Abstract
Inhibitor of apoptosis proteins (IAPs) such as XIAP subvert apoptosis by binding and inhibiting caspases. Because occupation of the XIAP BIR3 peptide binding pocket by Smac abolishes the XIAP–caspase 9 interaction, it is a proapoptotic event of great therapeutic interest. An assay for pocket binding was developed based on the displacement of Smac 7-mer from BIR3. Through the physical and biochemical analysis of a variety of peptides, we have determined the minimum sequence required for inhibition of the Smac–BIR3 interaction and detailed the dimensions and topology of the BIR3 peptide binding pocket. This work describes the structure–activity relationship (SAR) for peptide inhibitors of Smac-IAP binding.
Keywords: BIR, cancer, caspases, inhibitors of apoptosis protein (IAP), peptidomimetics, protein–protein interaction, Smac/DIABLO
Introduction
Programmed cell death plays a critical role in regulating cell number and in eliminating stressed or damaged cells from normal tissues. Indeed, the network of apoptotic signalling mechanisms inherent in most cell types provides a major barrier to the development and progression of human cancer. Since most commonly used chemo and radiation therapies rely on activation of apoptotic pathways to kill cancer cells, tumor cells which are capable of evading programmed cell death are often difficult to treat.
As the molecular circuitry underlying apoptotic signalling is elucidated, the nature of these desensitizing alterations are becoming better understood. Apoptosis signalling networks are classified as either intrinsic and mediated by cellular stress and mitochondrial permeabilization or extrinsic and mediated by death receptor–ligand interactions (Ashkenazi and Dixit, 1998; Green and Reed, 1998). Both pathways ultimately converge on caspases. Once activated, caspases cleave a number of cell death-related substrates, effecting destruction of the cell.
One strategy for evading apoptosis involves upregulation of Inhibitor of Apoptosis (IAP) proteins (Tamm et al., 2000, 2004; Hofmann et al., 2002; Krajewska et al., 2003; Shiraki et al., 2003). Eight IAP family members have thus far been described: XIAP, CIAP1, CIAP1, Apollon, Livin, Survivin, NAIP and ILP-2. Inclusion in the IAP family requires the presence of one or more baculoviral IAP repeat (BIR) domains. CIAP1, CIAP2 and XIAP each contain three BIR domains whereas ML-IAP and Apollon contain only one (Reviewed in Nachmias et al., 2004).
Caspase inhibitory properties have been described for at least five IAP family members – CIAP1, CIAP2, XIAP, ML-IAP (Livin) and Apollon. In experimental systems, overexpression of IAP proteins enhances resistance to radiation and a range of chemotherapeutic drugs (Liston et al., 1996; Duckett et al., 1998; Tamm et al., 1998; Jin et al., 2004). Furthermore, increased expression levels of IAP proteins correlates with poor prognostic outcomes in multiple neoplastic diseases (Tamm et al., 2000, 2004).
IAP proteins inhibit caspases through multiple potential mechanisms. XIAP, CIAP1, CIAP2 and Livin each possess a RING domain which may act as an E3 ligase to facilitate the proteosomal degradation of protein partners such as caspases, Smac and Omi/Htra2 (reviewed in Vaux and Silke, 2005). The factors governing this indirect caspase repression mechanism are poorly understood. In contrast, the molecular basis for direct inhibition of caspases has been methodically delineated through a combination of structural and biochemical studies (Chai et al., 2000, 2001; Liu et al., 2000; Wu et al., 2000; Huang et al., 2001; Riedl et al., 2001; Shiozaki et al., 2003).
The linker-BIR2 domain from XIAP has high affinity (Kd = 35 nM) for caspases 3 and 7 and inhibits these caspases by binding across their active sites and denying substrate access (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001). XIAP BIR3 inhibits caspase 9 by binding caspase 9 monomers and interfering with dimerization-based caspase 9 activation (Shiozaki et al., 2003). This inhibition is mediated through the interaction of a hydrophobic pocket on the surface of XIAP BIR3 with the IAP binding motif (IBM) or ATPF which becomes exposed upon proteolytic activation of caspase 9 (Srinivasula et al., 2001). While a second interface between caspase 9 and XIAP BIR3 has been described, the interaction between the IBM and the BIR3 peptide binding pocket is critical (Shiozaki et al., 2003). Active caspase 9 that is engaged by BIR3 is unable to bind and cleave substrate.
XIAP is by far the best characterized IAP family member and most effective of all the IAPs at caspase inhibition. Indeed, the caspase inhibitory properties intrinsic to the BIR domains of CIAP1, CIAP2, Apollon and Livin are controversial. The affinity of CIAP1 and CIAP2 BIR2 and BIR3 domains for caspase 3/7 and caspase 9 respectively are at least an order of magnitude less than that of XIAP (Deveraux et al., 1998). The single BIR domain from ML-IAP (Livin) reportedly can inhibit caspases 3, 7 and 9 (Ashhab et al., 2001; Kasof and Gomes, 2001; Vucic et al., 2000, 2002, 2005). Apollon reportedly can directly inhibit caspase 9 and may regulate the turnover of caspases 3/7 via proteosomal degradation (Qiu et al., 2005).
The ability of IAP proteins to buffer an apoptotic signal is subject to regulation by Smac/Diablo, a mitochondrial protein initially discovered as an XIAP interacting protein partner (Du et al., 2000; Verhagen et al., 2000). One well defined interaction interface maps to the amino terminus of Smac and the caspase 9-binding surface groove of XIAP BIR3. The minimal BIR3-binding region of Smac has been localized to the extreme amino terminus of proteolytically processed Smac, a region which shares homology to the IBM of caspase 9. Biochemical and protein–protein interaction assays demonstrated that occupancy of the BIR3 surface groove by Smac or Smac-derived peptides prohibits the BIR3–caspase 9 interaction (Srinivasula et al., 2001).
