Abstract
Background
We studied Dicer and Drosha, components of the RNA-interference machinery, in ovarian cancer.
Methods
We measured messenger RNA (mRNA) levels of Dicer and Drosha in specimens of invasive epithelial ovarian cancer from 111 patients, using a quantitative reverse-transcriptase–polymerase-chain-reaction assay, and compared the results with clinical outcomes. Validation was performed with the use of published microarray data from cohorts of patients with ovarian, breast, and lung cancer. Mutational analyses of genomic DNA from the Dicer and Drosha genes were performed in a subgroup of ovarian-cancer specimens. Dicer-dependent functional assays were performed by means of in vitro transfection with small interfering RNA (siRNA) and short hairpin RNA (shRNA).
Results
Levels of Dicer and Drosha mRNA correlated with the levels of expression of the corresponding protein and were decreased in 60% and 51% of ovarian-cancer specimens, respectively. Low Dicer expression was significantly associated with advanced tumor stage (P = 0.007), and low Drosha expression with suboptimal surgical cytoreduction (P = 0.02). Cancer specimens with both high Dicer expression and high Drosha expression were associated with increased median survival (>11 years, vs. 2.66 years for other subgroups; P<0.001). We found three independent predictors of reduced disease-specific survival in multivariate analyses: low Dicer expression (hazard ratio, 2.10; P = 0.02), high-grade histologic features (hazard ratio, 2.46; P = 0.03), and poor response to chemotherapy (hazard ratio, 3.95; P<0.001). Poor clinical outcomes among patients with low Dicer expression were validated in additional cohorts of patients. Rare missense mutations were found in the Dicer and Drosha genes, but their presence or absence did not correlate with the level of expression. Functional assays indicated that gene silencing with shRNA, but not siRNA, may be impaired in cells with low Dicer expression.
Conclusions
Our findings indicate that levels of Dicer and Drosha mRNA in ovarian-cancer cells have associations with outcomes in patients with ovarian cancer.
The discovery that gene expression can be altered through RNA interference1 has stimulated research on the role of RNA interference in the development of cancer. Targeting specific genes by RNA-interference molecules allows for the identification of regulators of angiogenic, proliferative, and survival pathways in cancer cells. Furthermore, RNA-interference molecules that silence specific genes are being tested in preclinical studies as a treatment for cancer.2,3
Regulation of gene expression through RNA interference occurs by means of microRNA (miRNA) or small interfering RNA (siRNA) (Fig. 1). In the nucleus, endogenous double-stranded RNA segments are cut into short, hairpin-shaped double-stranded RNA precursor structures (of approximately 60 to 70 nucleotides each)4,5 by the RNase III enzyme Drosha. These precursors move to the cytoplasm, where Dicer, also an RNase III enzyme, cleaves them into mature double-stranded RNA fragments (miRNA), 19 to 21 nucleotides each.6 Translational repression or degradation of messenger RNA (mRNA) occurs when miRNA binds to the RNA-induced silencing complex (RISC).7,8 The siRNA production occurs in a similar manner, although processing by Drosha is not required.9
Figure 1. The RNA-Interference Cascade in Humans.
