Abstract
Modulation of intracellular protein–protein interactions has been – and remains – a challenging goal for the discovery and development of small-molecule therapeutic agents. Progress in the pharmacological targeting and understanding at the molecular level of one such interaction that is relevant to cancer drug research, viz. that between the tumour suppressor protein p53 and its negative regulator HDM2, is reviewed here. The first X-ray crystal structure of a complex between a small peptide from the trans-activation domain of p53 and the N-terminal domain of HDM2 was reported almost 10 years ago. The nature of this interaction, which involves just three residue side chains in the p53 peptide ligand and a compact hydrophobic binding pocket in the HDM2 receptor, together with the attractive concept of reactivating the anti-proliferative functions of p53 in tumour cells, has spurned a great deal of effort aimed at finding drug-like antagonists of this interaction. A variety of approaches, including both structure-guided peptidomimetic and de novo design, as well as high through-put screening campaigns, have provided a wealth of leads that might be turned into actual drugs. There is still some way to go as far as optimisation and preclinical development of such leads is concerned, but it is clear already now that antagonists of the p53–HDM2 protein–protein interaction have a good chance of ultimately being successful in providing a new anti-cancer therapy modality, both in monotherapy and to potentiate the effectiveness of existing chemotherapies.
Keywords: Cancer, oncology, protein–protein interaction, drug discovery, structure-based design, peptidomimetics, p53, MDM2, HDM2, p53–HDM2 inhibitors
Introduction
Despite the fact that there are currently several polypeptide drugs in clinical use – and many more under development (Gadek and Nicholas, 2003) – the main thrust of drug discovery is still aimed at small-molecule agents, because these are generally more favourable in terms of biopharmaceutical properties, especially absorption, disposition, stability to metabolism, and lack of immunogenicity. Therapeutic polypeptides such as insulin, erythropoietin, humanised antibodies – to name just a few – all target cell surface receptors. However, most promising drug targets are actually found within – rather than on the external surface or outside – of the cells that make up diseased tissues. These intracellular targets cannot be addressed effectively with polypeptide agents that are generally incapable of traversing biological membranes. For the same reason such compounds cannot usually be administered orally but have to be given parenterally.
There is a long and successful history of the discovery and development of small-molecule agents against intracellular targets, especially enzymes and other receptors or proteins that naturally bind small ligands. The drawback here is that it has been notoriously difficult to find sufficiently potent drug-like leads and drug candidates that exert their pharmacological activity as a result of attenuating other target classes, particularly protein–protein interactions (PPIs). Nevertheless, drug modulation of PPIs is a highly desirable strategy, since it holds much promise, not only because there are presumably many more PPIs than enzyme and classical small-molecule receptor targets, but also in terms of specificity and selectivity of drug action. Unlike e.g. enzymes, which fall into groups of closely related members with highly conserved ligand-binding motifs (e.g. the ATP-binding pocket in protein kinases), PPIs are structurally more diverse and individual PPIs are therefore comparatively unique.
The main perceived difficulty in targeting PPIs with small molecules concerns the size and shape of the interfaces between associating proteins. The buried surface area in PPIs is typically much larger than the solvent-accessible surface area of even the largest potential inhibitor molecules that are still sufficiently membrane-permeable. Furthermore, PPIs tend to be shallower than enzyme active sites and it is therefore conceptually difficult to find shape-complementary small inhibitor molecules that can exclude sufficient water of hydration from a protein surface to form strong hydrophobic interactions, a prerequisite for affinity. New concepts are emerging, however, that suggest that the situation is not completely hopeless. It is now believed that most of the binding energy of PPIs is localised to so-called hot spots, involving just a few amino acid residues in either binding partner (Bogan and Thorn, 1998). These hot spots are not unlike enzyme active sites and might be able to be addressed with drug-like small molecules. Pioneer studies show that this can indeed be the case for certain PPIs (reviewed in Fischer, 2005). Importantly, blocking a hot spot with a small molecule can be sufficient to prevent formation of the much larger overall PPI. Exploitation of adventitious allosteric sites, present in individual members of protein conformational ensembles, is another powerful new concept in PPI inhibitor design (Arkin and Wells, 2004). In this approach small-molecule binding sites are targeted that may not have any natural ligands or physiological significance. If the conformational state of the target protein that is stabilised in this manner does not associate with the partner protein, then an indirect way of blocking a PPI has been achieved.
The need for new disease-associated targets and new therapeutic strategies is nowhere more important than in oncology, where the medical need is urgent and where drug approval productivity has dipped alarmingly over the last few years (Lengauer et al., 2005). Traditional cancer treatments rely heavily on chemotherapeutic agents that target the mechanics of DNA replication. The use of such drugs is limited by the side effects emanating from toxicity to normal proliferating cells. More recently, strategies have been adopted where deregulated components of the signal transduction pathways of cancer cells are targeted, especially protein kinases (Dancey and Sausville, 2003). A few of these agents are now in clinical use, but the validity and overall effectiveness of these newer treatment modalities remain to be established. For this reason alternative strategies are of interest. PPIs are especially important in the cellular processes that govern oncogene and tumour suppressor functions in general, as well as regulation and deregulation of the cell proliferation cycle and the cell death machinery (Oltersdorf et al., 2005). PPI antagonists are therefore of tremendous potential as a molecular strategy for the discovery of new cancer drugs. Here I shall review progress to date on the most advanced PPI drug target in oncology: the interaction between the tumour suppressor protein p53 and its negative regulator, the human double minute-2 (HDM2) oncogene. Several recent reviews on the topic of the p53–HDM2 interaction have been presented (Chene, 2003; Zheleva et al., 2003; Chene, 2004; Fischer and Lane, 2004; Lane and Fischer, 2004; Sunder-Plassmann and Giannis, 2004; Buolamwini et al., 2005; Fotouhi and Graves, 2005); here I shall put emphasis on the most recent work.
The interaction between p53 and HDM2: target rationale
The p53 protein integrates cellular stress signals and functions as a transcription factor with many target genes whose transcripts are involved in DNA repair, cell cycle arrest, and activation of the apoptotic programme. So central is p53 to the life and death decisions of every cell that its functions are ablated in practically all malignant cells (Lane, 2001). This can happen in different ways: typically through direct loss-of-function mutation in the p53 gene in about half of all tumours, or indirectly, frequently by amplification or over-expression of the HDM2 gene (Momand et al., 1998). HDM2 encodes a 491-amino acid residues polypeptide that contains a p53-binding domain, an acidic region, as well as zinc- and ring-finger domains. HDM2 is a p53-specific ubiquitin E3 ligase and thus promotes the proteasomal degradation of p53. Furthermore, it binds to the N-terminal transactivation domain of p53 and therefore blocks the latter’s transcriptional activity. A third mechanism by which HDM2 regulates p53 activity is by promoting the latter’s nuclear export. HDM2 contains a signal sequence that is similar to nuclear export signals of various viral proteins. When bound to HDM2, p53 is thus deactivated by removal from the nucleus, the site of transcription factor activity (Tao and Levine, 1999). There exists a negative feedback loop between HDM2 and p53: following genotoxic stress to normal cells, the ability of p53 to bind to HDM2 is blocked through various post-translational regulatory modifications, thereby preventing HDM2-mediated inactivation and degradation of p53. Consequently, p53 levels rise, causing cell cycle arrest or apoptosis. Over-expression of HDM2 is therefore an efficient way that tumour cells use to prevent accumulation and activation of p53.
It follows that reactivation of p53 in tumours is an attractive therapeutic strategy. Depending on whether or not p53 is functional in a tumour, various strategies can be proposed (Zheleva et al., 2003). If p53 is non-functional, e.g. reintroduction of p53 through gene therapy or pharmacological rescue of mutant p53 could be envisaged (Foster et al., 1999). On the other hand, if p53 is functional in the tumour cells, then inhibiting the ubiquitin ligase activity of HDM2, or blocking the interaction between p53 and HDM2, should be viable. Progress has recently been made in the discovery of HDM2 ligase inhibitors (Lai et al., 2002; Yang et al., 2005) and other ways of interfering with p53-specific HDM2 functions (Issaeva et al., 2004), but here we shall confine our in-depth discussion to inhibition of the p53–HDM2 PPI.
An important question for any new cancer therapy strategy is that of therapeutic margin, i.e. will a drug against the new target be able to distinguish between malignant and normally proliferating cells? It could be argued that attenuation of HDM2 might result in promiscuous toxicity on the basis that MDM2 (mouse double minute 2) knock-out mice are not viable (Montes de Oca Luna et al., 1995). However, gene knock-out is not the same as pharmacological inhibition of the corresponding gene product. Thus mice with a hypomorphic MDM2 allele produce only about 30% of the normal levels of MDM2. Such mice are viable, however, suggesting that attenuation of HDM2 in normal tissues is by no means invariably lethal (Mendrysa et al., 2003). There are clearly important differences between the p53 response in normal versus tumour cells. In normal cells HDM2 levels do not depend on the transcriptional activity of p53, whereas they do in cancer cells. Additionally, in normal cells another tumour suppressor protein, p14Arf, does not control HDM2, whereas in tumour cells p14Arf is involved in the negative regulation of HDM2. One can therefore expect that cancer cells with functional p53 should be selectively sensitive to blockade of the p53–HDM2 interaction, and reacquire the ability to die through p53-mediated apoptosis (O’Leary et al., 2004). The inherent safety of p53 reactivation in cancer cells is implied by several findings, e.g. the recent approval of the first gene therapy in China based on adenoviral delivery of p53 (Surendran, 2004). Furthermore, in vivo studies using HDM2 gene silencing mediated by antisense oligonucleotides, show selective and efficacious anti-tumour effects (Zhang et al., 2005).