The observation that short Smac-derived peptides could inhibit the caspase 9–BIR3 interaction in vitro hinted at a therapeutic utility for Smac. Agents which, like Smac, could disengage IAPs from caspases would be predicted to hypersensitize cancer cells to apoptotic stimuli. Indeed, delivery of Smac peptides into tumor cell lines or tumor xenografts through the use of Tat and Penetratin peptide fusions was found to exacerbate apoptosis induced by a spectrum of agents including Taxol, Doxorubicin and Trail (Arnt et al., 2002; Fulda et al., 2002; Vucic et al., 2002; Yang et al., 2003).
While these proof of concept studies galvanized interest in Smac as a therapeutic approach, these early reagents were inadequate as clinical entities. Efforts are in place in many laboratories to distill the biologic properties of Smac down to a drug-like small molecule for cancer therapy. The complexities of this challenge are emblematic of protein–protein interaction targets. Unlike typical enzyme/substrate interactions, protein–protein interactions generally span large flat surfaces and lack deep, defined binding pockets (Sharma et al., 2002). Disruption of protein–protein interactions is thus a formidable task for a small molecule.
Protein–protein targets of biological relevance are “drugable” if they are amenable to intervention by a small molecule resulting in a desired biological response. This novel small molecule must be potent, specific, metabolically stable, and preferably orally active with good pharmacokinetics and no toxicity. Structural information of the involved protein/protein interface is necessary not only to design a small molecule inhibitor of protein-protein interaction but also for providing further insight in the development of antagonist molecules by optimization of HTS hits. Even though it is relatively easy to identify peptide leads (8–12 amino acids), these leads usually suffer from poor stability and bioavailability. Furthermore, converting these peptide leads to small drug-like molecules presents a huge undertaking.
It appears from the published structures of BIR3-XIAP bound to either Smac peptide or protein, that inhibition of IAPs is indeed feasible using small peptides, as binding involves relatively few interface residues (Liu et al., 2000; Wu et al., 2000). Here, we describe a systematic study that was undertaken to explore the physical and chemical binding characteristics of the Smac peptide–BIR3 interaction. We defined the smallest possible biologically active peptide capable of high affinity binding to the BIR3 peptide pocket. The development and characterization of the first generation of small molecule Smac mimetics is described. This work represents the primary SAR for the future design of Smac mimetics.
Results: Smac/XIAP interaction: determination of optimal ligands for the BIR3 peptide binding pocket
Peptide–BIR3 Binding Assay
To rapidly profile compounds for their ability to bind the BIR3 peptide-binding pocket, a solution phase assay was developed based on FMAT technology (Lee et al., 2003). Briefly, biotinylated Smac 7-mer peptide (AVPIAQK, lysine ɛ-amino group is biotinylated) was immobilized on streptavidin coated beads. GST–BIR3 fusion protein was precipitated with FMAT beads in a manner strictly dependent on Smac peptide and could be detected using fluorescent tagged anti-GST antibodies. Importantly, non-biotinylated Smac peptide was highly effective at competing GST–BIR3 off the FMAT beads. The IC50 for non-biotinylated Smac was 400 nM. IC50 values for synthetic peptides were determined using this assay.
Minimum Sequence Requirement
Several shorter Smac-derived peptides starting from AVPIAQK (7-mer down to 3-mer) were synthesized and tested as described above (Table I). In all of these peptides, the N-terminal alanine was kept invariant and the length was changed at the C-terminal end. In addition, 4-mer peptides including Reaper, Hid, and Grim (derived from Drosophila) were also synthesized. In order to facilitate the rapid synthesis of these peptides, a solid phase approach was employed starting with Fmoc-Rink amide resins. Final cleavage of the peptide from the resin provides compounds containing C-terminal amides instead of acids, thus it should be noted that all peptides prepared contain this C-terminal amide unless otherwise noted. Our results indicate that inhibitory activity of peptides is maintained with N-terminal 4 amino acid containing peptides (IC50 range 270–580 nM) and confirm the observations made earlier by structural studies (Chai et al., 2000, 2001). The N-terminal tripeptide, however, loses its ability to inhibit Smac–BIR3 interaction (AVP, IC50 > 10 μM).
Table I.
Minimum Inhibitory Sequence
| Compound # | Sequence* | Source protein | IC50 (μM) |
|---|---|---|---|
| 1 | AVPIAQK | Smac | 0.39 |
| 2 | AVPIAQ | Smac | 0.27 |
| 3 | AVPIA | Smac | 0.30 |
| 4 | AVPI | Smac | 0.58 |
| 5 | AVP | Smac | >10.00 |
| 6 | AVAF | Reaper | 0.75 |
| 7 | AVPF | Hid | 0.12 |
| 8 | AIAY | Grim | 0.34 |
*Indicates sequences terminate as C-terminal primary amide.
**Indicates the highest concentration tested.
An alignment of all IAP binding proteins, including from Drosophila, provided a consensus sequence AVPF (Table II) with Phe as the consensus residue at the 4th position. It was no surprise that this 4-mer had the best inhibitory activity against the Smac–BIR3 interaction. Therefore, amino acid replacements were performed using the consensus sequence AVPF with N-terminal carboxamide.
Table II.
Consensus Sequence
| IAP binding protein | Sequence |
|---|---|
| Smac | A–V–P–I |
| Hid | A–V–P–F |
| Grim | A–I–A–Y |
| Reaper | A–V–A–F |
| Sickle | A–I–P–F |
| Consensus | A–V–P–F |
Alanine Replacements in AVPF
Alanine was replaced with other carefully chosen hydrophobic amino acids containing various size side chains including α-aminobutyric acid (Abu), serine (Ser), α,α-dimethylglycine (Dmg), α-cyclopropylglycine (Cpg), α-cyclohexylglycine (Chg) (Table III). We observed loss of inhibitory activity for all synthesized analogs except when Ala was replaced with Abu. These results suggest that the binding pocket around the methyl side chain of Ala is quite small. This observation is consistent with previous structural studies that showed that alanine replacement with methionine causes loss of inhibitory activity (Liu et al., 2000; Wu et al., 2000). However, substituting an ethyl to methyl side chain (Abu) resulted in enhanced inhibition suggesting that additional space exists in the pocket around this residue. Added van der Waals interaction with the extra methylene group in the hydrophobic pocket may be responsible for increased inhibitory activity. In addition, the functional role of amino group of Ala was assessed by the replacement of the α-amino group with a hydroxyl group (Lac for Ala), compound 14. This substitution for the α-amino group resulted in total loss of inhibitory activity of the 4-mer suggesting the critical role of the α-amino group in forming three hydrogen bonds with BIR3 pocket residues (Chai et al., 2001).