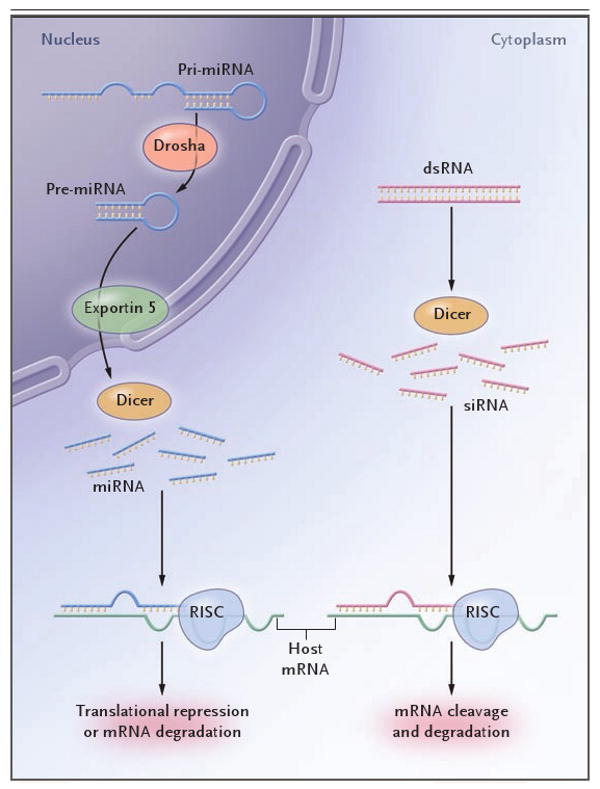
Long precursor microRNA (miRNA) segments, called pri-miRNA, are first cleaved in the nucleus by Drosha, an RNase III endonuclease, into segments of approximately 70 nucleotides each (called pre-miRNA). Transportation into the cytoplasm by means of exportin 5 leads to cleavage by Dicer, another RNase III endonuclease, which produces mature miRNA segments. Host degradation of messenger RNA (mRNA) and translational repression occurs after miRNA binds to the RNA-induced silencing complex (RISC). Cytoplasmic long double-stranded RNA (dsRNA) is cleaved by Dicer into small interfering RNA (siRNA), which is incorporated into RISC, resulting in the cleavage and degradation of specific target mRNA.
Alterations of miRNAs in human cancers have been reported, but the regulation of these molecules is unclear. In ovarian tumors, decreased expression of a substantial proportion of miRNAs has been found,10-13 but the downstream effects of this decrease are not known. Nevertheless, these findings support the hypothesis that miRNAs have an underlying role in cancer progression.10-12 We investigated whether altered levels of Dicer and Drosha mRNA, components of the RNA-interference machinery, are associated with clinical outcome in ovarian cancer.
Methods
Cell Lines
The derivation, sources, and maintenance of the ovarian-cancer cell lines used in this study have been reported previously.14 The lines were HeyA8, SKOV3ip1, A2780-Par, IGROV, EG, 222, OVCAR3, and OVCAR420 (for sources, see the Supplementary Appendix, available with the full text of this article at www.nejm.org).
Tumor Samples
After the study was approved by the Institutional Review Board and written informed consent was obtained from the patients for the use of clinical specimens for research, we obtained 111 specimens of invasive epithelial ovarian cancer from the M.D. Anderson Cancer Center Tumor Bank and the Brigham and Women's Gynecologic Oncology Tumor Bank. Before collection of these samples, 11 benign ovarian epithelial samples were obtained as controls from microdissected paraffin-embedded specimens or epithelial scrapings taken after surgical removal.
Data on clinical outcome were obtained from patients' records. Responses to initial chemotherapy were recorded as either sensitive (if there was normalization of the CA-125 level, a negative finding on second-look laparotomy, or both within 6 months after completion of initial chemotherapy) or refractory or resistant (if there was progression or recurrence within 6 months after completion of initial chemotherapy).
Dicer and Drosha mRNA
A quantitative reverse-transcriptase–polymerase-chain-reaction (RT-PCR) assay was used to measure Dicer and Drosha mRNA in RNA extracted from ovarian cell lines and ovarian tumors. Protein expression was examined in all cell lines by means of Western blots (see the Supplementary Appendix) and in ovarian tumors by means of immunohistochemical methods. Quantitative RT-PCR was performed with the use of the TaqMan gene-expression assay kit (Applied Biosystems), with either Dicer, Drosha, or 18s RNA primers (Applied Biosystems), as previously described.15,16 The final mRNA levels were converted to ratios of decreased expression (≤1) or increased expression (>1) relative to levels of Dicer and Drosha mRNA in normal ovarian epithelium.
Immunohistochemical Analysis
Slides were deparaffinized and rehydrated with sequential washes in xylene and ethanol. Antigen retrieval was performed in a pressure cooker at 90°C with the use of Borg Decloaker solution (Biocare Medical). Slides were then incubated with primary antibodies against Dicer (1:100 dilution, Sigma) or Drosha (1:200, Abcam) overnight, followed by incubation with a mouse–rabbit–horseradish peroxidase polymer (MR-HRP, Biocare Medical) and 3,3′-diaminobenzidine substrate (Phoenix Biotechnologies). The stained slides were scored by two investigators who were unaware of the RT-PCR results, on the basis of the histochemical score (with a score of >100 defined as high expression and ≤100, low expression), according to the method described by McCarty et al.,17,18 which considers both the intensity of staining and the percentage of cells stained.
