Summary
Circadian systems are comprised of multiple proteins functioning together to produce feedback loops driving robust, approximately 24 hour rhythms. In all circadian systems, proteins in these loops are regulated through myriad physically and temporally distinct post-translational modifications (PTMs). To better understand how PTMs impact a circadian oscillator we implemented a proteomics-based approach by combining purification of endogenous FREQUENCY (FRQ) and its interacting partners with quantitative mass spectrometry (MS). We identify and quantify time-of-day specific protein-protein interactions in the clock and show how these provide a platform for temporal and physical separation between the dual roles of FRQ. Additionally, by unambiguously identifying over 75 phosphorylated residues, following their quantitative change over a circadian cycle, and examining the phenotypes of strains that have lost these sites, we demonstrate how spatially and temporally regulated phosphorylation has opposing effects directly on overt circadian rhythms and FRQ stability.
Introduction
Central to the evolution of nearly all life is the persistent influence of environmental light and dark cycles. A common strategy that evolved to anticipate these rhythms is the circadian clock, a dynamic molecular oscillator that integrates multiple signals from the environment. Believed to be common to all circadian clocks described at the molecular level is the integration of time-dependent information in the form of phosphorylation (Gallego and Virshup, 2007; Merrow et al., 2006).
In all fungal and animal circadian systems analyzed to date, conserved PAS-domain containing transcription factors in the positive arm of the feedback loop are directly responsible for driving expression of genes whose products ultimately impinge on their own synthesis (Dunlap, 1999; Young and Kay, 2001). The products of these genes, components of the feedback loop's negative arm, have distinctive similarities in their inhibitory function within the loop and are rhythmically phosphorylated (Gallego and Virshup, 2007; Heintzen and Liu, 2007). Like many proteins, the biology of circadian clock proteins, be it stability, cellular localization, or protein-protein interaction, is likely regulated directly via phosphorylation (Krebs, 1993).
In Neurospora crassa the clock gene frequency (frq) is a central component in the negative arm of the feedback loop and is necessary for maintaining circadian rhythms (Dunlap, 1999). frq promoter is rhythmically bound and expressed by the activity of the positive arm, a heterodimeric transcription factor consisting of WHITE COLLAR-1 and 2 (the white-collar complex, WCC, Belden et al., 2007; Froehlich et al., 2002; He et al., 2006). FRQ undergoes dual molecular rhythms in abundance and phosphorylation, both of which are responsible for establishing the turnover kinetics of FRQ, the primary determinant of circadian period length (Garceau et al., 1997; Liu et al., 2000; Ruoff et al., 2005). The temporal phosphorylation status of FRQ is mediated by several kinases and phosphatases including the Casein kinases 1 and 2 (CK1 and CK2), Checkpoint Kinase 2 (PRD-4), Protein kinase A (PKA), and protein phosphatase 1, 2A, and 4 (PP1, PP2A, and PP4, Brunner and Schafmeier, 2006; Cha et al., 2008; Dunlap, 2006; Huang et al., 2007; Pregueiro et al., 2006). FRQ is targeted for degradation through the activity of the SCF-ubiquitin ligase recruiting protein FWD-1 (Heintzen and Liu, 2007).
FRQ has no known enzymatic activity and its primary function may be to act as a platform for recruiting, possibly in a temporal fashion, other factors involved in the negative feedback process required to sustain rhythms. Most clock components in Neurospora physically associate with FRQ including the WCC, FRQ-Interacting RNA Helicase (FRH), and CK1. Other regulators known or hypothesized to physically interact with FRQ include PRD-4, FWD-1, and CKII (Brunner and Schafmeier, 2006; Dunlap, 2006; Heintzen and Liu, 2007). This plethora of FRQ-interactions and regulatory enzymes has begun to describe a dynamic environment surrounding FRQ and its regulation through phosphorylation.
To fully understand the molecular biology of the circadian clock, it is necessary to explore the extent to which FRQ is phosphorylated and catalog the FRQ interactome. To this end, we first developed a static phosphomap and interactome of endogenous FRQ using high performance tandem MS, and informed by this we employed methods in stable isotope dilution to quantitatively track how steady-state levels of protein interaction and phosphorylation of FRQ change over the course of a day. Together these studies show that FRQ can be phosphorylated at >75 residues, has two distinct regions that are maximally phosphorylated at different phases, and that phosphorylation of these regions leads to opposing effects on stability. Furthermore, we show that the FRQ interactome is dynamic and that most FRQ-protein interactions show phase-specific profiles.