Table III.
Amino Acid Replacements
| Compound # | Sequence* | IC50 (μM) |
|---|---|---|
| Ala replacements | ||
| 9 | Abu–VPF | 0.12 |
| 10 | SVPF | 1.00 |
| 11 | Dmg–VPF | >1.6 |
| 12 | Cpg–VPF | >1.6 |
| 13 | Chx–VPF | >1.6 |
| 14 | Lac–VPF | >1.6 |
| Val replacements | ||
| 15 | ALPF | 0.18 |
| 16 | AIPF | 0.08 |
| 17 | ATPF | 0.20 |
| 18 | A–Cpg–PF | >1.6 |
| 19 | AFPF | 0.19 |
| Pro replacements | ||
| 20 | AV–Hyp–F | 1.0 |
| 21 | AV–Pip–F | 0.71 |
| Phe replacements | ||
| 4 (Smac 4-mer) | AVPI | 0.58 |
| 7 (Hid 4-mer) | AVPF | 0.12 |
| 8 (Grim 4-mer) | AIAY | 0.34 |
| N-Methylamides | ||
| 22 | A–(N-Me)VPF | >1.6** |
| 23 | AVP–(N-Me)F | 0.38 |
*Indicates sequences terminate as C-terminal primary amide.
**Indicates the highest concentration tested.
Valine Replacements in AVPF
Previously, alanine scans of Smac-derived peptides indicated that Val could be replaced by other hydrophobic amino acids (Liu et al., 2000; Wu et al., 2000). The isopropyl side chain of Val makes important hydrophobic contacts with W310 residue of BIR3 domain. We synthesized and analyzed several 4-mer peptides using other hydrophobic amino acids to maximize this interaction. In all cases with the exception of Cpg, the inhibitory activity of the parent 4-mer was maintained (IC50 range 0.08 vs. 0.20 μM) with Ile as the best replacement (Table III). This result with Cpg is not surprising as it involves a α,α-disubstituted amino acid (steric bulk at α-carbon). Recently, Arg was reported as the best replacement at this position (Kipp et al., 2002).
Proline Replacements in AVPF
Proline seems to play an important role in establishing the β-strand conformation of Smac peptides when bound to the BIR3 domain (Chai et al., 2000, 2001). Thus, we made a few conservative Pro replacements in this study (Table III). Only two replacements were used for optimization, including Hyp and Pip (a 6-membered homolog of Pro). Replacement of Pro to Pip and Hyp in the parent 4-mer resulted in loss of some inhibitory activity (IC50 0.12 vs. 0.75 and 1.0 μM, respectively). Our Smac–BIR3 binding model suggests that the introduction of a hydroxyl group on the proline ring may disturb the hydrophobic contact established with the nearby W310. Even though this loss of activity is significant, Ala is present in this position in other natural IAP inhibitors, including Grim and Reaper, suggesting changes at this position would be allowed.
Phenylalanine Replacements in AVPF
Phe, the consensus residue at the 4th position, forms van der Waals contacts with BIR3 pocket residues. This implies that any aliphatic or aromatic amino residue at this position would be allowed. In addition, Fesik and co-workers showed that inhibitory activity of a 7-mer peptide is retained when Ala is substituted for Phe (Liu et al., 2000). Analogs synthesized from this series included naturally occurring 4-mer peptides derived from either Smac (AVPI), Hid (AVPF), and Grim (AIAY) (Table III). Although all 4-mer analogs showed good inhibition of Smac–BIR3 interaction, our results further confirmed that Phe at this position is the most optimal residue.
N-Methyl Amide Analogs of AVPF
The consensus 4-mer peptide AVPF has two secondary amide bonds. Two analogs with (N-Me)-Val and (N-Me)-Phe (Table III) were synthesized in a similar manner as described for AVPF using Fmoc-(N-Me)Val or Fmoc-(N-Me)Phe, respectively. With these modifications, we analyzed the significance of H-bond contributions of the amide NHs. An alkylation of these NHs without loss of inhibitory activity would facilitate further mimetic design. N-methylation of Val resulted in total loss of inhibitory activity of the parent compound (IC50 0.12 vs. >1.6 μM) indicating the critical role of this NH in forming a H-bond with BIR3 pocket residues. However, N-methylation of Phe retained significant activity (loss of activity by 3-fold, IC50 0.12 vs. 0.38 μM). Though this amide NH is involved in H-bond, our result suggests that this amide can be replaced, if desired, for mimetic designs.
Manipulation of the N-terminus in AVPF
Several analogs of the AVPF 4-mer in which substitution on the N-terminus of alanine was varied were synthesized in a similar manner as described for AVPF. With these modifications we examined the significance of the H-bond contributions of the terminal amine. N-methylation of the alanine residue provided 24 which exhibited a 2-fold increase in inhibitory activity of the parent compound (IC50 0.12 vs. 0.075 μM). On the other hand, single alkylation of the parent with other groups resulted in precipitous loss of activity (Et: IC50 = 4 μM; cyclopropyl methyl: IC50 = 1.5 μM; cyclohexyl methyl: IC50 = 2 μM). It was also observed that double alkylation of the alanine resulted in complete loss of activity (N,N-dimethyl Ala IC50 > 10 μM), suggesting that this residue is involved in crucial hydrogen bonds in the BIR3 pocket.
An overview of the observed SAR is presented in Fig. 1, and a working model for the interactions of the AVPI fragment based on published X-ray and NMR structural data (Chai et al., 2000, 2001) is shown in Fig. 2.