Radiotherapy and most current forms of chemotherapy, which damage DNA, also induce the p53 response. The negative regulation of p53 by HDM2 limits the extent of p53 activation and therefore the therapeutic effectiveness of DNA-damaging agents. If the HDM2 feedback inhibition of p53 is interrupted, an increase in functional p53 levels should augment the therapeutic effectiveness of such agents by restoring p53 functions that lead to apoptosis, or by reversing p53-associated drug resistance. Again this expectation has been borne out by studies with HDM2-targeted antisense oligonucleotides, which have been observed to potentiate the effects of both radio and chemotherapy in vitro and in vivo (Bianco et al., 2005).
Structural characterisation of the p53–HDM2 interaction
The region of HDM2 that binds to p53 was originally narrowed down to residues 19–102, whereas the minimal HDM2-binding domain of p53 was identified as a 15 residue sequence (amino acids 15–29) from the transactivation domain. It was realised that the transactivation and HDM2-binding sites of p53 must overlap, since e.g. residues F19 and W23 were important in both respects. An X-ray crystal structure of the complex between HDM2 and the 15-residue transactivation domain peptide of p53 (Kussie et al., 1996) showed that HDM2 possesses a deep hydrophobic cleft into which the p53 peptide binds as an amphipathic α-helix (Fig. 1). Residues F19, W23, and L26 of p53 insert deeply into a compact binding pocket composed of 14 conserved hydrophobic and aromatic amino acids that make multiple van-der-Waals contacts to p53. The p53–HDM2 interface buries a total of 1498 Å2 of surface area (690 Å2 thereof in HDM2), most of which is hydrophobic. There are only two intermolecular hydrogen bonds: one between the F19 backbone amide of p53 and the Q72 side chain of HDM2 at the entrance of the cleft, the other between the p53 W23 indole NH and the HDM2 L54 backbone carbonyl deep inside the cleft. Consistent with p53 peptide structure–activity relationships (SARs) and protein mutagenesis studies, the p53–HDM2 interaction is dominated by only three residues in p53: F19, W23, and L26, which together bury no more than about 500 Å2 of surface area. The p53-binding site of HDM2 thus possesses all the hallmarks of a PPI hot spot (Bogan and Thorn, 1998).
Fig. 1.
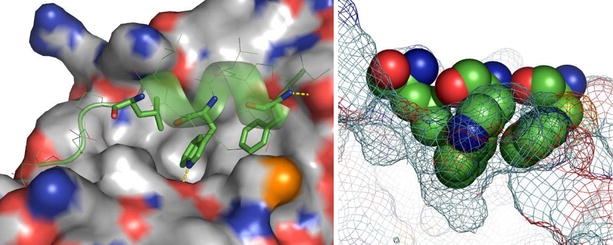
The X-ray crystal structure complex between a p53 peptide (residues 17–29; green CPK sticks and ribbon indicating secondary structure) and HDM2 (grey CPK surface). The three main p53 residue side chains (F19, W23, and L26) involved in the interaction are shown with solid sticks. Hydrogen bonds are indicated by dashed yellow lines (left). The p53-binding hot spot is illustrated on the right, where the three p53 residues are shown as a space-filled model, with the HDM2 surface as a mesh. Constructed from PDB # 1YCR. 3D structural illustrations were created using the programme PyMOL (DeLano, W.L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org).
The HDM2-binding site on p53 was mapped with the aid of overlapping synthetic peptides and the minimal sequence was found to be 18TFSDLW23 (Picksley et al., 1994). Although longer peptides encompassing this sequence were potent inhibitors of p53–HDM2 complex formation, the 18–23 hexapeptide itself had little affinity, presumably due to conformational flexibility (Böttger et al., 1997). Screening of phage-displayed peptide libraries also revealed sequences containing the HDM2 binding motif (Böttger et al., 1996). Here the starting 12mer peptide MPRFMDYWEGLN had sub-micromolar affinity and was 28-fold more potent than the corresponding wild-type p53-derived peptide 16QETFSDLWKLLF27. Substitution and truncation studies revealed that the 8mer peptide FMDYWEGL was the minimal active sequence retaining micromolar affinity for HDM2. Based on the known binding mode of the corresponding p53 sequence (Fig. 1), the helical structure of this peptide was stabilised by introduction of the α,α-disubstituted amino acid residues α-aminoisobutyric acid (Aib) and 1-aminocyclopropanecarboxylic acid (Ac3c) in place of the Asp and Gly residues, respectively. Molecular modelling suggested proximity of the Tyr side chain to the ε-amino group of the HDM2 K94 residue and a phosphonomethylphenylalanine (Pmp) residue was used to replace Tyr. The resulting peptide was about 7-fold more potent, suggesting that the hypothetical stabilising salt bridge between the phosphonate and amino groups was in fact operating. Finally, inspection of the binding pocket for W23 showed incomplete occupancy, suggesting substituents at the indole 6-position would improve binding. This was the case and substantial potency gain was obtained. Thus starting with the wild-type p53 12mer sequence, affinity was increased by >1700-fold and the optimised peptide (Fig. 2) inhibited full-length p53 binding to GST-HDM2 with an IC50 of 5 nM (García-Echeverría et al., 2000). Interestingly, this peptide was found to possess cellular activity, presumably due to its high solubility and extremely high affinity, and despite poor permeability (even lipophilic peptides rarely show high membrane permeability). The peptide was capable of inducing p53 activation and apoptosis selectively in HDM2-overexpressing cancer cells in vitro (Chène et al., 2000, 2002).
Fig. 2.
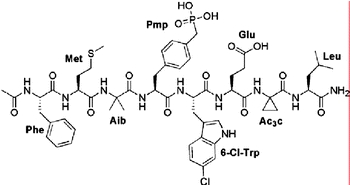
An optimised p53-derived peptide inhibitor of the p53–HDM2 interaction.
The p53–HDM2 interaction has also been studied by NMR (Stoll et al., 2000; Schon et al., 2002, 2004; McCoy et al., 2003; Fry et al., 2004). Overall the HDM2 N-terminal domain constructs that have been used to date in structural studies are comparatively flexible and their solution conformations are stabilised upon ligand binding. The first experimental NMR structure of uncomplexed HDM2 has recently been determined (Uhrinova et al., 2005). This structure is interesting because it sheds light on the likely conformational changes in HDM2 upon p53 binding. It had previously been suggested that the N-terminal 15 or 20 residues of HDM2 occupy the hydrophobic p53-binding cleft in the apo-form (McCoy et al., 2003; Liu et al., 2004; Yin et al., 2005a). It thus appeared that the N-terminus of HDM2 forms an intramolecular lid, which is opened upon p53 binding. The apo-HDM2 structure partially confirms this picture: a part of the N-terminus of HDM2 does indeed occupy one side of the p53-binding site, and major conformational changes in HDM2 upon ligand binding are thus likely (Fig. 3).
Fig. 3.
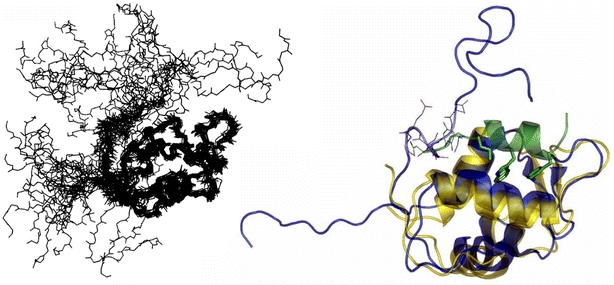
An ensemble of 23 NMR solution structures of HDM2 (Uhrinova et al., 2005), illustrating the flexibility of the termini (left). A comparison between the p53 peptide-bound HDM2 structure (HDM2 yellow, p53 green) and a representative NMR apo-HDM2 conformation (blue) reveals that overall the two are very similar but that the former displays a more open p53-binding cleft (right). Furthermore, in the apo structure a part of the N-terminus (residues 18–24; blue CPK sticks) occupies part of the p53-binding cleft taken up by the p53 residues 27–29 (green CPK sticks) in the complex structure. Constructed from PDB # 1YCR & 1Z1M.
Peptidomimetic inhibitors
β-Peptides
Peptides composed of β-amino acids, i.e. homologues of natural α-amino acids containing an additional backbone CH2 group, are of great interest because of their conformational versatility and in vivo stability compared to natural peptides (Seebach and Matthews, 1997; Seebach et al., 1998b; Cheng et al., 2001). Depending on the substitution patterns at C2 (β2) and C3 (β3) of β-amino acid residues in β-peptides, the latter take up a host of different helical and sheet-like conformations (Cheng et al., 2001). Of particular importance are peptides composed of β3-amino acids, because these can readily be obtained by various synthetic methods, including homologation of natural α-amino acids (Juaristi, 1997; Seebach et al., 1998a). Depending on the stereochemistry of the β-amino acids, either a left-handed or a right-handed 14-helix is formed. Peptides consisting of β3-amino acids that are derived from naturally occurring l-amino acids adopt left-handed 14-helices. The 14-helix is stabilised by hydrogen bonds between backbone amides at position i and carbonyls at i+2, forming a series of intercatenated 14-membered rings (Cheng et al., 2001). Whereas the α-helix has a 3.6-residue repeat, the 14-helix repeats approximately every 3 residues, which positions the side chains of every third residue along one of three faces of the helix. It was shown that the 14-helix, which in many cases is not very stable in water, can be stabilised with oppositely charged residues positioned to form intramolecular salt bridges between residues at the i and i+3 positions along one or two of the three helical faces (Hart et al., 2003).