Results
In vivo phosphorylation of FRQ
Leveraging recent improvements in the ability to engineer strains (Larrondo et al., 2009) a tandem V5-epitope 6X-His C-terminal encoding cassette was knocked-in at the frq locus, creating the frqV5H6 allele. Neurospora strains expressing this allele retain wild-type (WT) clock properties (Figure 1A, B). Endogenously produced FRQV5H6 could then be isolated from liquid cultures to near-purity following a simple two-step, non-denaturing purification. Due to the gentle nature of the enrichment protocol, shotgun proteomic analysis of proteins co-purified with FRQ allowed identification of all anticipated Neurospora circadian clock proteins FRH, WC-1, WC-2, CK1 – as well as now providing biochemical evidence for direct, albeit weak, interactions between FRQ and CK2 (cka is the catalytic subunit of CK2, Figure 1C, Table S1).
Figure 1. Affinity-tagged FRQ maintains physiological and molecular rhythms and reveals FRQ interactome and phosphorylation.

(A) Race tube assay of wild-type (WT) and the frqV5H6 strain (τ = period in hours, σ = standard deviation, n = number of race tubes). (B) Western blot of frqV5H6 showing circadian rhythmic abundance and phosphorylation of FRQ over 48h detected with α-V5. Coomassie stained membranes serve as loading controls. (C) FRQ interactome identified by MS/MS after purification for 2 biological replicates. Listed is the number of unique peptides sequenced and their combined percent (%) mass; see Table S1. (D) Schematic of FRQ showing the position of translational start sites, domain structure, position and period length of previously characterized alleles, and newly identified in vivo phosphorylation sites. Regions of FRQ in green were either directly sequenced by MS/MS or do not contain S/T. Seventy-five S/T residues were unambiguously identified as phosphorylated (blue lines) and positions with low confidence are labeled ambiguous (purple lines). Sites marked by a red asterisk (*) are conserved over 120 million years of evolution. CC – coiled-coiled domain, NLS – nuclear localization signal, FCD – FRQ-CKI interacting domain, FFD – FRQ-FRH interacting domain, PEST-# - pest domains, frq# - frq alleles followed by period length.
Protein isolated from frqV5H6 grown in constant light (LL) was used to develop an in vivo phosphomap of FRQ (Figure 1D, Table S2) covering 92% of the FRQ protein. Phosphorylation events at 75 serine and threonine (S/T) residues were unambiguously identified along with between 7 and 18 additional ambiguous sites. Ambiguous sites are found in complex S/T peptides containing phosphorylated residues for which the spectra identifying the precise acceptor S or T is not assigned with high confidence (Table S2).
Previous analyses of FRQ have allowed biological functions to be assigned for many regions within the 92% of FRQ examined. These include the coiled-coil domain for forming FRQ dimers (CC), a nuclear localization signal (NLS), the FRQ-CKI interaction domain (FCD), FRQ-FRH interaction domain (FFD), and the PEST-1 domain important for FRQ turnover (Dunlap, 2006; Heintzen and Liu, 2007). Of these domains, only the PEST-1 is phosphorylated even though peptides corresponding to all were routinely sequenced, demonstrating that phosphorylation sites on FRQ are not evenly distributed over the protein but appear clustered. Moreover, despite the large size of FRQ, phosphorylation is absent in only a few extended regions of unknown function (i.e. ∼287-383). Thus FRQ comprises a highly heterogeneous population of structures seeming to have mostly exposed, kinase-accessible, residues.
Development of Neurospora crassa for quantitative proteomics
The discrete mobility shifts observed for FRQ on Western blots (Figure 1B, Garceau et al., 1997) indicated that time-dependent phosphorylation of FRQ might integrate phase-specific information. To address this we adapted Neurospora to the quantitative MS method SILAC (Ong et al., 2002). SILAC utilizes the in vivo incorporation of amino acids labeled with stable heavy isotopes of 13C and 15N into endogenously translated protein to allow determination of relative protein abundance among two or more biological states.