Fig. 1.
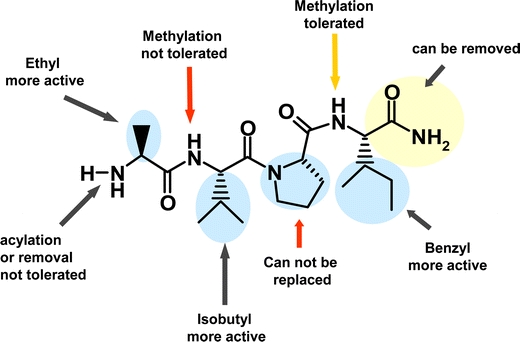
SAR of Smac 4-mer, AVPI.
Fig. 2.
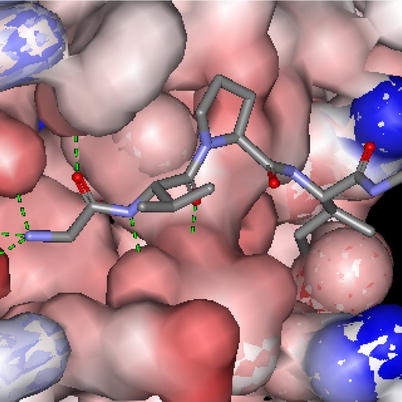
Binding of Smac 4-mer AVPI in BIR3 pocket. The green dotted lines indicate H-bonding.
Functional Analysis of 4-mer Peptides
Results described thus far suggest that Smac’s BIR3 binding activity is preserved in a 4-mer peptide (relative to Smac 7-mer) provided that Phe was substituted for Ile at position 4. In order to assess whether these alterations had functional consequence, DEVD cleavage assays were performed in the presence of recombinant GST–BIR3. In these assays, caspase 3 activity present in 293 cell cytoplasmic extracts was monitored using the fluorescent-tagged synthetic substrate DEVD-AMC. While this is the optimal peptidic caspase 3 substrate, DEVD cleavage still requires caspase 3 processing by active caspase 9. BIR3 inhibits caspase 9 and this results in the inhibition of DEVD cleavage. As shown in Table IV, this inhibition is overcome by Smac 7-mer (AVPIAQK, IC50 = 3 μM). Substitution of Phe for Ile slightly enhanced potency (AVPFAQK, IC50 = 2 μM). Interestingly, 4-mer peptide AVPF was as active as Phe-containing Smac 7-mer (AVPIAQK) (Table IV).
Table IV.
Functional Assay with IAP Binding Peptides
| Sequence | IC50 (μM) |
|---|---|
| AVPIAQK | 3.0 |
| AVPFAQK | 2.0 |
| AVPF | 2.0 |
Optimization for Cellular Activity
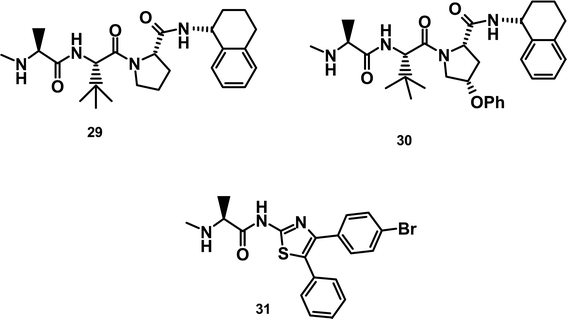
At this point, although we had demonstrated potent activity against the target in binding assays, these compounds were devoid of any significant cellular activity (i.e., in proliferation assays). Suspecting that the lack of activity related to the inability of these molecules to penetrate the cells, we undertook an effort to remove dispensable charged residues and introduce hydrophobic fragments into the molecules where tolerated. Our first step was to remove the primary amide present on the Phe residue, giving compound 25 (Fig. 3). This resulted in a significant increase in cellular activity from >10 to ~1 μM. Previously discussed SAR suggested portions of the molecule that could be altered to introduce hydrophobicity. This led to the synthesis of compound 26 in which the proline was substituted with a cyclohexyl methyl amine at the 4 position, and compound 27 in which the phenethyl amine was changed to a 1-benzyl 2-phenethyl amine. These alterations changed the cLogP of the molecules from 1.1 for 24 and 2.2 for 25 to 3.9 and 4.4 for 26 and 27 respectively. This resulted in cellular activity of 0.6 μM for 26 and 0.25 μM for 27 (Table V). Anti-proliferative activity was abrogated by co-incubation of compounds with a pan-caspase inhibitor, consistent with cell death occurring via apoptosis (data not shown).
Fig. 3.
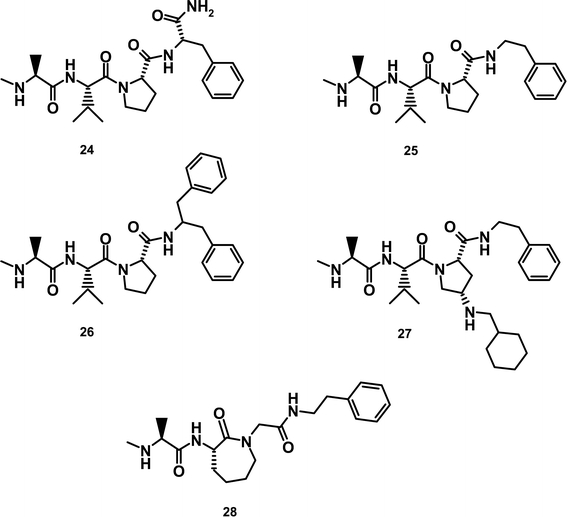
Structures of Novartis inhibitors.
Table V.