Such 14-helical β3-peptides have now been used as a peptidomimetic scaffold for the three p53 residue side chains that are responsible for the interaction with HDM2 (Kritzer et al., 2004). As discussed above, naturally these three residues are found on one face of an α-helix at positions i, i+4, and i+7. It turns our that this geometry is very similar in a β3-peptide containing the corresponding β3-residues at positions i, i+3, and i+6 (Kritzer et al., 2005). Peptides of the form 1 (Fig. 4) were prepared and studied in a competitive binding assay based on fluorescence polarisation (Lai et al., 2000; Knight et al., 2002). Only the variant where β3-Xaa, β3-Yaa, and β3-Zaa corresponded to β3-Leu, β3-Trp, and β3-Phe, respectively, was found to inhibit the p53–HDM2 interaction selectively, with an IC50 of about 100 μM, i.e. with an affinity that was about 2.5-fold lower than the native p53(15–31) peptide (Kritzer et al., 2004).
Fig. 4.
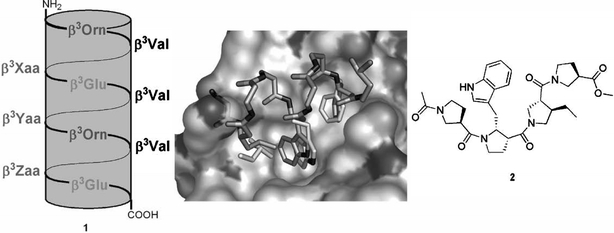
Macrodipole-stabilised 14-helical β3-peptides 1 as p53–HDM2 antagonists. Different combinations of the F19, W23, and L26 side chains were introduced at the positions β3-Xaa, β3-Yaa, and β3-Zaa. The best solution was Leu, Trp, and Phe at Xaa, Yaa, and Zaa, respectively (see text). A model of this peptide, assuming an ideal 14-helical structure, was build (dark sticks; only interacting side chains shown) and overlaid with the known interaction of a p53 peptide (only key side chains shown as light sticks) with HDM2 (surface). The residue preceding ß3-Leu clashes with the H96 imidazole, although it can easily be imagined that the H96 side chain moves to allow access. Constructed from PDB # 1YCR. A hypothetical peptidomimetic of the p53 peptide residue triad based on a β-Pro oligomer (2) has also been proposed.
Oligomers of β-proline, i.e. (S)-pyrrolidine-3-carboxylic acid, and related cyclic β-amino acids, have also been suggested as potential scaffolds for p53 mimetics (Zhong and Carlson, 2005), and generally as a good choice for a short helical scaffold (Huck et al., 2003; Sandvoss and Carlson, 2003). Specifically, compound 2 (Fig. 4) was designed based on known HDM2 inhibitors; as far as can be ascertained this compound has not actually been synthesised and tested, however (Zhong and Carlson, 2005).
As with many polypeptide and peptidomimetic compounds, membrane permeability is a problem and the appeal of the β3-peptide strategy as a therapeutic approach towards p53–HDM2 inhibition may therefore be limited. It should be noted, however, that several vectors are known that can be conjugated with impermeable effector molecules to effect delivery into cells (Fischer et al., 2001). These delivery vectors are peptides themselves and therefore still limited in terms of physiological stability and thus in vivo use. It turns out, however, that such delivery peptides can also be mimicked by β3-peptide analogues (Umezawa et al., 2002). It might therefore be possible to develop entirely β3-peptidic p53–HDM2 inhibitors for in vivo use.
β-Hairpin Peptidomimetics
The β-turn is a structural motif that is frequently used by proteins to display localised groups of amino acid residues for recognition of partner proteins (Ball et al., 1993) and β-hairpin mimetics of the β-turn, including e.g. cyclic peptides incorporating the l-Pro–d-Pro dipeptide unit, have been designed as scaffolds for the synthesis of conformationally constrained peptides and peptidomimetics (Favre et al., 1999; Jiang et al., 2000; Descours et al., 2002; Robinson et al., 2005). The distance between the Cα atoms of F19 and W23 on one face of the HDM2-bound p53 α-helix happens to be close to the distance expected between the Cα atoms of two residues (i and i+2) along one strand of a β-hairpin. The three key p53 residues were therefore mounted on a β-hairpin core in order to mimic the natural spatial arrangement (Fasan et al., 2004). An initial lead compound (3a in Fig. 5) was thus obtained. However, this showed only modest inhibitory activity (IC50=125 μM) in a biosensor assay that employed a streptavidin-immobilised biotinylated p53 peptide (residues 15–29). The HDM2 protein used (residues 17–126) bound to the p53 peptide–sensor surface with a Kd value of 670 nM. Lead optimisation afforded peptide 3b, with almost 1000-fold higher affinity (IC50=140 nM). Based on NMR studies it seemed likely that the weak inhibitory activity of 3a was due to the absence of a stable β-hairpin solution conformation. Peptide SAR studies revealed that the introduction of aromatic side chains (R4 & R5 side chains in 3) stabilised the hairpin conformation and enhanced binding to HDM2. Furthermore, introduction of a 6-chloro group in the Trp indole ring (R2 in 3b) further increased potency, analogously to what had been observed earlier in the context of linear peptides (García-Echeverría et al., 2000). The complex crystal structure between HDM2 and peptide 3b (Fig. 5) illustrates that the side chain of 6-Cl-Trp penetrates deeply into the core of HDM2 and brings the chlorine atom into close contact with the aromatic side chain of F86.
Fig. 5.
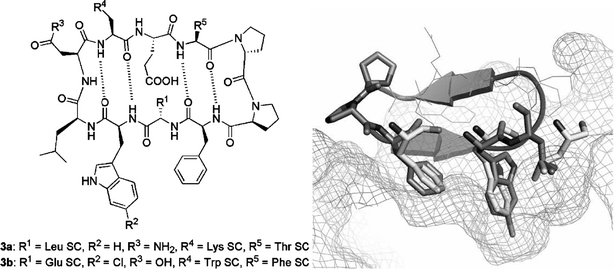
β-Hairpin mimetics of α-helical p53 peptides (left; SC indicates side chain). The X-ray crystal structure of the inhibitor 3b–HDM217-125 complex at 1.4 Å resolution is shown on the right. The Phe, 6-Cl-Trp, and Leu residues of the peptidomimetic are depicted as sticks, the remainder as a secondary structure cartoon only; the corresponding p53 peptide residue side chains (alignment with PDB # 1YCR) are shown as light sticks, and the HDM2 surface is depicted as a mesh. Constructed from a structure co-ordinate file kindly provided by Prof. J.A. Robinson (University of Zürich) ahead of publication.
Chlorofusin
This new fungal metabolite was discovered as a result of a screening programme that involved testing over 53,000 microbial extracts for the presence of inhibitors of the binding of p53 to HDM2 (Duncan et al., 2001). The structure of chlorofusin was subsequently elucidated (Duncan et al., 2001, 2002; Desai et al., 2003; Malkinson et al., 2003): it consists of a 9-residue cyclic peptide that contains both common and unusual l- and d-amino acids; the Orn side chain amino group forms part of a highly functionalised tricyclic chromophore (Fig. 6). It was demonstrated by surface plasmon resonance that chlorofusin binds the N-terminal domain of HDM2 containing residues 1–126, with a Kd of 4.7 μM (Duncan et al., 2003). Because chlorofusin analogues lacking the chromophore do not inhibit the p53–HDM2 interaction (Desai et al., 2003; Malkinson et al., 2003), it is likely that the chromophore, which contains novel fused 1-oxa-6-aza-spiro[4.5]dec-7-en-10-ol and butyric acid 3-chloro-1-methyl-2,6-dioxo-cyclohex-3-enyl ester systems, is involved directly in the recognition of HDM2, perhaps at the p53 F19-W23-L26 site. Apart from the facts that the chromophore is essential for biological activity and that the stereochemistry of the d-Ade-8 residue is important for overall conformation, no SARs are known at present (Desai et al., 2003; Malkinson et al., 2003).
Fig. 6.
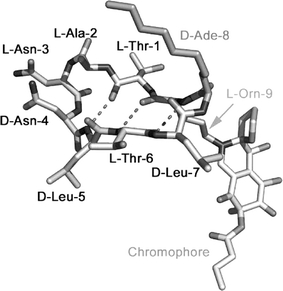
A plausible modelled solution conformation [based on structural studies (Duncan et al., 2001, Malkinson et al., 2003)] of chlorofusin is depicted. Amino acids are labelled, including the 2-aminodecanoyl (Ade) and ornithine (Orn) residues. The side chain of the latter bears the chromophore unit, whose absolute stereochemistry is still unknown.
Terphenyls
Terphenyls exhibit low-energy structures in which the phenyl rings adopt staggered conformations where substituents (e.g. R1 and R2 in 4; Fig. 7) approximate the position and angular orientation of amino acid residues on one face of an α-helix (Orner et al., 2001). Because α-helical secondary structures play an important role in the way that non-contiguous amino acid residue side chains are displayed for PPI hot spots, non-peptidic scaffolds that mimic this structural motif have wide potential application in drug design (Yin and Hamilton, 2005). In fact different terphenyl derivatives have been designed successfully as inhibitors of PPIs where helical conformations are important in one of the interacting partners. Examples include pro-apoptotic antagonists of the BCL-xL–Bak BH3 domain (Yin et al., 2005b) and inhibitors of HIV gp41 assembly and viral fusion (Ernst et al., 2002).
