JC, Jr, Kao PC, Hu DC, Heser DW, Heublein D, Granger JP, Opgenorth TJ, Reeder GS. Atrial natriuretic peptide elevation in congestive heart failure in the human. Science. 1986;231:1145–1147. doi: 10.1126/science.2935937. [DOI] [PubMed] [Google Scholar]
- 5.Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N Engl J Med. 1998;339:321–328. doi: 10.1056/NEJM199807303390507. [DOI] [PubMed] [Google Scholar]
- 6.Morrison LK, Harrison A, Krishnaswamy P, Kazanegra R, Clopton P, Maisel A. Utility of a rapid B-natriuretic peptide assay in differentiating congestive heart failure from lung disease in patients presenting with dyspnea. J Am Coll Cardiol. 2002;39:202–209. doi: 10.1016/s0735-1097(01)01744-2. [DOI] [PubMed] [Google Scholar]
- 7.Troughton RW, Frampton CM, Yandle TG, Espiner EA, Nicholls MG, Richards AM. Treatment of heart failure guided by plasma aminoterminal brain natriuretic peptide (N-BNP) concentrations. Lancet. 2000;355:1126–1130. doi: 10.1016/s0140-6736(00)02060-2. [DOI] [PubMed] [Google Scholar]
- 8.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 10.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 11.Sucharov CC, Mariner P, Long C, Bristow M, Leinwand L. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J Biol Chem. 2003;278:31233–31239. doi: 10.1074/jbc.M301917200. [DOI] [PubMed] [Google Scholar]
- 12.Mariner PD, Luckey SW, Long CS, Sucharov CC, Leinwand LA. Yin Yang 1 represses alpha-myosin heavy chain gene expression in pathologic cardiac hypertrophy. Biochem Biophys Res Commun. 2005;326:79–86. doi: 10.1016/j.bbrc.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Lee TC, Zhang Y, Schwartz RJ. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- 14.Bhalla SS, Robitaille L, Nemer M. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J Biol Chem. 2001;276:11439–11445. doi: 10.1074/jbc.M100208200. [DOI] [PubMed] [Google Scholar]
- 15.Austen M, Luscher B, Luscher-Firzlaff JM. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 16.LaPointe MC, Wu G, Garami M, Yang XP, Gardner DG. Tissue-specific expression of the human brain natriuretic peptide gene in cardiac myocytes. Hypertension. 1996;27:715–722. doi: 10.1161/01.hyp.27.3.715. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Garami M, Cheng T, Gardner DG. 1,25(OH)2 vitamin D3, and retinoic acid antagonize endothelin-stimulated hypertrophy of neonatal rat cardiac myocytes. J Clin Invest. 1996;97:1577–1588. doi: 10.1172/JCI118582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crittenden JR, Heidersbach A, McManus MT. Chapter 5: Lentiviral strategies for RNAi knockdown of neuronal genes. Curr Protoc Neurosci. 2007 doi: 10.1002/0471142301.ns0526s39. Unit 5 26. [DOI] [PubMed] [Google Scholar]
- 19.Liang F, Schaufele F, Gardner DG. Functional interaction of NF-Y and Sp1 is required for type a natriuretic peptide receptor gene transcription. J Biol Chem. 2001;276:1516–1522. doi: 10.1074/jbc.M006350200. [DOI] [PubMed] [Google Scholar]
- 20.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Liu H, Wang Z, Liu X, Guo L, Huang L, Gao L, McNutt MA, Li G. The role of transcription factors Sp1 and YY1 in proximal promoter region in initiation of transcription of the mu opioid receptor gene in human lymphocytes. J Cell Biochem. 2008;104:237–250. doi: 10.1002/jcb.21616. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, Kawachi Y, Nakamura Y, Sakurai H, Hirota A, Banno T, Takahashi T, Roop DR, Otsuka F. Yin-yang 1 negatively regulates the differentiation-specific transcription of mouse loricrin gene in undifferentiated keratinocytes. J Invest Dermatol. 2004;123:1120–1126. doi: 10.1111/j.0022-202X.2004.23492.x. [DOI] [PubMed] [Google Scholar]
- 24.Bushmeyer S, Park K, Atchison ML. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 25.Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, Kim JK, Kim KK, Epstein JA, Kook H. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 28.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 29.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 30.Sucharov CC, Helmke SM, Langer SJ, Perryman MB, Bristow M, Leinwand L. The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression. Mol Cell Biol. 2004;24:8705–8715. doi: 10.1128/MCB.24.19.8705-8715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLellan WR, Lee TC, Schwartz RJ, Schneider MD. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 32.Schwartz K, de la Bastie D, Bouveret P, Oliviero P, Alonso S, Buckingham M. Alpha-skeletal muscle actin mRNA's accumulate in hypertrophied adult rat hearts. 1986;Vol 59:551–555. doi: 10.1161/01.res.59.5.551. [DOI] [PubMed] [Google Scholar]
- 33.Krippner-Heidenreich A, Walsemann G, Beyrouthy MJ, Speckgens S, Kraft R, Thole H, Talanian RV, Hurt MM, Luscher B. Caspase-dependent regulation and subcellular redistribution of the transcriptional modulator YY1 during apoptosis. Mol Cell Biol. 2005;25:3704–3714. doi: 10.1128/MCB.25.9.3704-3714.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, McKinsey TA, Zhang C-L, Olson EN. Regulation of Skeletal Myogenesis by Association of the MEF2 Transcription Factor with Class II Histone Deacetylases. Molecular Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 36.Sucharov CC, Langer S, Bristow M, Leinwand L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am J Physiol Cell Physiol. 2006;291:C1029–C1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- 37.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II Histone Deacetylases Act as Signal-Responsive Repressors of Cardiac Hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3[beta] activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]