For metabolic labeling, an auxotrophic Neurospora strain was engineered with knock-out (KO) alleles in lysine and arginine biosynthesis (lys-4 and arg-12, respectively), and arginase (aga), an enzyme in the Arg catabolism pathway (Perkins et al., 2001). Using this strain the quantitative nature of SILAC in Neurospora is demonstrated by mixing lysate prepared from heavy and light Arg and Lys supplemented cultures in known ratios and analyzing a sample by MS (Figure S1A and B). Additionally, we observe nearly 100% incorporation of heavy Lys and Arg (Figure S1C). The Δaga allele was included to prevent the Arg-to-Pro metabolic conversion reported in Saccharomyces cerevisiae (Gruhler et al., 2005) and mammalian tissue culture (Bendall et al., 2008) which can reduce the accuracy of downstream quantification (Figure S1D). Both WT frq and frqV5H6 in the SILAC background display robust circadian rhythms with slightly increased periods (23.7±0.2 and 24.1±0.2 hours respectively) due to incorporation of the Δlys-4 locus (data not shown). These strains also have a delayed phase of conidiation that correlates with a delayed rhythm in FRQ abundance and turnover (Figure S2). For the purposes of this work using SILAC, growth times are reported in hours of constant dark (DD) corresponding to circadian times (CT) where CT0 is defined as subjective dawn. To compare the late phase of conidiation and molecular rhythmicity of FRQV5H6 to WT, we adjust the subjective time of the SILAC strain by aligning the of the peak of FRQ rhythmicity.
Quantitative analysis of FRQ-interacting proteins
A pooled reference sample approach (Eisen et al., 1998) was used to test whether protein interaction and FRQ phosphorylation in the Neurospora clock are time-of-day specific (Figure 2A). Six isotopically heavy and six isotopically light amino acid supplemented cultures were inoculated and grown in LL at 25°C. These cultures were transferred to DD to initiate rhythmicity, and whole-cell protein lysates prepared every 4h. A pooled reference sample was prepared by mixing all isotopically light lysates together and therefore contained all time-dependent FRQ isoforms. Each time specific heavy labeled lysate was mixed 1:1 with the pooled reference prior to FRQ purification. A heavy-to-light (H/L) ratio representing the relative change in the abundance of any given peptide is calculated from the MS1 spectra by comparing the abundance of the pooled reference peptide to the heavy time-point specific peptide (Figure 2B top) and sequenced in the MS2. For each protein, relative changes in total abundance at a given time are calculated by averaging log2 (H/L) for all sequenced peptides. For example, FRQ abundance increases by nearly 2-fold when compared to the reference at DD24 (early subjective night, Figure 2B bottom). To correct for mixing errors, a random sample of >1250 peptides from proteins in the 1:1 mixture, prior to FRQ purification, were measured for each time (Figure 2B, Table S3). In total, an average of 103 ± 15 FRQ, 171 ± 21 FRH, 24 ± 8 WC-1, 10 ± 3 WC-2, and 39 ± 3 CK1 individual peptides were surveyed for each DD time (Table S4). Figure 2C reports how the relative levels of these proteins change over the course of a day. Since FRH, WC-1, WC-2 and CK1 were co-purified through interaction with FRQ, their values were normalized by subtracting the FRQ value at the same DD (Figure 2C right). In LL, all FRQ interacts with FRH to form the FRQ-FRH complex (FFC, Cheng et al., 2005). These data show that the H/L ratios of FRH mirror FRQ, confirming continuous strong interaction throughout the entire day. In contrast, while both WC-1 and WC-2 ratios track each other, together they interact with FRQ largely in the early part of the circadian day and this interaction is severely reduced later, even though total FRQ abundance increases. The dynamic nature of this interaction suggests WCC preferentially interacts with hypophosphorylated FRQ, while FRH interacts with most FRQ phosphoisoforms (Figure 2C, Figure S3, and Cheng et al., 2001). CK1's steady state interaction with FRQ is stronger than any other kinase identified, based on total number of peptides sequenced. CK1 is consistently associated with FRQ and this interaction appears cyclic with > 4-fold ratios in the early subjective morning/day at DD16 (CT4, when the WCC-FFC interaction is declining) and a minimum at DD24-28 (CT12-16, early subjective night). Similar to results from LL cultures, peptides corresponding to both the alpha (CKA) and beta I (CKB-1) subunits of CK2 were detected (Figure S3C and data not shown). However, because few peptide pairs were quantified, precise determination of their abundance in relation to FRQ was difficult.
Figure 2. Dynamic changes in the FRQ-interactome provide a temporal framework for regulation of the WCC.
(A) Schematic of the pooled reference sample design for the complete circadian day SILAC experiment (see text). Proteins were purified via FRQ and split to separately follow the interactome and FRQ phosphorylation. (B) Top – Representative MS spectra for doubly charged heavy and light FRQ peptides (387-DNGSASNSGGDQTELGGTGTGSGDGSGSGGR-417) show an observed m/z difference of 5.01 (6-13C and 4-15N in Arg) and a H/L ratio of ∼2. Bottom – Distribution of log2 (H/L) for all sequenced FRQ and a sample of the 1:1 mix peptide pairs at CT12 (n = number of H/L ratios measured, μ = mean log2 (H/L), σ = SD). (C) The FFC interacts with the WCC primarily in the early circadian day, phase-leading the interaction with CK1. Left – Mean log2 (H/L) of all peptides sequenced for FRQ (diamond at CT12 = μFRQ-μ1:1 from B), FRH, WC-1, WC-2, and CK1 at each time (± SD). Right – Same data in left panel normalized to FRQ. That levels of FRH closely track FRQ is seen by the near flat FRH line. Note: CT, circadian time, is calculated here sensu stricto, but period and phase differences arising from use of the SILAC strain means that the circadian biochemistry seen at CT4 (DD16) more closely matches that typically seen in the late subjective night in WT cultures (Figure S2).