Fig. 7.
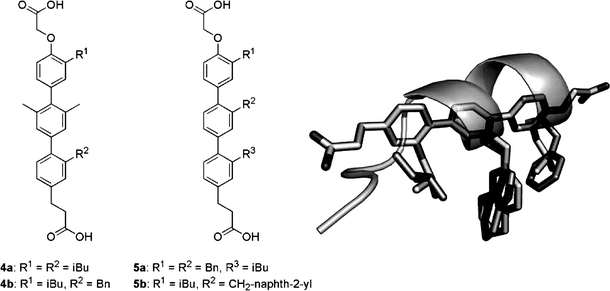
Substituted terphenyls 4 and 5 mimic the α-helical i, i+4, and i+7 side-chain arrangement found for the F19, W23, and L26 p53 residues (dark sticks and ribbon) in the HDM2-bound conformation. A likely conformation of terphenyl 5b (light sticks) was modelled and aligned with the three p53 residue side chains (PDB # 1YCR).
This concept has now also been applied for the design of peptidomimetics of the three key p53 residues implicated in HDM2-binding (Chen et al., 2005; Yin et al., 2005a). A fluorescence polarisation competition assay (Kritzer et al., 2004) with a labelled p53 peptide (C-terminal Cys-fluorescein 15SQETFSDLWKLLPENNV31; Kd=233 nM) was used to determine the binding affinities of the terphenyl derivatives to HDM2 (residues 21–188). Compounds with a bismethylated central phenyl, e.g. 4a and 4b (Fig. 7) showed Ki values of around 2–3 μM. Analogues with aromatic substituents on the central ring were more potent, e.g. for the benzyl and methylnaphth-2-yl congeners 5a and 5b, Ki values of ca. 1 μM and 0.2 μM, respectively, were recorded. Clearly the naphthyl side chain is a comparatively close mimic of the Trp indole system.
The HDM2 binding surface of the terphenyl compounds was mapped using 15N-HSQC NMR experiments. E.g. compound 5b induced chemical shift changes at residues V28, F55, L57, G58, V93, and K94, which, with the exception of V28, line the p53 binding pocket. Similar chemical shift changes were observed in the binding of the p53 peptide to HDM2 (Schon et al., 2004). It therefore seems highly likely that active terphenyls recognise the same HDM2 site as p53.
At present nothing is known about the cellular effects and mode of action for the potent terphenyls 5. Analysis of the earlier compounds 4, on the other hand, has been presented (Chen et al., 2005). These induce nuclear p53 accumulation and transcriptional activation in cell culture at 15–40 μM. Furthermore, anti-proliferative effects were observed in the colon carcinoma cell line HCT116, which harbours wild-type p53, but not in a derivative cell line with targeted deletion of p53 (Bunz et al., 1998).
Tryptophan-Based Peptidomimetics
The first comparatively small peptidomimetic p53–HDM2 antagonists to be reported were Nα-acyl-tryptophanyl-piperazides from AstraZeneca (Luke et al., 1999). These compounds were discovered using a combination of structure-based design and combinatorial chemistry. Compound 6 (Fig. 8), e.g., had a potency of 4 μM in a competitive p53 binding assay (Luke et al., 2000). A related approach has been adopted more recently by workers at Schering–Plough, who developed indole-substituted Nα-(2-phenoxybenzoyl)tryptophan derivatives, again in a combined design and library synthesis and deconvolution strategy (Zhang et al., 2004). Here a high trough-put assay was developed and applied. This was based on fluorescence polarisation anisotropy and used the 2-nM Kd peptide Ac-Phe-Arg-Dpr(fluorophore)-Ac6c-(6-Br-Trp)-Glu-Glu-Leu-NH2 (where the fluorophore was 6-carboxyfluorescein; Dpr, 2,3-diaminopropionic acid; Ac6c, α-aminocyclohexyl carboxylic acid) and HDM2(17–125). A library of (2-phenoxybenzoyl)tryptophan-based HDM2 inhibitors was synthesised and tested in order to explore the SARs of C5/C6/N1 substitutions. The phenoxy moiety was intended to mimic the p53 F19 side chain and the C5/C6-substituted Trp residues were used to mimic W23. The N1 substitutions were intended to target the adjacent p53 L26 pocket in HDM2. Several compounds with low micromolar to sub-micromolar potency were found, e.g. for compounds 7a, 7b, and 7c (Fig. 8) Ki values of 0.4, 0.1, and 0.6 μM were measured, respectively.
Fig. 8.
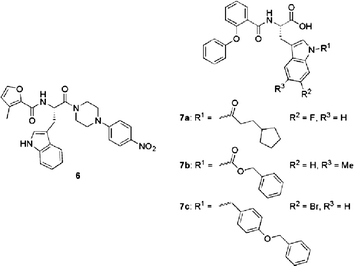
Acyltryptophanylpiperazides and indole-substituted Nα-(2-phenoxybenzoyl)tryptophan peptidomimetic p53–HDM2 inhibitors.
Small molecule inhibitors
Chalcones
Chalcones have long been known as flavonoid precursor compounds with anti-proliferative properties emanating from various biological activities, including disruption of the cell cycle, inhibition of angiogenesis, mitochondrial uncoupling, or induction of apoptosis (Go et al., 2005). Certain chalcones were also found to interfere with the p53–MDM2 interaction (Stoll et al., 2001). Using a two-site ELISA assay based on a biotinylated p53-derived peptide with the sequence MPRFMDYWEDL (Böttger et al., 1996), chalcone carboxylic acids 8a–8d (Fig. 9) were shown to be inhibitors with mid-micromolar potency. Furthermore, an electrophoretic gel mobility shift assay (Böttger et al., 1997) was used to ascertain the ability of test compounds to restore DNA binding of full-length tetrameric p53 following release from the complex with HDM2. Compounds that were active in the competitive binding ELISA had some effect but only compound 8c completely resolved the super-shift induced by HDM2-binding and released active p53. NMR using a recombinant HDM2(1–118) fragment was also used to verify binding affinities and binding modes of the active chalcones (Stoll et al., 2000, 2001). The Kd derived for e.g. chalcone 8d was 250 μM (244 μM by ELISA). The NMR shift perturbation pattern observed suggested that this compound – and other active chalcones – recognised the p53 binding site (Fig. 9), but in a different way to the natural ligand. The chlorophenyl group of 8d binds somewhat less deeply than the p53 W23 indole into the Trp sub-pocket, whereas the F19 and L23 sub-sites are not occupied by 8d. Instead, the anisole ring stacks against F55 (perturbations were also observed at the neighbouring Y56) of HDM2, which brings the carboxylate group into proximity with the K51 side chain amino group. In the p53–HDM2 complex crystal structure (Kussie et al., 1996) the side chain amino group of K51 forms a salt bridge with E25; possibly this salt bridge is broken upon binding of 8d and a new bridge is formed between the chalcone carboxylate and K51.
Fig. 9.
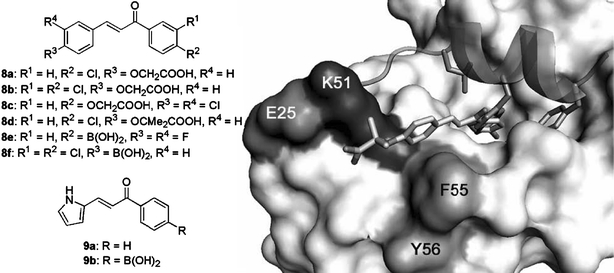
Chalcones 8 and the related pyrrole derivatives 9 inhibit the p53–HDM2 interaction. The modelled binding mode of 8d [based on NMR studies (Stoll et al., 2001)] into the structure of the p53–HDM2 complex (PDB # 1YCR) is shown on the right. The HDM2 surface is shown in light grey, with the exception of the labelled residues K51, E52, and F55 & Y56; the p53 peptide is shown as a ribbon with the F19, W23, and L26 side chains as sticks; chalcone 8d is shown as a stick model.
No cellular activity was discussed in the original report of the p53 antagonist chalcone carboxylic acids (Stoll et al., 2001). Compounds with carboxylic acid functions that are ionised at physiological pH often display poor membrane permeability. It was subsequently suggested that corresponding boronic acid chalcones remain unionised at physiological pH and that they would be capable of forming more favourable interactions with the side chain amino group of K51 in HDM2 (Kumar et al., 2003). However, a comparison between e.g. carboxylic acid 8c and boronic acids 8e and 8f in terms of anti-proliferative potency against a range of breast cancer cell lines, did not reveal an appreciable difference in potency (Kumar et al., 2003). Because no biochemical potency data for the boronic acid derivatives has been reported, this result is difficult to interpret. However, both in the phenyl 8 and pyrrole 9 series (e.g. 9a versus 9b), cytotoxic activity did not apparently depend on the presence of the boronate function. Subsequently it was reported that the cellular mode of action of boronic acid chalcones is consistent with inhibition of the p53–HDM2 complex. Both apoptosis analysis and colony formation assays of p53-isogenic cell lines showed that the p53+/+ cells were more sensitive to the active boronic chalcones than the p53−/−cancer cells (Modzelewska et al., 2004).