Validation
The relation between levels of Dicer and Drosha mRNA and survival among patients with ovarian cancer (GEO accession number, GSE314919), breast cancer (ArrayExpress accession number, E-TABM-15820; and GEO accession numbers, GSE145621 and GSE492222), and lung cancer (GEO accession number, GSE314119) was examined in existing microarray data sets of samples that had been pro-filed with an Affymetrix GeneChip assay (either HG-U133A or HG U133 Plus 2.0). Probe sets 212888_at and 218629_at were used to measure Dicer and Drosha expression, respectively.
Mutational Analysis
Genomic DNA extracted from four ovarian cell lines and 37 previously examined ovarian tumors was sequenced for the Dicer gene DICER1 (NM_177438) and the Drosha gene RNASEN (NM_013235) coding exons and their flanking splice sites to assess for potential mutations, as previously described.23 All sequence variants were verified through manual inspection of the chromatograms.
Transfection with Small Interfering and Short Hairpin RNA
The anti–galectin-3 oligonucleotide (target sequence, GTACAATCATCGGGTTAAATT; Dharmacon) and control nontargeting oligonucleotides (target sequence, UUCUCCGAACGUGUCACGU; Qiagen) were used for transfection of siRNA and short hairpin RNA (shRNA). The shRNA was prepared using a lentivirus gene-transfer vector (containing green fluorescent protein), as previously described24; shRNA induces stable genetic silencing.
For siRNA and shRNA transfections, 2×105 cells per well were plated and 5 μg of galectin-3 or nontargeting siRNA was added according to the manufacturer's protocol. Transfection was considered successful if a transfection rate of more than 90% was achieved. Gene silencing was assessed by means of Western blot analysis.
Statistical Analysis
To determine the distribution of Dicer and Drosha levels around cutoff points, histograms were created on a log2 scale of the expression ratio and tested for normality with the Kolmogorov–Smirnov test. The Wilcoxon test was used to compare rank distributions of continuous variables not conforming to the assumptions of normality. Contingency tables and Fisher's exact test were used to evaluate the relationship between death and categorical variables. Kaplan–Meier plots were constructed and a log-rank test was used to determine differences among survival curves according to Dicer and Drosha expression level. Multivariate analyses were performed with the use of a Cox proportional-hazards model to examine the effects of Dicer and Drosha expression on death from disease while adjusting for other covariates. Bonferroni corrections were used in analyses involving multiple comparisons.
The relation between the expression of the Dicer and Drosha genes and survival was explored in microarray data sets by separating the cases from each cohort into a group with a high median level of expression and a group with a low median level of expression. The statistical significance of the Cox hazard ratio was assessed by means of Wald's test (using the “survival” package [version 2.34] in the R language for statistical computing [version 2.6.1]).25 A kappa statistic was calculated to assess the agreement between RT-PCR results and histochemical scores. All P values were two-sided, and a P value of less than 0.05 was considered to indicate statistical significance.
Results
Dicer and Drosha Expression
We examined 111 ovarian-cancer specimens and 11 benign epithelial-ovarian specimens by using quantitative RT-PCR for mRNA and calculated the ratios of the expression in the tumors and the expression in the benign specimens. The distribution of Dicer mRNA levels in the ovarian-cancer specimens was bimodal (P = 0.002 by the Kolmogorov–Smirnov test for normality) with two peaks in the ratio (0.43 and 4.25). The division between these two subpopulations corresponded to a Dicer mRNA ratio of 1.20. Levels of Drosha mRNA in cancer specimens followed a normal distribution (P = 0.15 by the Kolmogorov–Smirnov test for normality), with a peak corresponding to a median ratio of 1.00. These findings support our decision to use 1 as the cutoff value for high and low Dicer and Drosha mRNA levels in subsequent analyses.