Cellular Proliferation Assay, SKOV3 Cells
| Compound # | IC50 (μM) |
|---|---|
| 24 | >10.0 |
| 25 | 0.9 |
| 26 | 0.6 |
| 27 | 0.25 |
Analysis of the X-ray Structures of 26 with XIAP BIR3
Co-crystallization yielded a 1.6 Å resolution X-ray structure of a truncated XIAP BIR3 domain in complex with 26 (Fig. 4). This structure confirmed much of the SAR that had been obtained in our efforts to minimize and optimize the 7-mer peptide. The N-terminus Ala is involved in a potent hydrogen bond to Glu314. The increase in activity against XIAP seen with the methylated peptide appears to be the result of the increase of the pKa of this residue when it is alkylated, while the decrease in activity seen with larger alkyl substituents is most likely due to steric interference with Lys311, bordering this section of the pocket. The methyl group of the Ala residue fills a hydrophobic pocket defined by Gln319, Trp310 and Leu307. The small size of this pocket explains the rather limited substitutions tolerated in this portion of the molecule. The Val nitrogen is hydrogen bonded to Thr308, while the side chain i-Pr group is solvent exposed, thus suggesting another portion of the molecule that could be altered to provide improved cellular penetration. The Pro residue has edge on hydrophobic contact with Trp323 and Tyr324 and also provides the β-turn required to orient the phenyl group of the C-terminus of both molecules into the hydrophobic pocket defined by Gly305 and Gly306. The second phenyl ring of 26 is exposed to solvent, allowing for the substitution of other hydrophobic groups to further improve cellular penetration. It also appears that cis substituents at the 4 position of the proline ring are exposed to solvent, thus explaining the increased cellular activity seen with 27.
Fig. 4.
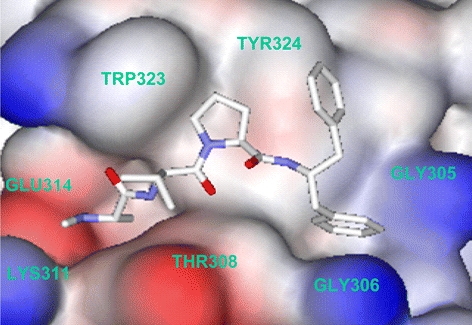
X-ray structure of compound 26 co-crystallized with the BIR3 domain of XIAP.
Smac Mimetics Inhibit Assembly of Native Caspase 9–XIAP Complexes
Having firmly established that these analogs were capable of binding to the hydrophobic pocket of XIAP BIR3, we next assessed whether compound 27 could interfere with the formation of native XIAP–caspase 9 complexes in cell extracts. In hypotonic 293 extracts, activation of caspase 9 is triggered by the addition of cytochrome C and dATP (lanes 1 and 2). As reported previously (Srinivasula et al., 2001) only the cleaved form of caspase 9 coimmunoprecipitated with XIAP (Fig. 5, lanes 3 and 4) and this was dependent on cytochrome C stimulation. Addition of 27 to the extracts inhibited the recovery of caspase 9 from XIAP immunoprecipitates. 27 was slightly more effective than Smac 7-mer in blocking the XIAP–caspase 9 interaction (compare lanes 6 and 8). A related 4-mer peptide (28) lacking detectable BIR3 binding activity (IC50 > 50 μM), was without effect in XIAP–caspase 9 coimmunoprecipitations (lane 9). Thus, these first generation Smac mimetic compounds can compete with activated caspase 9 for occupancy of the XIAP BIR3 surface groove.
Fig. 5.
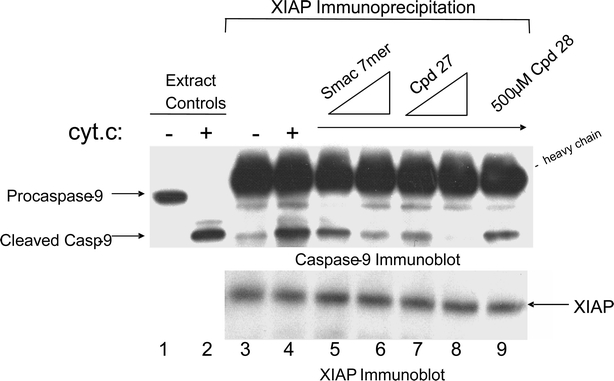
Disruption of native XIAP–caspase 9 complexes.
Discussion and conclusions
Target-based therapeutic strategies for cancer treatments continue to evolve. Many protein–protein interaction targets have been identified for cancer therapy including apoptotic signaling targets. Recent structural studies have identified inhibition of the caspase–IAP interaction as an attractive target for cancer intervention. The BIR2 and BIR3 domains of XIAP directly interact with and inhibit caspase 3/7 and caspase 9, respectively. Smac is a naturally occurring antagonist of the caspase–IAP interaction. Full length Smac appears necessary for neutralizing BIR2, while short 4–7-mers derived from the Smac amino terminus are sufficient for overcoming BIR3-mediated repression. As inhibition of protein–protein interactions often requires larger peptides unamenable to small molecule conversion, targeting BIR3-caspase 9 represents a unique opportunity for drug developers. This almost certainly explains why although several small molecule BIR2 inhibitors have been described (Wu et al., 2003; Schimmer et al., 2004), the effort to develop BIR3 inhibitors has been more intense thus far.
The structural features underlying Smac–BIR3 binding have been elucidated with both full length Smac protein and Smac 9-mer peptide by X-ray and NMR, respectively (Liu et al., 2000; Wu et al., 2000). Smac binding to the BIR3 domain involves two distinct binding interfaces. The first interface involves the N-terminal four residues of Smac (small interface, size 892 Å2), while the second involves the middle portion of Smac (large interface, size >2000 Å2). Mutational studies have determined that the “hot spot” residues – those which contribute to maximal binding energy (Sharma et al., 2002) – are localized at the N-terminus of Smac (Liu et al., 2000; Wu et al., 2000).
Using X-ray crystallography, NMR binding analyses, CAMM, and a systematic, position by position chemical optimization strategy we and others have extended these studies in pursuit of a biostable, cell permeable, small molecular weight Smac mimetic compounds (Kipp et al., 2002; Franklin et al., 2003; Li et al., 2004; Oost et al., 2004; Sharma et al., 2004; Sun et al., 2004a, 2004b).