Aryl Sulphonamides
A virtual screening approach was taken by workers at Cyclacel for the identification of p53–HDM2 inhibitors (Wang et al., 2004; Zheleva et al., 2004). A high through-put docking programme known as LIDAEUS (Wu et al., 2003) was employed in an in silico screen of small-molecule databases, configured from the co-ordinates of the known p53–HDM2 complex X-ray crystal structure (Kussie et al., 1996). Proposed hits included phenyl thienylsulfonates such as 10 (Fig. 10) and structurally related extended phenyl thienylsulfonamides 11. Verification of hits was accomplished using a competitive binding assay between HDM2 and a fluorescently labelled p53 peptide (Böttger et al., 1996), similar to the assay used by the 3D Pharmaceuticals/J&J group as described below. Lead optimisation was aimed at designing out the potential physiological reactivity of the 2-chloro-3-nitro-thiophene system present in 10, as well as potency optimisation. The former objective was accomplished with analogues such as 12, and potency gains were achieved by introduction of a third aryl group on the sulphonamide N, e.g. compound 13, with target-consistent cellular activity at low micromolar potency.
Fig. 10.
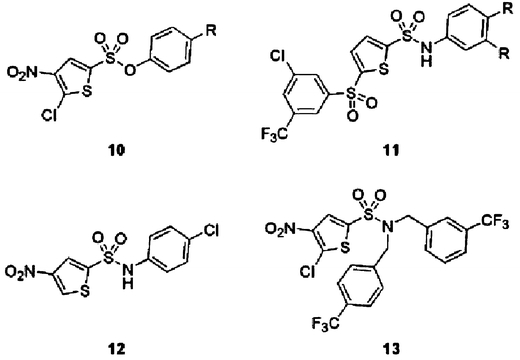
Bisaryl-sulfonate and -sulfonamide p53–HDM2 inhibitors 10–13. The binding modes of the lead compounds suggested from the docking screens were subsequently confirmed using macromolecular NMR methods. It was found that compounds such as 13 did indeed bind the HDM2 site occupied by the natural p53 ligand (Fig. 1).
The arylsulfonamide p53–HDM2 antagonists were found significantly to induce p53 transcriptional activity, as measured in a luciferase-based p53 reporter gene assay, and the p21WAF1 gene product, which is transcriptionally regulated by p53, was induced in cancer cells treated with lead compounds. Furthermore, these compounds potently and selectively killed cancer cells through induction of apoptosis. Interestingly, the effects of the compounds were not limited to cells containing wild-type p53, suggesting that there is another target of HDM2 that is affected. It is known that the regulation of the E2F-1 transcription factor involves HDM2 and that the PPIs of HDM2 with p53 and E2F-1 are probably related not only functionally, but also structurally (Martin et al., 1995; Loughran and La Thangue, 2000; Tortora et al., 2000).
1,4-Benzodiazepine-2,5-diones
1,4-Benzodiazepine-2,5-diones were discovered as p53–HDM2 antagonists using the so-called ThermoFluor® screening method (Pantoliano et al., 2001) developed by 3-Dimensional Pharmaceuticals (now Johnson & Johnson). This entails the use of fluorescent dyes to monitor protein unfolding as a function of temperature, allowing the detection of test compounds binding to target proteins. Fluorescence-based thermal shifts arise as a result of ligand-induced conformational stabilisation of the assayed protein and energetic coupling of ligand binding and protein melting reactions. This method is especially useful to identify small-molecule modulators of proteins for which little functional or structural information is available. Application to HDM2 of this method took the form of a screen of some 338,000 compounds and provided 1216 hits, of which 116 originated from a benzodiazepinedione library (Grasberger et al., 2005). One of these hits, a racemic benzodiazepinedione methyl ester (14a), is shown in Fig. 11. A lead optimisation programme subsequently established SARs, including the importance of stereochemistry (Parks et al., 2005). This provided benzodiazepinedione carboxylic acid compounds such as 14b and 14c, with low to sub-micromolar potency in a conventional p53–HDM2 fluorescence polarisation assay. In this competitive binding assay a p53-derived 9mer peptide [N-terminal fluorescein-RFMDYWEGL; (Böttger et al., 1997)] and an HDM2 fragment (residues 17–125) were used. Similar to a number of other p53–HDM2 inhibitor pharmacophores, the benzodiazepinediones are highly lipophilic (e.g. CLogP for 14c is 5.9). Hit compounds such as 14a were carboxylic acid esters. As the carboxylate group is not directly involved in binding (refer Fig. 11), solubility could be improved by hydrolysis of the synthesis precursor esters. The compounds were prepared using the Ugi four-component condensation reaction (Gokel et al., 1971) between an anthranilic acid (R1 component), an amino acid ester (R3,4 component), an aldehyde (R2 component), and 1-isocyanocyclohexene, followed by acid-catalysed cyclisation (Hulme et al., 1998). Most of the active compounds appear to possess an iodo group at C7. Since aryl iodides can undergo facile in vivo de-iodination (Goodman et al., 1992), alternative substituents are desirable. This seems to be possible and the isopropyl analogue 14e was only marginally less potent than the corresponding iodo compound 14d.
Fig. 11.
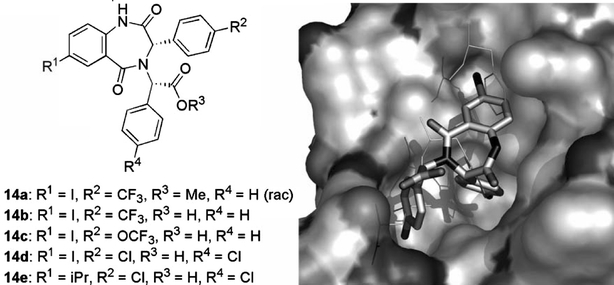
3,4-Dihydro-1H-benzo[e][1,4]diazepine-2,5-diones 14 block p53 binding to HDM2 by occupying the hydrophobic pocket normally recognised by the side chains of p53 residues F19, W23, and L26 (thin sticks) in HDM2-p53 complexes. A view of the binding pose of 14d (solid sticks) to HDM2 (surface) is shown on the right. Constructed from PDB # 1T4E & 1T4F.
Isoindolinones
Compounds 15a and 15b (Fig. 12), amongst others, were discovered by workers at the University of Newcastle and De Novo Pharmaceuticals as mid-micromolar potency p53–HDM2 inhibitors in a biochemical screen, and were subsequently used as seeds in a structure-guided optimisation strategy (Hardcastle et al., 2005). These compounds apparently displayed inhibitory activity in the NCI 60 cell-line screen and were COMPARE negative with respect to any known class of anti-tumour agents, suggesting a novel mechanism of action (Paull et al., 1989).
Fig. 12.
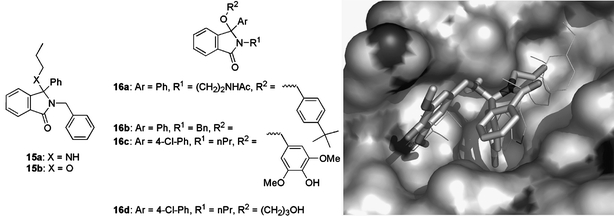
Isoindolinone seed compounds 15 and optimised analogues 16 inhibit the p53–HDM2 interaction. The modelled binding mode, based on the pharmacophore model presented (Hardcastle et al., 2005), of compound 16c (solid sticks) in the p53 peptide (F19, W23, L26 side chains shown as thin sticks) binding pocket of HDM2 (surface, from PDB # 1YCR) is shown on the right.
The compounds were docked into the p53-binding pocket of the published complex structure of HDM2 (Kussie et al., 1996) using a new hybrid optimisation method called quantum stochastic tunnelling, and implemented in a modelling programme known as EasyDock (Mancera et al., 2004). A plausible binding mode was proposed and used as the basis for a subsequent virtual screen of variously substituted hypothetical isoindolinones. The possible binding interactions between ligands and receptor were explored using a simulated annealing optimisation of an empirical free-energy function using the programme Skelgen (Stahl et al., 2002). Analogues able to form at least one additional hydrogen bond with target residues of HDM2 were selected as virtual hits and were synthesised and tested. The binding mode determination was then revisited with the six most active analogues. This showed that it was impossible to distinguish a single preferred binding mode from the many docking solutions. It had previously been shown that the probability of predicting an actual binding mode correctly increases significantly when multiple binding modes are considered (Kallblad et al., 2004). Therefore, several high-scoring, unique binding modes were chosen as starting points for a second round of virtual screening. In order to validate this approach, some 57 hit compounds, including representatives unique to each binding mode, were synthesised and assayed for inhibition of p53–HDM2 binding using a competitive p53-binding ELISA format assay with a luminometric detection end-point. In this assay immobilised streptavidin was complexed with a biotinylated p53-derived peptide [biotin-MPRFMDYWEGLN; (Böttger et al., 1996)]. The peptide Ac-Phe-Met-Aib-Pmp-(6-Cl-Trp)-Glu-Ac3c-Leu-NH2 served as a positive control with an IC50 of 5 nM (García-Echeverría et al., 2000).