Levels of mRNA varied among cancer specimens; 60% had decreased Dicer mRNA and 51% had decreased Drosha mRNA (Table 1 in the Supplementary Appendix). In 39% of specimens, there were decreased levels of both Dicer and Drosha mRNA. The median ratio of Dicer expression in cancer specimens with decreased Dicer mRNA levels was 0.27 (range, 0.01 to 1.00; three specimens with undetectable levels) and in those with decreased Drosha mRNA levels was 0.52 (range, 0.02 to 1.00; one specimen with un-detectable levels). Specimens with increased mRNA levels had a median ratio for Dicer of 3.38 (range, 1.13 to 10.41) and a median ratio for Drosha of 1.98 (range, 1.02 to 18.85).
To determine whether mRNA levels reflected protein expression, a subgroup of 32 ovarian tumors was also examined through immunohistochemical methods (Fig. 1 in the Supplementary Appendix). The histochemical score was in agreement with the quantitative RT-PCR results for both Dicer (kappa = 1.00; 95% confidence interval [CI], 1.00 to 1.00) and Drosha (kappa = 0.86; 95% CI, 0.68 to 1.00).
We also examined Dicer and Drosha mRNA levels in a panel of ovarian-cancer cell lines. As compared with nontransformed ovarian surface-epithelial cells, 50% of ovarian-cancer cell lines had reduced levels of both Dicer mRNA (to between 50 to 8% of the control level) and Drosha mRNA (to between 91 and 7% of the control level). Similar to ovarian tumors, the ovarian-cancer cell lines had mRNA levels that were concordant with protein levels (Fig. 2 in the Supplementary Appendix).
Clinical Associations
Table 1 lists the baseline characteristics of all 111 patients (mean age, 62.5 years) with invasive epithelial ovarian carcinoma. Most of these patients had advanced-stage, poorly differentiated tumors, and 77.5% had undergone optimal surgical reduction of the primary tumor (residual tumor, ≤1 cm in diameter). Most patients (53.2%) had tumors that were sensitive to initial chemotherapy, whereas 33.3% had refractory or resistant disease (with data missing for 13.5% of patients). In univariate analyses, neither Dicer nor Drosha mRNA levels were significantly associated with age, tumor grade, or response to chemotherapy (Table 2). Low Dicer mRNA levels were, however, significantly associated with advanced tumor stage (P = 0.007), as were low Drosha mRNA levels with suboptimal cytoreductive surgery (P = 0.02).
Table 1. Baseline Characteristics of the 111 Patients with Invasive Epithelial Ovarian Carcinoma.
| Characteristic | Value |
|---|---|
| Age — yr | |
| Mean | 63 |
| Range | 25–96 |
| Tumor stage — no. (%) | |
| I or II | 8 (7.2) |
| III or IV | 103 (92.8) |
| Cytoreduction — no. (%)☆ | |
| Optimal | 86 (77.5) |
| Suboptimal | 25 (22.5) |
| Tumor grade — no. (%) | |
| Low (1 or 2) | 16 (14.4) |
| High (3) | 95 (85.6) |
| Response to initial chemotherapy — no. (%) | |
| Sensitive | 59 (53.2) |
| Resistant or refractory | 37 (33.3) |
| Missing data | 15 (13.5) |
| Disease status — no. (%) | |
| Alive with disease | 14 (12.6) |
| Alive without disease | 33 (29.7) |
| Dead from disease | 64 (57.7) |
Optimal cytoreduction was defined as cytoreduction resulting in residual tumor of 1 cm or less in diameter.