The molecules which have emerged from these efforts share numerous structural features (see Figs. 3, 6 and 7). The amino-terminal alanine is essentially invariant. This residue makes several critical H-bonds with BIR3 residues (Fig. 2). Methylation of the amino terminal nitrogen increases the BIR3 binding potency by filling a mini-hydrophobic pocket (Gln319, Trp310 and Leu307) and may have an overall stabilizing effect on the peptidomimetic by reducing exopeptidase access (Kipp et al., 2002). The Val–Pro sequence in position 2–3 has been shown to be part of an antiparallel β-sheet binding conformation resulting from H-bonds formed with the β-sheet residues of the BIR3 pocket. Pro forms a kink in the peptide backbone and directs the hydrophobic side chain of the 4th residue (Ile) of Smac into the hydrophobic cleft of BIR3, defined by Gly305 and Gly306. Lack of activity of the tripeptide (AVP) suggests this hydrophobic interaction is essential for binding potency.
Fig. 6.
Structures of Abbott IAP inhibitors.
Fig. 7.
Structures of University of Michigan IAP inhibitors.
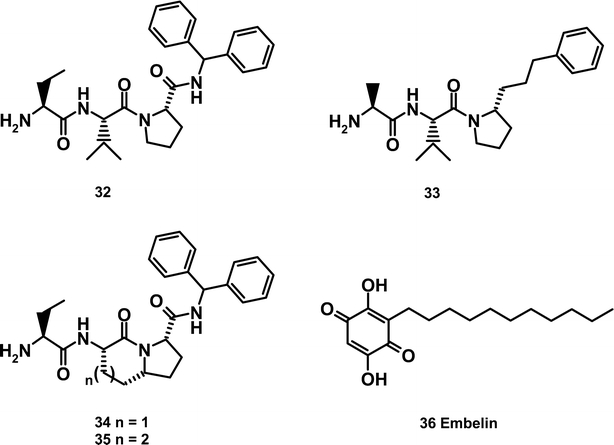
Sun et al. succeeded in further depeptidizing the mimetic by cyclizing the valine and proline residues to give the fused bicyclic core of structures 34 and 35 (Sun et al., 2004a, 2004b). Application of the SAR developed previously resulted in 34 which exhibits a Ki of 350 nm as determined in a fluorescence polarization (FP) assay. The decrease in binding affinity vs. 32 was ascribed to conformational differences the bicyclic scaffold imparts on the side chains when bound to XIAP. Increasing the size of the 6-membered ring by one carbon provided the 7/5 fused structure 35 which perfectly mimics the confirmation of the Smac tetrapeptide. This alteration resulted in a Ki of 25 nM, a 14-fold increase in the binding affinity vs. 34 and a 4-fold increase vs. AVPI. It is important to note that only the indicated stereochemistry at the proline junction is tolerated, as this provides the proper orientation of the side chains to preserve the hydrogen bonding and hydrophobic interactions of the Smac/XIAP interface.
The greatest diversity among Smac mimetics involves variation at the C-terminus. Substituents at this position have the potential to pick up hydrophobic interactions with BIR3 and may also be solvent exposed, allowing for attachment of substituents which can modulate cellular permeability. Whereas we found phenethyl to provide a balance of BIR3 binding and cellular potency, others obtained similar results with a tetrahydronapthyl group (Oost et al., 2004) or an α-phenylbenzylamine (Sun et al., 2005).
Interestingly, dimerization of Smac mimetic compounds was found to significantly enhance BIR3 binding and cellular potency (Li et al., 2004). This is perhaps not surprising as natural Smac was found to homodimerize (Chai et al., 2000). What remains unclear is the precise binding mode for small molecular weight dimers. Does the dimer engage BIR3 domains from two separate IAP molecules or does the dimer induce a conformational change in a single IAP molecule allowing simultaneous binding to BIR2 and BIR3? A crystal structure of these dimers in complex with native XIAP is necessary to resolve this question.
The structural homogeneity of the compounds referenced above stems from the commonality of the approach used for their derivation. Two noteworthy approaches which deviated from this strategy yielded highly unique BIR3-binding compounds.
Wang and coworkers exploited the BIR3–Smac X-ray structure by employing it in a virtual docking screen of a 3D structure database comprising 8200 small organic molecules isolated from traditional Chinese medicinal herbs. Using a consensus scoring program in conjunction with the DOCK program they narrowed the possibilities to 200 high scoring compounds. The most potent compound tested was Embelin, exhibiting an IC50 of 4.1 μM. Binding of Embelin to the XIAP BIR3 domain was confirmed via HSQC NMR studies with 15N labeled protein (Nikolovska-Coleska et al., 2004).
Fesik and colleagues similarly embraced a non-traditional peptidomimetic approach by synthesizing several libraries in which the terminal alanine residue was held constant and linked to fragments that could form hydrogen bonds with the XIAP BIR3 domain (Park et al., 2005). The most potent leads (determined by both NMR binding studies and the FP assay) were further optimized based on the structural information known about peptides bound to the BIR3 domain. This approach led to the synthesis of 31 with a Kd of 740 nM. NMR binding studies revealed a binding motif for the N-terminal alanine which was similar to that seen with the tripeptides above. In addition, it was observed that the amide between the alanine and the thiazole provides contact to the carbonyl of Thr308, while the phenyl group in the 5 position of the thiazole fills the hydrophobic pocket occupied by the proline of the tripeptides and the 4-bromo-phenyl group is pointing towards the hydrophobic grove filled by the isoleucine of Smac.
Based on their assumed mechanism of action, Smac mimetics would be predicted to lower the threshold for apoptotic cell death. Here 4-mer peptides were capable of restoring DEVD cleavage activity to BIR3-inhibited cytoplasmic extracts. Smac mimetics from other laboratories have been found to synergize with TNF and Trail (Li et al., 2004; Sun et al., 2004a, 2004b) and in certain cases, to induce cell death as single agents (Oost et al., 2004). Compound 29 was reported to display nanomolar potency in breast (BT-549, MDA-MB-231), leukemia (HL-60) and renal (RXF-393) lines, and low micromolar activity in melanoma (SK-MEL-5), ovarian (SK-OV-3) and the NSCLC lines NCI-H23 and NCI-H522 (Oost et al., 2004). Furthermore, several of the optimized compounds displayed efficacy in a subcutaneous MDA-MB-231 xenograft model when dosed daily at 20–40 mg/kg/day (Oost et al., 2004). Interestingly, no correlation between the observed potency and XIAP expression exists, thus other factors may be involved. Understanding the contextual basis for these responses may well be the secret to successful implementation of this therapeutic strategy.