This allowed the identification of a number of compounds that displayed improved activity, including congeners 16a–16d with IC50 values of 14, 18, 5.3, and 16 μM, respectively (Fig. 12). The increased potency observed for the 4-chlorophenyl compound 16c was proposed to be consistent with the predicted binding mode for the parent 16b, which binds HDM2 with the phenyl ring occupying the Trp binding pocket, the N-benzylisoindolinone in contact with the broad, shallow, hydrophobic cleft (F19 site), and the phenolic OH of the syringic alcohol making a hydrogen bond to the backbone of Y100 on HDM2. Modelling suggests that this hydrogen bond from he Y100 backbone NH is more likely to involve one of the methoxy groups, however (Fig. 12). It should be noted that all the isoindolinones were isolated and tested as racemic mixtures. Providing the proposed pharmacophore model is correct, one would predict that the (R)-enantiomers are mainly responsible for biological activity.
The most potent compound identified, 16c, was examined for cellular activity. SJSA cells, in which the HDM2 gene is amplified, were treated with the compounds and cell extracts analysed by Western immunoblotting for p53 and p21WAF1. A dose-dependent increase in HDM2 and p21, consistent with p53 activation, was observed.
Nutlins
Of all the small-molecule p53–HDM2 antagonists, the nutlins, named geographically after Hoffmann–La Roche’s research site in Nutley, N.J., USA, where they were discovered, are the most advanced, because in vivo proof of concept has been shown with one of these compounds (Vassilev et al., 2004). Nutlins are piperazin-1-yl-(2,4,5-triphenyl-4,5-dihydro-imidazol-1-yl)-methanones 17 (Fig. 13). Cis-imidazolines were originally identified as leads in a high through-put screening campaign and were subsequently optimised to afford the nutlins. These compounds displaced recombinant p53 protein from its complex with HDM2 with IC50 values in the 100–300 nM range. The imidazoline compounds were synthesised as racemic mixtures. Enantiomers could be resolved and it was found that e.g. with 17c, one enantiomer was 150-fold more potent than the other. Judging from the complex crystal structure of 17b with HDM2, the active enantiomers are those with the 4-(S),5-(R)-imidazoline stereochemistry (Fig. 13; right panel).
Fig. 13.
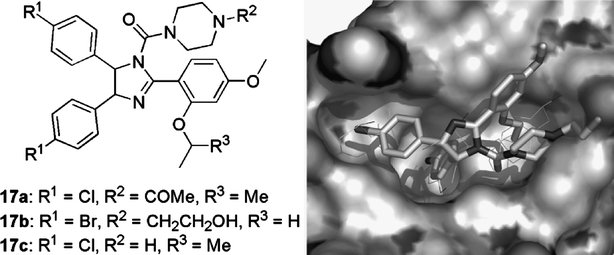
Nutlins 17 bind in the p53 pocket of HDM2. A complex crystal structure of nutlin-2 (17b; solid sticks) and HDM2 (surface) shows that all three sub-pockets for the p53 residues F19, W23, and L26 (thin sticks) are occupied by the aryl substituents of the cis-imidazoline core present in the nutlins. Constructed from PDB # 1RV1.
The HDM2-binding modes of nutlin-2 (17b), determined by X-ray crystallography (Vassilev et al., 2004), and nutlin-3 (17c), determined by NMR (Fry et al., 2004), are similar. In both cases one of the halophenyl rings reaches deeply into the W23 pocket, while the other occupies the shallower L26 site. In both cases the alkyloxy substituent at the anisole ortho position binds into the F19 site, whereas the piperazine solubilising appendages face away from the binding pocket into the solvent (Fig. 13; right panel).
As far as cellular mode of action is concerned, treatment with nutlin-1 (17a) of exponentially growing tumour cell lines with wild-type p53 led to a dose-dependent increase in the levels HDM2, p53, and p21WAF1 proteins, as expected from the release of p53 transcriptional activity. By contrast, cell lines in which p53 is disabled by mutations or deletions, when exposed to the same conditions, showed high basal levels of p53, but no detectable HDM2 or p21. Furthermore, it was shown that accumulation of p53 was due to decreased degradation of the protein rather than elevated expression of the p53 gene. Cell cycle analysis of HDM2-overexpressing cells treated with nutlin-1 revealed increased G1- and G2-phase fractions and nearly complete depletion of the S-phase compartment, consistent with induction of the cell-cycle inhibitor p21. The anti-proliferative potency of nutlins on tumour cell lines could be clearly delineated on the basis of p53 status and the possibility that activation of p53 by nutlins was independent of their effect on HDM2 was excluded: the mechanism by which DNA-damaging agents activate p53 involves phosphorylation of the protein at specific serine residues near the HDM2 binding domain. Of these, S15 is phosphorylated most frequently (Martinez et al., 1997). Analysis of S15 phosphorylation in p53 from lysates of wild-type p53 cells treated with nutlin-1 and two genotoxic drugs, doxorubicin and etoposide, revealed that all three compounds induced accumulation of p53, but only doxorubicin and etoposide caused phosphorylation of S15. These results suggested that a genotoxic mechanism is unlikely to contribute to activation of p53 by nutlin-1. Next it was demonstrated that the anti-proliferative effects of nutlins emanate from induction of apoptosis; co-treatment with caspase inhibitors reduced cell death, showing the involvement of caspase activation. Perhaps the most exciting finding was that human and mouse normal diploid fibroblasts with a functioning p53 pathway were far less responsive to nutlins than transformed cell lines.
This selectivity subsequently translated into an apparent therapeutic margin in a xenograft experiment with the human osteosarcoma cell line SJSA-1, which over-expresses HDM2. Nutlin-3 was well tolerated upon oral administration of 200 mg/kg twice a day for 20 days and achieved plasma levels above in vitro IC90. Treatment of mice bearing established tumours resulted in 90% inhibition of tumour growth relative to a vehicle-treated control group. The treated animals did not lose significant weight and did not show any gross abnormalities. By comparison, doxorubicin administered intravenously at the maximal tolerated dose inhibited the growth of SJSA-1 xenografts by 81%.
Other Small-Molecule p53–HDM2 Inhibitors
A molecular modelling programme known as Hydropathic INTeractions (HINT) was originally used to derive a pharmacophore model that integrated the large body of SAR information from peptide-based p53–HDM2 inhibition studies (Galatin and Abraham, 2001). This model (Fig. 14; left panel) has now been applied as a search query for a 3D screen of the National Cancer Institute (NCI) chemical database (Galatin and Abraham, 2004). Compounds corresponding to high-ranking hits were subsequently obtained and screened in a binding assay based on the p53 peptide 16QETFSDLWKLLP27 (IC50=13 μM). It was found that most of the NCI compounds did not demonstrate a dose-dependent inhibition response, with the exception of the 1,2-diphenyl-3,5-dioxopyrazolidine derivative 18 (Wrzeciono and Szpak-Wojsznis, 1976), for which an IC50 of 32 μM was measured. Furthermore, at high concentrations this compound induced p53 transcriptional activity in a reporter gene assay using the HDM2-overexpressing SJSA-1 osteosarcoma cell line stably transfected with the firefly luciferase gene under a p53-dependent promoter (Galatin and Abraham, 2004).
Fig. 14.
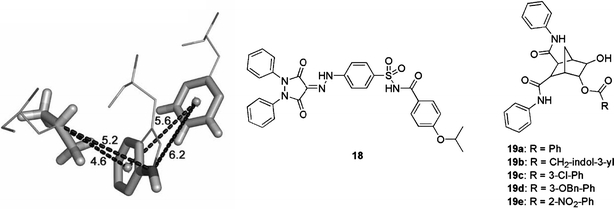
A pharmacophore model for the design of small-molecule p53 mimics (left). This can be used in the reduction of large peptide structures down to small molecules maintaining the proper spatial arrangement of key functional groups (solid sticks). The HDM2-bound (PDB # 1YCR) relative conformations of the p53 residues F19, W23, and L26 are shown. Distance constraints (in Å) derived from this model are indicated. Compound 18 was discovered as an actual p53–HDM2 inhibitor using this model. Compounds 19 show target-consistent anti-proliferative effects in cancer cells.
Finally, the aromatic 6-esters of 5,6-dihydroxy-bicyclo[2.2.1]heptane-2,3-dicarboxylic acid bis-phenylamide (19a–19e; Fig. 14) were reported as inhibitors of the p53–HDM2 interaction (Zhao et al., 2002). Despite modest biochemical potency, e.g. compound 19a was shown to induce apoptotic death of some tumour cells, which express wild-type p53. Similar effects with unidentified analogues from the same chemical series have also very recently been reported (Li et al., 2005).
Conclusions
The interaction between p53 and HDM2 is one of the best-understood PPIs in terms of both structure and inhibitor design. It represents a convincing example of the current hypothesis that blocking PPI hot spots with small molecules is an effective way of modulating protein surface interactions. Several series of drug-like small molecules have been discovered and are currently being optimised. It is hoped that this work will eventually lead to new mechanism-and target-specific cancer drugs.
Abbreviations
- Non-standard abbreviations are defined at the first occurrence. Amino acid and peptide nomenclature conforms to IUPAC-IUB guidelines [Eur. J. Biochem.