Table 2. Correlation of Clinical and Pathological Features with Drosha and Dicer Messenger RNA (mRNA) Levels in the 111 Patients with Invasive Epithelial Ovarian Carcinoma.☆.
| Variable | Drosha mRNA Level | Dicer mRNA Level | ||||
|---|---|---|---|---|---|---|
| Low (N = 57) | High (N = 54) | P Value | Low (N = 66) | High (N = 45) | P Value | |
| Age — yr | 61.2±13.0 | 63.8±11.7 | 0.27 | 63.1±11.7 | 61.6±13.3 | 0.54 |
| Tumor stage — no. (%) | ||||||
| I or II | 2 (3.5) | 6 (11.1) | 0.15 | 1 (1.5) | 7 (15.6) | 0.007 |
| III or IV | 55 (96.5) | 48 (88.9) | 65 (98.5) | 38 (84.4) | ||
| Tumor grade — no. (%) | ||||||
| Low | 7 (12.3) | 9 (16.7) | 0.59 | 9 (13.6) | 7 (15.6) | 0.79 |
| High | 50 (87.7) | 45 (83.3) | 57 (86.4) | 38 (84.4) | ||
| Cytoreduction — no. (%) | ||||||
| Optimal | 39 (68.4) | 47 (87.0) | 0.02 | 47 (71.2) | 39 (86.7) | 0.07 |
| Suboptimal | 18 (31.6) | 7 (13.0) | 19 (28.8) | 6 (13.3) | ||
| Response to initial chemotherapy — no. (%)† | ||||||
| Sensitive | 27 (47.4) | 32 (59.3) | 0.53 | 31 (47.0) | 28 (62.2) | 0.40 |
| Resistant or refractory | 20 (35.1) | 17 (31.5) | 23 (34.8) | 14 (31.1) | ||
| Missing data | 10 (17.5) | 5 (9.3) | 12 (18.2) | 3 (6.7) | ||
Plus–minus values are means ±SD.
P values were calculated after missing values were excluded.
The median overall survival was substantially reduced among women whose tumor had low levels of Dicer mRNA (2.33 years, vs. 9.25 years for high levels; P<0.001) and Drosha mRNA (2.74 years, vs. 7.92 years for high levels; P = 0.008) (Fig. 2). As compared with other subgroups (tumors with low Dicer and low Drosha expression, high Dicer and low Drosha expression, and low Dicer and high Drosha expression), tumors with high levels of both Dicer and Drosha mRNA were associated with an increased median survival (>11 years [median survival not reached], vs. 2.66 years; P<0.001) (Fig. 3 in the Supplementary Appendix). In univariate analyses, death from ovarian cancer was associated with low levels of both Dicer and Drosha mRNA (P = 0.01 and P = 0.007, respectively).
Figure 2. Kaplan–Meier Survival Curves for Patients According to Tumor Expression of Dicer and Drosha.
For patients with invasive epithelial ovarian cancer, curves are shown for Dicer and Drosha expression (Panel A), with curves for Dicer and Drosha combined shown for comparison in Fig. 3 in the Supplementary Appendix. Curves from validation analyses are also shown for the expression of Dicer and Drosha in independent cohorts of patients with ovarian cancer (Panel B) and lung cancer (Panel C), as well as Dicer expression in two cohorts of patients with breast cancer (Panels D and E).
In multivariate analyses (including age, tumor stage and grade, Dicer and Drosha mRNA levels, optimal or suboptimal cytoreduction, and response to initial chemotherapy), poorly differentiated tumors (P = 0.03) and a resistant or refractory chemo-response (P<0.001) were associated with poor survival (Table 3). A decreased Dicer mRNA level was an indicator of a poor prognosis (hazard ratio, 2.10; 95% CI, 1.15 to 3.85; P = 0.02). A low Drosha mRNA level, however, was not an independent predictor of survival (hazard ratio, 1.22; 95% CI, 0.69 to 2.16; P = 0.50). When paired in an interaction model, low Dicer and low Drosha mRNA levels had a greater association with decreased survival (hazard ratio, 4.00; 95% CI, 1.82 to 9.09; P<0.001) than either one alone.