Future challenges will be to convert these peptides to small drug-like molecules for cancer therapy. We are confident that by utilizing a peptidomimetic approach, we can produce bioavailable molecules which will potently inhibit IAP proteins, thereby providing a novel method of enhancing the effectiveness of many of the currently available therapies for treating cancer.
Acknowledgements
We gratefully acknowledge the contributions of Kirk Clark, Yigong Shi, Ken Bair, and Mary Tran, Mark Palermo, and Nagarajan Chandramouli.
Abbreviations
- Abu
α-aminobutyric acid
- Ala
alanine
- BIR
baculovirus IAP repeat
- BSA
bovine serum albumin
- Chg
α-cyclohexylglycine
- Cpg
α-cyclopropylglycine
- DIC
N,N-diisopropylcarbidiimide
- DMEM
Dulbecco’s modified Eagle’s medium
- Dmg
α,α-dimethylglycine
- FP
fluorescence polarization
- HOBt
N-hydroxybenztriazole
- HSQ
cheteronuclear single quantum coherence spectroscopy
- HTS
high throughput screening
- Hyp
hydroxyproline
- IAP
inhibitor of apoptosis protein
- Ile
isoleucine
- Lac
lactic acid
- Leu
leucine
- Phe
phenylalanine
- Pip
pipecolic acid
- Pro
proline
- SAR
structure–activity relationship
- Ser
serine
- Smac
secondary mitochondrial activator of caspase
- TFA
trifluoroacetic acid
- Val
valine
References
- Arnt C. R., Chiorean M. V., Heldebrant M. P., Gores G. J., Kaufmann S. H., (2002). J. Biol. Chem. 277: 44236–44243 [DOI] [PubMed]
- Ashhab Y., Alian A., Polliack A., Panet A., Ben Y. D., (2001). FEBS Lett. 495: 56–60 [DOI] [PubMed]
- Ashkenazi A., Dixit V. M., (1998). Science 281: 1305–1308 [DOI] [PubMed]
- Chai J., Du C., Wu J. W., Kyin S., Wang X., Shi Y., (2000). Nature 406: 855–862 [DOI] [PubMed]
- Chai J., Shiozaki E., Srinivasula S. M., Wu Q., Datta P., Alnemri E. S., Shi Y., (2001). Cell 104: 769–780 [DOI] [PubMed]
- Deveraux Q. L., Roy N., Stennicke H. R., Van A. T., Zhou Q., Srinivasula S. M., Alnemri E. S., Salvesen G. S., Reed J. C. (1998). EMBO J. 17: 2215–2223 [DOI] [PMC free article] [PubMed]
- Du C., Fang M., Li Y., Li L., Wang X., (2000). Cell 102: 33–42 [DOI] [PubMed]
- Duckett C. S., Li F., Wang Y., Tomaselli K. J., Thompson C. B., Armstrong R. C., (1998) Mol Cell. Biol.18608–615 [DOI] [PMC free article] [PubMed]
- Franklin M. C., Kadkhodayan S., Ackerly H., Alexandru D., Distefano M. D., Elliott L. O., Flygare J. A., Mausisa G., Okawa D. C., Ong D., Vucic D., Deshayes K., Fairbrother W. J., (2003). Biochemistry (Mosc.) 42: 8223–8231 [DOI] [PubMed]
- Fulda S., Wick W., Weller M., Debatin K. M., (2002). Nat. Med. 8: 808–815 [DOI] [PubMed]
- Green D. R., Reed J. C., (1998). Science 281: 1309–1312 [DOI] [PubMed]
- Hofmann H. S., Simm A., Hammer A., Silber R. E., Bartling B., (2002). J. Cancer Res. Clin. Oncol. 128: 554–560 [DOI] [PubMed]
- Huang Y., Park Y. C., Rich R. L., Segal D., Myszka D. G., Wu H., (2001). Cell 104: 781–790 [PubMed]
- Jin Y., McEwen M. L., Ghandour M. S., Springer J. E.. (2004). Cell. Mol. Neurobiol. 24: 853–863 [DOI] [PMC free article] [PubMed]
- Kasof G. M., Gomes B. C., (2001). J. Biol. Chem. 276: 3238–3246 [DOI] [PubMed]
- Kipp R. A., Case M. A., Wist A. D., Cresson C. M., Carrell M., Griner E., Wiita A., Albiniak P. A., Chai J., Shi Y., Semmelhack M. F., McLendon G. L., (2002). Biochemistry (Mosc.)417344–7349 [DOI] [PubMed]
- Krajewska M., Krajewski S., Banares S., Huang X., Turner B., Bubendorf L., Kallioniemi O. P., Shabaik A., Vitiello A., Peehl D., Gao G. J., Reed J. C. (2003). Clin. Cancer Res. 9: 4914–4925 [PubMed]
- Lee J. Y., Miraglia S., Yan X., Swartzman E., Cornell-Kennon S., Mellentin-Michelotti J., Bruseo C., France D. S., (2003). J. Biomol. Screen. 8: 81–88 [DOI] [PubMed]
- Li, L., Thomas, R. M., Suzuki, H., De Brabander, J. K., Wang, X. and Harran, P. G.: 2004. Science 305(5689), 1471–1474 [DOI] [PubMed]
- Liston P., Roy N., Tamai K., Lefebvre C., Baird S., Cherton-Horvat G., Farahani R., McLean M., Ikeda J. E., MacKenzie A., Korneluk R. G., (1996). Nature 379: 349–353 [DOI] [PubMed]