138 (1984) 9]
References
- Arkin M. R., Wells J. A., (2004) Nat. Rev. Drug. Discov. 3: 301–317 [DOI] [PubMed]
- Ball J. B., Hughes R. A., Alewood P. F., Andrews P. R., (1993) Tetrahedron 49: 3467–3478
- Bianco R., Ciardiello F., Tortora G., (2005) Curr. Cancer Drug Targets 5:51–56 [DOI] [PubMed]
- Bogan A. A., Thorn K. S., (1998) J. Mol. Biol. 280:1–9 [DOI] [PubMed]
- Böttger A., Böttger V., Garcia-Echeverria C., Chène P., Hochkeppel H. K., Sampson W., Ang K., Howard S. F., Picksley S. M., Lane D. P., (1997) J. Mol. Biol. 269:744–756 [DOI] [PubMed]
- Böttger V., Böttger A., Howard S. F., Picksley S. M., Chene P., Garcia-Echeverria C., Hochkeppel H. K., Lane D. P., (1996) Oncogene 13:2141–2147 [PubMed]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B., (1998) Science 282:1497–1501 [DOI] [PubMed]
- Buolamwini J. K., Addo J., Kamath S., Patil S., Mason D., Ores M., (2005) Curr. Cancer Drug Targets 5:57–68 [DOI] [PubMed]
- Chen L., Yin H., Farooqi B., Sebti S., Hamilton A. D., Chen J., (2005) Mol. Cancer Ther. 4:1019–1025 [DOI] [PubMed]
- Chene P., (2003) Nat. Rev. Cancer 3:102–109 [DOI] [PubMed]
- Chene P., (2004) Mol. Cancer Res. 2:20–28 [PubMed]
- Chène P., Fuchs J., Bohn J., García-Echeverría C., Furet P., Fabbro D., (2000) J. Mol. Biol. 299:245–253 [DOI] [PubMed]
- Chene P., Fuchs J., Carena I., Furet P., Garcia-Echeverria C., (2002) FEBS Lett. 529:293–297 [DOI] [PubMed]
- Cheng R. P., Gellman S. H., DeGrado W. F., (2001) Chem. Rev. 101:3219–3232 [DOI] [PubMed]
- Dancey J., Sausville E. A., (2003) Nat. Rev. Drug. Discov. 2:296–313 [DOI] [PubMed]
- Desai P., Pfeiffer S. S., Boger D. L., (2003) Org. Lett. 5:5047–5050 [DOI] [PubMed]
- Descours A., Moehle K., Renard A., Robinson J. A., (2002) ChemBioChem 3:318–323 [DOI] [PubMed]
- Duncan S. J., Cooper M. A., Williams D. H., (2003) Chem. Commun. 7:316–317 [DOI] [PubMed]
- Duncan S. J., Grueschow S., Williams D. H., McNicholas C., Purewal R., Hajek M., Gerlitz M., Martin S., Wrigley S. K., Moore M., (2001) J. Am. Chem. Soc. 123:554–560 [DOI] [PubMed]
- Duncan S. J., Williams D. H., Ainsworth M., Martin S., Ford R., Wrigley S. K., (2002) Tetrahedron Lett. 43:1075–1078
- Ernst J. T., Kutzki O., Debnath A. K., Jiang S., Lu H., Hamilton A. D., (2002) Angew. Chem. Int. Ed. 41:278–281 [DOI] [PubMed]
- Fasan R., Dias R. L. A., Moehle K., Zerbe O., Vrijbloed J. W., Obrecht D., Robinson J. A., (2004) Angew. Chem. Int. Ed. 43:2109–2112 [DOI] [PubMed]
- Favre M., Moehle K., Jiang L., Pfeiffer B., Robinson J. A., (1999) J. Am. Chem. Soc. 121:2679–2685
- Fischer P. M., (2005) Drug Des. Rev. Online 2:179–207
- Fischer P. M., Krausz E., Lane D. P., (2001) Bioconjugate Chem. 12:825–841 [DOI] [PubMed]
- Fischer P. M., Lane D. P., (2004) Trends Pharmacol. Sci. 25:343–346 [DOI] [PubMed]
- Foster B. A., Coffey H. A., Morin M. J., Rastinejad F., (1999) Science 286:2507–2510 [DOI] [PubMed]
- Fotouhi N., Graves B., (2005) Curr. Topics Med. Chem. 5:159–165 [DOI] [PubMed]
- Fry D. C., Emerson S. D., Palme S., Vu B. T., Liu C.-M., Podlaski F., (2004) J. Biomol. NMR 30:163–173 [DOI] [PubMed]
- Gadek T. R., Nicholas J. B., (2003) Biochem. Pharmacol. 65:1–8 [DOI] [PubMed]
- Galatin P. S., Abraham D. J., (2001) Proteins 45:169–175 [DOI] [PubMed]
- Galatin P. S., Abraham D. J., (2004) J. Med. Chem. 47:4163–4165 [DOI] [PubMed]
- García-Echeverría C., Chène P., Blommers M. J. J., Furet P., (2000) J. Med. Chem. 43:3205–3208 [DOI] [PubMed]
- Go M. L., Wu X., Liu X. L., (2005) Curr. Med. Chem. 12:481–499 [DOI] [PubMed]
- Gokel, G., Luedke, G. and Ugi, I.: 1971 Isonitrile Chem. 145–199
- Goodman, M. M., Kabalka, G. W., Meng, X., Waterhouse, R. N., Knapp, F. F., Jr. and Longford, C. P. D.: 1992 Appl. Isot. Labelled Compd. 1991, Proc. Int. Symp., 4th, 353–358
- Grasberger B. L., Lu T., Schubert C., Parks D. J., Carver T. E., Koblish H. K., Cummings M. D., LaFrance L. V., Milkiewicz K. L., Calvo R. R., Maguire D., Lattanze J., Franks C. F., Zhao S., Ramachandren K., Bylebyl G. R., Zhang M., Manthey C. L., Petrella E. C., Pantoliano M. W., Deckman I. C., Spurlino J. C., Maroney A. C., Tomczuk B. E., Molloy C. J., Bone R. F., (2005) J. Med. Chem. 48:909–912 [DOI] [PubMed]
- Hardcastle I. R., Ahmed S. U., Atkins H., Calvert A. H., Curtin N. J., Farnie G., Golding B. T., Griffin R. J., Guyenne S., Hutton C., Kaellblad P., Kemp S. J., Kitching M. S., Newell D. R., Norbedo S., Northen J. S., Reid R. J., Saravanan K., Willems H. M. G., Lunec J., (2005) Bioorg. Med. Chem. Lett. 15:1515–1520 [DOI] [PubMed]
- Hart S. A., Bahadoor A. B. F., Matthews E. E., Qiu X. J., Schepartz A., (2003) J. Am. Chem. Soc. 125:4022–4023 [DOI] [PubMed]
- Huck B. R., Fisk J. D., Guzei I. A., Carlson H. A., Gellman S. H., (2003) J. Am. Chem. Soc. 125:9035–9037 [DOI] [PubMed]
- Hulme C., Peng J., Tang S.-Y., Burns C. J., Morize I., Labaudiniere R., (1998) J. Org. Chem. 63:8021–8023
- Issaeva N., Bozko P., Enge M., Protopopova M., Verhoef L. G. G. C., Masucci M., Pramanik A., Selivanova G., (2004) Nat. Med. 10:1321–1328 [DOI] [PubMed]
- Jiang L., Mochle K., Dhanapal B., Obrecht D., Robinson J. A., (2000) Helv. Chim. Acta 83:3097–3112
- Juaristi E., (1997). Enantioselective Synthesis of β-Amino Acids, Wiley-VCH, New York
- Kallblad P., Mancera R. L., Todorov N. P., (2004) J. Med. Chem. 47:3334–3337 [DOI] [PubMed]
- Knight S. M. G., Umezawa N., Lee H.-S., Gellman S. H., Kay B. K., (2002) Anal. Biochem. 300:230–236 [DOI] [PubMed]
- Kritzer J. A., Hodsdon M. E., Schepartz A., (2005) J. Am. Chem. Soc. 127:4118–4119 [DOI] [PMC free article] [PubMed]
- Kritzer J. A., Lear J. D., Hodsdon M. E., Schepartz A., (2004) J. Am. Chem. Soc. 126:9468–9469 [DOI] [PubMed]
- Kumar S. K., Hager E., Pettit C., Gurulingappa H., Davidson N. E., Khan S. R., (2003) J. Med. Chem. 46:2813–2815 [DOI] [PubMed]
- Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P., (1996) Science 274:948–953 [DOI] [PubMed]
- Lai Z., Auger K. R., Manubay C. M., Copeland R. A., (2000) Arch. Biochem. Biophys. 381:278–284 [DOI] [PubMed]
- Lai Z., Yang T., Kim Y. B., Sielecki T. M., Diamond M. A., Strack P., Rolfe M., Caligiuri M., Benfield P. A., Auger K. R., Copeland R. A., (2002) Proc. Natl. Acad. Sci. USA 99:14734–14739 [DOI] [PMC free article] [PubMed]
- Lane D., (2001) Nature 414:25–27 [DOI] [PubMed]
- Lane D. P., Fischer P. M., (2004) Nature 427:789–790 [DOI] [PubMed]
- Lengauer C., Diaz L. A., Saha S., (2005) Nat. Rev. Drug. Discov. 4:375–380 [DOI] [PubMed]
- Li W. D., Wang M. J., Ding F., Yin D. L., Liu Z. H., (2005) World J. Gastroenterol. 11:2927–2931 [DOI] [PMC free article] [PubMed]
- Liu Z., Olejniczak E. T., Fesik S. W., (2004) Protein Expr. Purif. 37:493–498 [DOI] [PubMed]
- Loughran O., La Thangue N. B., (2000) Mol. Cell. Biol. 20:2186–2197 [DOI] [PMC free article] [PubMed]