Table 3. Results of Multivariate Analyses of Independent Prognostic Factors in Patients with Invasive Epithelial Ovarian Carcinoma.
| Factor | Hazard Ratio for Death (95% CI) | P Value |
|---|---|---|
| Low Dicer expression | 2.10 (1.15–3.85) | 0.02 |
| Low Drosha expression | 1.22 (0.69–2.16) | 0.50 |
| Increased age | 1.01 (0.99–1.03) | 0.43 |
| Tumor stage III or IV | 4.40 (0.59–33.15) | 0.15 |
| High-grade tumor | 2.46 (1.10–5.52) | 0.03 |
| Suboptimal cytoreduction | 1.27 (0.68–2.38) | 0.46 |
| Resistant or refractory response to chemotherapy | 3.95 (2.01–7.52) | <0.001 |
| Low Dicer and low Drosha expression☆ | 4.00 (1.82–9.09) | <0.001 |
A separate analysis was performed that included the original factors in addition to a combined Dicer and Drosha variable.
To validate our findings, we used previously reported microarray data to compare expression of Dicer and Drosha genes with survival in 132 patients with ovarian cancer.19 Similar to our initial findings, increased survival was associated with high expression of Drosha mRNA (hazard ratio, 0.55; 95% CI, 0.34 to 0.89; P = 0.02) and Dicer mRNA (hazard ratio, 0.53; 95% CI, 0.33 to 0.85; P = 0.008) (Fig. 2B).
To examine whether this association also holds for other tumors, we measured the relative expression ratios for Dicer and Drosha in microarrays with cohorts of 91 patients with lung cancer19 and 129 patients with breast cancer.20 Increased survival in the lung-cancer cohort was associated with high levels of Dicer mRNA (hazard ratio, 0.43; 95% CI, 0.23 to 0.80; P = 0.008) but not Drosha mRNA (hazard ratio, 1.34; 95% CI, 0.74 to 2.40; P = 0.33) (Fig. 2C). Similarly, increased disease-free survival in the breast-cancer cohort was associated with high levels of Dicer mRNA (hazard ratio, 0.32; 95% CI, 0.14 to 0.72; P = 0.006) but not of Drosha mRNA (hazard ratio, 0.93; 95% CI, 0.45 to 1.92; P = 0.84), as was overall survival (data not shown). A relationship between high Dicer mRNA levels and increased disease-free survival was also found from microarray analysis of specimens obtained from two other cohorts of patients with breast cancer: one with 159 patients21 (hazard ratio, 0.33; 95% CI, 0.17 to 0.66; P = 0.002) (Fig. 2D) and one with 249 patients22 (hazard ratio, 0.64; 95% CI, 0.42 to 0.97; P = 0.04) (Fig. 2E).
Mutations of Dicer and Drosha
We next investigated whether the variable levels of Dicer and Drosha mRNA could be explained by the presence of mutations. Initial studies, performed in cell lines with high or low gene expression, revealed several missense mutations for both genes and one splice-site mutation for the Drosha gene (see the Supplementary Appendix). Next, genomic DNA from 37 ovarian tumors previously analyzed for Dicer and Drosha mRNA was examined for the Dicer and Drosha gene mutations. For the Dicer gene, two missense mutations were discovered in two tumors. Similarly, two missense mutations were revealed for the Drosha gene. These mutations were not, however, associated with alterations in levels of mRNA of Dicer or Drosha.
Functional Analysis of Decreased Dicer Expression
To elucidate the functional consequences of low Dicer mRNA levels, we compared the silencing of a constitutively expressed gene, the galectin-3 gene, between ovarian-cancer cell lines with high Dicer expression and those with low Dicer expression. Specifically, we compared shRNA constructs (exhibiting stable gene silencing) with the shorter siRNA fragments (which have transient gene silencing) (Fig. 3). As compared with control (non-targeting) constructs, transfection of siRNA reduced galectin-3 levels in the cell lines with low Dicer expression: HeyA8, which showed a reduction by 78% from the control level, and SKOV3ip1, reduced by 95% from the control level. In contrast, poor silencing was noted in these cells with shRNA (8% and 4%, respectively). In the OVCAR3 and 222 cells, which have high Dicer expression, silencing of galectin-3 by 62% to 73% as compared with control levels was observed with both siRNA and shRNA constructs.
Figure 3. Transfection of Small Interfering RNA (siRNA) and Short Hairpin RNA (shRNA) Targeting Galectin-3 in Ovarian-Cancer Cell Lines with Low Dicer Expression and Those with High Dicer Expression.