Due to the complex nature of the full time-course experiment and the changing FRQ-interactome, the SILAC experiment was repeated with 3 independent biological samples using a two-state experimental design (Figure S4A). Cultures harvested at DD16 (when FRQ is at its minimum in this strain) were compared to cultures harvested at DD24 (when FRQ is at its maximum). While some variability exists in the amplitude of changes in protein level between the two experimental designs (see Supplemental Data), these experiments confirm the dynamic FRQ-interactome for all clock proteins: FRQ and FRH ratios increase at a similar rate, while the normalized amount of WCC and CKI ratios relative to FRQ decrease (Figure S4B).
Quantitative Analysis of FRQ phosphorylation
To better understand the time-dependent phosphorylation of FRQ, phosphopeptides from 90% of the isotopically-labeled purified FRQ were enriched on titania microspheres (Larsen et al., 2005; Pinkse et al., 2004) and analyzed by high performance tandem MS (Table S5). Relative levels of phosphorylation are calculated as described for the interactome for each individual phosphopeptide. To facilitate visualization of the large phosphopeptide data set individual ratios were log2 transformed and plotted graphically to represent changes over time (Figure 3). This reveals the relative under- or over-representation of phosphorylation at a specific FRQ residue. It is clear that many variables are changing simultaneously across this time series, including both the amount of FRQ as well as its phosphorylation profile, and by simply following H/L ratios, phosphorylation of individual sites merely appear as increasing across the day (Figure 3A). To account for some of this change, the H/L ratios were normalized to the average level of phosphorylated FRQ peptides at that time (Figure 3B). In this way, regions of FRQ that are qualitatively hyper- (yellow) or hypophosphorylated (blue) at specific times could be identified. This time-dependent phosphomap of FRQ reveals a number of facets that potentially impact the activities of this central clock protein.
Figure 3. Topological and phase specific changes in FRQ phosphorylation.
(A) Dual molecular rhythms in FRQ abundance and phosphorylation. Top – Graph showing change in total (i.e. Figure 3C) and phosphorylated FRQ (calculated by averaging all log2 (H/L) phosphopeptides). Early in the subjective day FRQ abundance and phosphorylation increase, while later phosphorylation plateaus as FRQ levels start to decline. Bottom – Heat map of individual FRQ phosphopeptides (rows) throughout the circadian day (CT in columns). (B) FRQ undergoes phase-specific phosphorylation in discrete domains. Left – Schematic of FRQ showing the position of phosphorylated residues identified in the full time course experiment. Right – Heat map representing the relative change in abundance of 63 phosphopeptides normalized to the average level of FRQ phosphorylation. Values tracking the mean appear black, while hyper- and hypophosphorylated residues are yellow and blue respectively. Numbers on the right of the heat map indicate the specific FRQ residue(s) phosphorylated for both (A) and (B).
First, locations of nearly all of the sites seen in LL are also seen in DD and fall into distinct clusters (Figure 3B). Second, there are conserved and dynamically phosphorylated sites in the N-terminal region that is specific to the long form of FRQ whose synthesis is enriched at higher temperatures. Third, only a few extended regions of the protein are not phosphorylated. Fourth, phosphorylation appears quickly across the protein and clusters of sites appear to change in a semi-coordinated manner. For example, a cluster of S/T residues near the C-terminus (795–929) that surrounds the PEST-2 region becomes hyperphosphorylated early in the cycle (by CT8) when FRQ levels are increasing. Conversely, residues in the PEST-1 domain (537–558) show a pronounced increase in phosphorylation later, maximally around CT12 when FRQ is at its peak and starts to decrease in abundance (compare to Figure 3A top). Some sites (e.g. 404-462 and 923–950) show little change, reflecting, perhaps, “recreational” phosphorylation having little or no phase-specific function (see Discussion). The relative abundance of phosphopeptides in some regions (e.g. 211–257) decreases at later times. This could either result from dephosphorylation reflecting a phase-specific phosphatase activity, or continuing de novo synthesis of FRQ bypassing early phosphorylation events through forming homodimers with advanced-phase FRQ.