- Liu Z., Sun C., Olejniczak E. T., Meadows R. P., Betz S. F., Oost T., Herrmann J., Wu J. C., Fesik S. W., (2000). Nature 408: 1004–1008 [DOI] [PubMed]
- Nachmias B., Ashhab Y., Ben-Yehuda D., (2004). Sem. Cancer Biol. 14(4): 231–243 [DOI] [PubMed]
- Nikolovska-Coleska Z., Xu L., Hu Z., Tomita Y., Li P., Roller P. P., Wang R., Fang X., Guo R., Zhang M., Lippman M. E., Yang D., Wang S., (2004). J. Med. Chem. 47(10): 2430–2440 [DOI] [PubMed]
- Oost T. K., Sun C., Armstrong R. C., Al-Assaad A. S., Betz S. F., Deckwerth T. L., Ding H., Elmore S. W., Meadows R. P., Olejniczak E. T., Oleksijew A., Oltersdorf T., Rosenberg S. H., Shoemaker A. R., Tomaselli K. J., Zou H., Fesik S. W., (2004). J. Med. Chem. 47(18): 4417–4426 [DOI] [PubMed]
- Park C. M., Sun C., Olejniczak E. T., Wilson A. E., Meadows R. P., Betz S. F., Elmore S. W., Fesik S. W., (2005). Bioorg. Med. Chem. Lett. 15: 771–775 [DOI] [PubMed]
- Qiu, X.-B. and Goldberg, A. L.: 2005, J. Biol. Chem. 280(1), 174–182 [DOI] [PubMed]
- Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. (2001). Cell 104: 791–800 [DOI] [PubMed]
- Schimmer A. D., Welsh K., Pinilla C., Wang Z., Krajewska M., Bonneau M. J., Pedersen I. M., Kitada S., Scott F. L., Bailly-Maitre B., Glinsky G., Scudiero D., Sausville E., Salvesen G., Nefzi A., Ostresh J. M., Houghten R. A., Reed J. C., (2004). Cancer Cell. 5(1): 25–35 [DOI] [PubMed]
- Sharma S. K., Ramsey T. M., Bair K. W., (2002). Curr. Med. Chem. – Anti-Cancer Agents 2: 311–330 [DOI] [PubMed]
- Sharma, S., Zawel, L., Palermo, M., Chandramouli, N. and Bair, K.: 2004, WO2004005248A1, Novartis AG
- Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y., (2003). Mol. Cell 11: 519–527 [DOI] [PubMed]
- Shiraki K., Sugimoto K., Yamanaka Y., Yamaguchi Y., Saitou Y., Ito K., Yamamoto N., Yamanaka T., Fujikawa K., Murata K., Nakano T., (2003). Int. J. Mol. Med. 12: 705–708 [PubMed]
- Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., Alnemri E. S., (2001). Nature 410: 112–116 [DOI] [PubMed]
- Sun, H., Nikolovska-Coleska, Z., Chen, J., Yang, C.-Y., Tomita, Y., Pan, H., Yoshioka, Y., Krajewski, K., Roller, P. P. and Wang, S.: 2005, Bioorg. Med. Chem. Lett. 15(3), 793–797 [DOI] [PubMed]
- Sun H., Nikolovska-Coleska Z., Yang C. Y., Xu L., Liu M., Tomita Y., Pan H., Yoshioka Y., Krajewski K., Roller P. P., Wang S., (2004a). J. Am. Chem. Soc. 126(51): 16686–16687 [DOI] [PubMed]
- Sun H., Nikolovska-Coleska Z., Yang C. Y., Xu L., Tomita Y., Krajewski K., Roller P. P., Wang S., (2004b). J. Med. Chem. 47(17): 4147–4150 [DOI] [PubMed]
- Tamm I., Kornblau S. M., Segall H., Krajewski S., Welsh K., Kitada S., Scudiero D. A., Tudor G., Qui Y. H., Monks A., Andreeff M., Reed J. C., (2000). Clin. Cancer Res. 6: 1796–1803 [PubMed]
- Tamm I., Richter S., Oltersdorf D., Creutzig U., Harbott J., Scholz F., Karawajew L., Ludwig W. D., Wuchter C. (2004). Clin. Cancer Res. 10(11): 3737–3744 [DOI] [PubMed]
- Tamm I., Wang Y., Sausville E., Scudiero D. A., Vigna N., Oltersdorf T., Reed J. C. (1998). Cancer Res. 58: 5315–5320 [PubMed]
- Vaux D. L., Silke J., (2005). Nature Rev. Mol. Cell Biol. 6: 287–297 [DOI] [PubMed]
- Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L., (2000). Cell 102: 43–53 [DOI] [PubMed]
- Vucic D., Deshayes K., Ackerly H., Pisabarro M. T., Kadkhodayan S., Fairbrother W. J., Dixit V. M., (2002) J. Biol. Chem. 277: 12275–12279 [DOI] [PubMed]
- Vucic, D., Franklin, M. C., Wallweber, H. J. A., Das, K., Eckelman, B. P., Shin, H., Elliott, L. O., Kadkhodayan, S., Deshayes, K., Salvesen, G. S. and Fairbrother, W. J.: 2005, Biochem. J. 385(1), 11–20 [DOI] [PMC free article] [PubMed]
- Vucic D., Stennicke H. R., Pisabarro M. T., Salvesen G. S., Dixit V. M., (2000). Curr. Biol. 10: 1359–1366 [DOI] [PubMed]
- Wu G., Chai J., Suber T. L., Wu J. W., Du C., Wang X., Shi Y., (2000). Nature 408: 1008–1012 [DOI] [PubMed]
- Wu T. Y., Wagner K. W., Bursulaya B., Schultz P. G., Deveraux Q. L. (2003). Chem. Biol. 10: 759–767 [DOI] [PubMed]
- Yang L., Mashima T., Sato S., Mochizuki M., Sakamoto H., Yamori T., Oh-Hara T., Tsuruo T., (2003). Cancer Res. 63: 831–837 [PubMed]