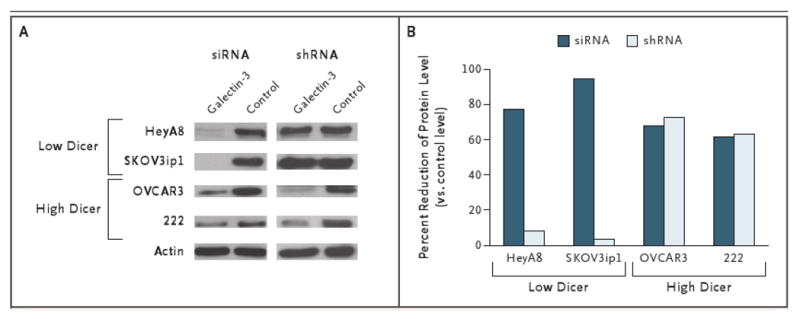
Western blot densitometry analysis was performed for the cell lines transfected with siRNA or shRNA or control (nontargeting) sequences. Actin was used for purposes of normalization.
Discussion
We found that Dicer and Drosha mRNA expression is variable among invasive epithelial ovarian cancer specimens and in ovarian-cancer cell lines and that they are significantly associated with survival, indicating that levels of mRNA of Dicer and Drosha in ovarian-cancer cells are clinically relevant. The association was validated in independent clinical data sets for ovarian cancer. We also sought mutations in the genomic sequences of the Dicer and Drosha genes in a cohort of patients with variable gene expression and did not find a consistent trend. Furthermore, the results of our functional assays indicate that cells with low Dicer expression could not effectively silence genes when synthetic shRNA constructs were transfected.
The production of mature endogenous interfering RNA involves a cascade of events that are inextricably linked to the functions of Dicer and Drosha. For example, Lee et al.5 demonstrated that in cells with silenced Dicer or Drosha expression, precursor and mature miRNA sequences were reduced. Loss of Dicer in mice disrupts embryonic stem-cell differentiation and is lethal during early development.26 Low levels of Dicer mRNA also affect normal cellular development and immune responses in preclinical models.27-29 Furthermore, abnormalities in the copy number of DNA of the Dicer gene and the argonaute 2 gene (a component of the RNA-induced silencing complex) have been described in human melanoma, breast, and ovarian cancers.13 It is therefore possible that deregulated miRNA expression, observed in several types of tumor,13 is secondary to defective RNA silencing machinery. Decreased Dicer mRNA has also been associated with decreased survival in patients with non–small-cell lung cancer.30 In addition, Dicer expression appears to be up-regulated in noninvasive precursors of invasive lung adenocarcinoma.31
Some findings in other tumor types are inconsistent with our findings. High Dicer expression and high Drosha expression were correlated with poor prognostic factors in prostate cancer and esophageal carcinoma.32,33 Furthermore, reduction of Drosha expression by means of siRNA reduced cellular proliferation has been reported in esophageal-cancer cell lines.33 There are several plausible explanations for the divergent expression patterns of Dicer and Drosha among different solid tumors and how they relate to clinical and pathologic variables. Direct interactions with other components of the RNA-interference cascade could result in compensatory alterations of Dicer or Drosha expression in the presence of mutated cofactors such as genes for the DiGeorge syndrome critical region gene 8 (DGCR8), exportin 5 (XPO5), and argonaute 2 (AGO2).31,34-36 In addition, miRNA could have varying regulatory effects independent of alterations in the RNA-interference processing machinery.37 In support of this contention is the association of altered Dicer protein expression (both overexpression and underexpression relative to controls) in mucoepidermoid carcinomas of the upper aerodigestive tract with overall survival.38
Despite growing evidence that Dicer and Drosha mRNA levels vary among tumor types, the regulation of these genes is unclear. Dicer gene mutations have been found in Caenorhabditis elegans39 and in humans, and deletions of the Dicer gene locus were detected in some precancerous and invasive lung adenocarcinomas.31 Our mutational analyses showed that alterations of genomic DNA probably do not account for the variability in Dicer and Drosha levels. We did find that single-nucleotide mutations can occur in both genes; however, the functional role of such mutations remains unclear. In breast-cancer cell lines, there are two forms of Dicer, due to alternative splicing mechanisms, which appear to affect the stability of the Dicer protein.40 DNA methylation of the Dicer gene was not found in a small subgroup of lung-cancer specimens.30 As the function of miRNAs in the genesis of tumors becomes clearer, further studies will be needed to elucidate the regulation and stability of the RNA-interference machinery.