- Luke, R. W. A., Hudson, K., Hayward, C. F., Fielding, C., Cotton, R., Best, R., Giles, M. B., Veldman, M. H., Griffiths, L. A., Jewsbury, P. J., Breeze, A. L., and Embrey, K. J., (1999) Proc. Am. Assoc. Cancer Res. 40, Abs. 4099
- Luke R. W. A., Jewsbury P. J., Cotton R., (2000) Preparation of amino acid and peptidyl piperazine-4-phenyl derivatives as inhibitors of the interaction between MDM2 and p53. PCT Int. Pat. Appl. Publ. WO 2000015657. Zeneca Ltd., UK
- Malkinson J. P., Zloh M., Kadom M., Errington R., Smith P. J., Searcey M., (2003) Org. Lett. 5:5051–5054 [DOI] [PubMed]
- Mancera R. L., Kallblad P., Todorov N. P., (2004) J. Comput. Chem. 25:858–864 [DOI] [PubMed]
- Martin K., Trouche D., Hagemeier C., Sorensen T. S., La Thangue N. B., Kouzarides T., (1995) Nature 375:691–694 [DOI] [PubMed]
- Martinez J. D., Craven M. T., Joseloff E., Milczarek G., Bowden G. T., (1997) Oncogene 14:2511–2520 [DOI] [PubMed]
- McCoy M. A., Gesell J. J., Senior M. M., Wyss D. F., (2003) Proc. Natl. Acad. Sci. USA 100:1645–1648 [DOI] [PMC free article] [PubMed]
- Mendrysa S. M., McElwee M. K., Michalowski J., O’Leary K. A., Young K. M., Perry M. E., (2003) Mol. Cell. Biol. 23:462–472 [DOI] [PMC free article] [PubMed]
- Modzelewska A., Geetha A., Ghosh M., Pettit C., Davidson N. E., Holak T., Huang P., Khan S. R., (2004) Eur. J. Cancer 2(8, Suppl.): 38
- Momand J., Jung D., Wilczynski S., Niland J., (1998) Nucl. Acids Res. 26:3453–3459 [DOI] [PMC free article] [PubMed]
- Montes de Oca Luna R., Wagner D. S., Lozano G., (1995) Nature 378:203–206 [DOI] [PubMed]
- O’Leary K. A., Mendrysa S. M., Vaccaro A., Perry M. E., (2004) Mol. Cell. Biol. 24:186–191 [DOI] [PMC free article] [PubMed]
- Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S. C., Nimmer P. M., O’Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H., (2005) Nature 435:677–681 [DOI] [PubMed]
- Orner B. P., Ernst J. T., Hamilton A. D., (2001) J. Am. Chem. Soc. 123:5382–5383 [DOI] [PubMed]
- Pantoliano M. W., Petrella E. C., Kwasnoski J. D., Lobanov V. S., Myslik J., Graf E., Carver T., Asel E., Springer B. A., Lane P., Salemme F. R., (2001) J. Biomol. Screen. 6:429–440 [DOI] [PubMed]
- Parks D. J., Lafrance L. V., Calvo R. R., Milkiewicz K. L., Gupta V., Lattanze J., Ramachandren K., Carver T. E., Petrella E. C., Cummings M. D., Maguire D., Grasberger B. L., Lu T., (2005) Bioorg. Med. Chem. Lett. 15:765–770 [DOI] [PubMed]
- Paull K. D., Shoemaker R. H., Hodes L., Monks A., Scudiero D. A., Rubinstein L., Plowman J., Boyd M. R. (1989) J. Natl. Cancer Inst. 81:1088–1092 [DOI] [PubMed]
- Picksley S. M., Voijtesek B., Sparks A., Lane D. P., (1994) Oncogene 9:2523–2529 [PubMed]
- Robinson J. A., Shankaramma S. C., Jetter P., Kienzl U., Schwendener R. A., Vrijbloed J. W., Obrecht D., (2005) Bioorg. Med. Chem. 13:2055–2064 [DOI] [PubMed]
- Sandvoss L. M., Carlson H. A., (2003) J. Am. Chem. Soc. 125:15855–15862 [DOI] [PubMed]
- Schon O., Friedler A., Bycroft M., Freund S., Fersht A., (2002) J. Mol. Biol. 323:0022–2836 [DOI] [PubMed]
- Schon O., Friedler A., Freund S., Fersht A. R., (2004) J. Mol. Biol. 336:197–202 [DOI] [PubMed]
- Seebach D., Abele S., Gademann K., Guichard G., Hintermann T., Jaun B., Matthews J. L., Schreiber J. V., Oberer L., Hommel U., Widmer H., (1998a) Helv. Chim. Acta 81:932–982
- Seebach D., Abele S., Schreiber J. V., Martinoni B., Nussbaum A. K., Schild H., Schulz H., Hennecke H., Woessner R., Bitsch F., (1998b) Chimia 52:734–739
- Seebach, D., and Matthews, J. L.: 1997 Chem. Commun. 2015–2022
- Stahl M., Todorov N. P., James T., Mauser H., Boehm H.-J., Dean P. M., (2002) J. Comp. Aided Mol. Des 16:459–478 [DOI] [PubMed]
- Stoll R., Renner C., Hansen S., Palme S., Klein C., Belling A., Zeslawski W., Kamionka M., Rehm T., Muehlhahn P., Schumacher R., Hesse F., Kaluza B., Voelter W., Engh R. A., Holak T. A., (2001) Biochemistry 40:336–344 [DOI] [PubMed]
- Stoll R., Renner C., Muhlhahn P., Hansen S., Schumacher R., Hesse F., Kaluza B., Engh R. A., Voelter W., Holak T. A., (2000) J. Biomol. NMR 17:91–92 [DOI] [PubMed]
- Sunder-Plassmann N., Giannis A., (2004) ChemBioChem 5:1635–1637 [DOI] [PubMed]
- Surendran A., (2004) Nat. Med. 10:9
- Tao W., Levine A. J., (1999) Proc. Natl. Acad. Sci. USA 96:3077–3080 [DOI] [PMC free article] [PubMed]
- Tortora G., Caputo R., Damiano V., Bianco R., Chen J., Agrawal S., Bianco A. R., Ciardiello F., (2000) Int. J. Cancer 88:804–809 [DOI] [PubMed]
- Uhrinova S., Uhrin D., Powers H., Watt K., Zheleva D., Fischer P., McInnes C., Barlow P. N., (2005) J. Mol. Biol. 350:587–598 [DOI] [PubMed]
- Umezawa N., Gelman M. A., Haigis M. C., Raines R. T., Gellman S. H., (2002) J. Am. Chem. Soc. 124:368–369 [DOI] [PubMed]
- Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A., (2004) Science 303:844–848 [DOI] [PubMed]
- Wang S., Gibson D., Duncan K., Bailey K., Thomas M., MacCallum D., Zheleva D., Turner N. J., Fischer P. M., (2004) Bisarylsulfonamide compounds and their use in cancer therapy. PCT Int. Pat. Appl. Publ. WO 2004005278. Cyclacel Limited, UK.
- Wrzeciono U., Szpak-Wojsznis E., (1976) Pharmazie 31:92–94 [PubMed]
- Wu S. Y., McNae I., Kontopidis G., McClue S. J., McInnes C., Stewart K. J., Wang S., Zheleva D. I., Marriage H., Lane D. P., Taylor P., Fischer P. M., Walkinshaw M. D., (2003) Structure 11:399–410 [DOI] [PubMed]
- Yang Y., Ludwig R. L., Jensen J. P., Pierre S. A., Medaglia M. V., Davydov I. V., Safiran Y. J., Oberoi P., Kenten J. H., Phillips A. C., Weissman A. M., Vousden K. H., (2005) Cancer Cell 7:547–559 [DOI] [PubMed]
- Yin H., Hamilton A. D., (2005) Angew. Chem. Int. Ed. 44:4130–4163 [DOI] [PubMed]
- Yin H., Lee G-i., Park H. S., Payne G. A., Rodriguez J. M., Sebti S. M., Hamilton A. D., (2005a) Angew. Chem. Int. Ed. 44:2704–2707 [DOI] [PubMed]
- Yin H., Lee G-i., Sedey K. A., Kutzki O., Park H. S., Orner B. P., Ernst J. T., Wang H.-G., Sebti S. M., Hamilton A. D., (2005b) J. Am. Chem. Soc. 127:10191–10196 [DOI] [PubMed]
- Zhang R., Mayhood T., Lipari P., Wang Y., Durkin J., Syto R., Gesell J., McNemar C., Windsor W., (2004) Anal. Biochem. 331:138–146 [DOI] [PubMed]
- Zhang R., Wang H., Agrawal S., (2005) Curr. Cancer Drug Targets 5:43–49 [DOI] [PubMed]
- Zhao J., Wang M., Chen J., Luo A., Wang X., Wu M., Yin D., Liu Z. (2002) Cancer Lett. 183:69–77 [DOI] [PubMed]
- Zheleva, D., McInnes, C., Baxter, C., Gibson, D., MacCallum, D., Powers, H., Duncan, K., Bailey, K., Cummings, L., Thomas, M., Wang, S., Turner, N., Uhrinova, S., Barlow, P., Taylor, P., Walkinshaw, M., Lane, D. and Fischer, P.: 2004 Proc. Am. Assoc. Cancer Res. 45, Abs. 5552
- Zheleva D. I., Lane D. P., Fischer P. M., (2003) Mini-Rev. Med. Chem. 3:257–270 [DOI] [PubMed]
- Zhong H., Carlson H. A., (2005) Proteins 58:222–234 [DOI] [PubMed]