Our findings have implications for the development of treatments for ovarian cancer that are based on RNA interference. Highlighting this point is the differential targeting efficiency of a constitutively expressed gene that we found through a functional assay of gene silencing. The shRNA that is complementary to the target gene has been tested in animal models and found to induce robust gene silencing. However, one study showed increased mortality among mice after delivery of multiple shRNA sequences.41 These results and our data suggest that miRNA processing may be hindered in tumors with low Dicer and low Drosha expression, which could lead to a poor clinical outcome.
Supplementary Material
Acknowledgments
Supported by grants from the National Cancer Institute (T32 Training Grant CA101642), the Ovarian Cancer Research Fund Program Project Development Grant, the University of Texas M.D. Anderson Cancer Center Ovarian Cancer Specialized Program of Research Excellence (P50 CA083639), the National Institutes of Health (CA110793, CA109298, P50 CA58207, and U54 CA112970), the Gynecologic Cancer Foundation, the Zarrow Foundation, the Betty Ann Asche Murray Distinguished Professorship, the Marcus Foundation, and the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (contract DE-AC03-76SF00098).
Footnotes
Dr. Gray reports receiving consulting fees from Agendia and Sirna. No other potential conflict of interest relevant to this article was reported.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Halder J, Kamat AA, Landen CN, Jr, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin Cancer Res. 2006;12:4916–24. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 4.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 7.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–89. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 9.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–47. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 10.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 11.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–8. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Coukos G. MicroRNAs: a new insight into cancer genome. Cell Cycle. 2006;5:2216–9. doi: 10.4161/cc.5.19.3319. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Huang J, Yang N, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sood AK, Seftor EA, Fletcher MS, et al. Molecular determinants of ovarian cancer plasticity. Am J Pathol. 2001;158:1279–88. doi: 10.1016/S0002-9440(10)64079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thaker PH, Han LY, Kamat AA, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat Med. 2006;12:939–44. doi: 10.1038/nm1447. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS. Estrogen receptor analyses: correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109:716–21. [PubMed] [Google Scholar]
- 18.Singh M, Zaino RJ, Filiaci VJ, Leslie KK. Relationship of estrogen and progesterone receptors to clinical outcome in metastatic endometrial carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2007;106:325–33. doi: 10.1016/j.ygyno.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 20.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Pawitan Y, Bjöhle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 23.Ahituv N, Kavaslar N, Schackwitz W, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–91. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The R Foundation for Statistical Computing. Vienna: The R Foundation; 2007. [November 24, 2008]. R: a language and environment for statistical computing. http://www.R-project.org. [Google Scholar]
- 26.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]; Nat Genet. 2008;35:287. Erratum. [Google Scholar]
- 27.Cobb BS, Hertweck A, Smith J, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–27. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damiani D, Alexander JJ, O'Rourke JR, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–87. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105:1949–54. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiosea S, Jelezcova E, Chandran U, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 32.Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of Dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugito N, Ishiguro H, Kuwabara Y, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–8. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 34.Qi HH, Ongusaha PP, Myllyharju J, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–4. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitter D, Filkowski J, Sewer A, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–15. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–44. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Chiosea SI, Barnes EL, Lai SY, et al. Mucoepidermoid carcinoma of upper aerodigestive tract: clinicopathologic study of 78 cases with immunohistochemical analysis of Dicer expression. Virchows Arch. 2008;452:629–35. doi: 10.1007/s00428-007-0574-5. [DOI] [PubMed] [Google Scholar]
- 39.Welker NC, Habig JW, Bass BL. Genes misregulated in C elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–102. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irvin-Wilson CV, Chaudhuri G. Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res. 2005;7:R563–R569. doi: 10.1186/bcr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