In addition to sites on FRQ several phosphopeptides were identified for other clock proteins (Figure S3C). Most of these sites were detected in few time-points. However, phosphorylation of S433 on WC-2 and T108 on FRH were consistently detected. While phosphorylated T108 on FRH changes at the same rate as the entire protein, the level of phosphorylation on S433 on WC-2 shows some time-dependence compared to total abundance of WCC (Figure S3D and E). Whether these sites have functional significance is yet to be determined.
Functional analysis of FRQ phosphorylation
Such diversity among the temporal changes in FRQ phosphorylation suggested functional significance of specific and/or clusters of sites. To examine this, a knock-in construct was designed to replace the Δfrq locus with the entire frqV5H6 sequence, effectively restoring frq into its endogenous locus and rescuing rhythmicity (Figure 4A). Using frqKI, a library of 51 site-directed point mutant cassettes was created changing 70 phosphorylation sites from S/T→A to prevent phosphorylation. Mutant strains were assayed for free running period (FRP) on race tubes and/or by following frq-promoter driven luciferase (Table S6).
Figure 4. Phosphorylation of FRQ leads to opposing effects on circadian rhythms and protein half-life.

(A) A frq knock-in cassette (frqKI) can restore rhythmicity to Δfrq. (B) Ablation of key phosphorylation sites in the PEST-1 domain results in lengthened periods. Representative luciferase traces show site-specific increases in period length of mutants. (C) Similar to (B) except these strains carry mutations in the early-phase phosphorylated C-terminal region of FRQ and result in decreased periods. (D) Summary of race tube (rt) and luciferase (luc) data showing the change (Δ) in period compared to a WT transformed control for the indicated phosphorylation site mutants. (E) Left – Representative Western blots of FRQ degradation after an LD transition for frqS900A, frqS548A, and frqKI probed with α-V5. Right – Densitometric analysis of (E) showing exponential fit used for FRQ half-life calculation. (F) Plot of FRQ half-life vs. period length. FRQ half-life correlates with period length where shorter half-lives result in shorter periods. The black line represents the predicted correlation at 25°C based on Ruoff et al. (2005). Error bars cover the range of two independent estimates of FRQ half-life and the standard deviation for period.
The salient model regarding FRQ modification is that phosphorylation leads to degradation, and a prediction of that model is that removal of phosphorylation sites should result in period increases (Liu et al., 2000; Ruoff et al., 2005). While none of the phosphorylation site mutants characterized displayed an absolute loss of rhythmicity, period increases are the most common phenotypic class (Table S6). The largest period increases were recorded when residues within the PEST-1 domain were altered (Figure 4B). For instance, mutation of S538 and S540 or S548, each of which becomes highly phosphorylated around DD24, results in a dramatic period lengthening between 3 and 5 hours (Figure 4B, D). However, strains bearing mutation of S558, a residue preferentially phosphorylated earlier, display WT periods (Table S6).
Period lengthening was not the only phenotype observed. A region of FRQ that generally increases in relative phosphorylation at an earlier phase is directly C-terminal to the PEST-2 domain (Figure 3B). Mutant alleles in this region yield only period decreases (Figure 4C). These data indicate that some FRQ phosphorylation may increase stability. To test this, we examined the steady-state degradation rate of FRQ following transfer from LL to DD (Figure 4E and 4F). Consistent with this interpretation, a clear decrease in protein stability is seen in the short period S900A mutant while stability of FRQ is increased in the long period S548A mutant. These data demonstrate the context-dependent nature of FRQ phosphorylation by showing an early-phase phosphorylated region can increase period length in part by stabilizing FRQ, while later phosphorylation of a physically separate region can decrease period length by decreasing FRQ half-life.
Appropriate timing of phosphorylation is critical for period maintenance
To further test the mechanism behind regulated protein turnover, several key phosphorylation sites in the PEST-1 domain were mutated to aspartate (D) to mimic the negative charge of phosphorylation (Figure 5). Mutation of S548 to D restored period length to WT (Figure 5B). This indicates that mutation per se is not stabilizing FRQ but rather altered surface charge at this residue is necessary, but not sufficient, for protein turnover. S548D alone might not drive precocious degradation because either the PEST-1 domain requires an accumulation of multiple charges prior to degradation and/or this region might remain inaccessible to the degradation machinery until later in the cycle. To test this, two new strains were created combining mutation of 5 phosphorylated sites in the PEST-1 domain to either Ala or Asp (frq5XS→A and frq5XS→D, Figure 5A). Both strains exhibited a loss of overt rhythmicity (Figure 5B) while the frq5XS→A has a long molecular rhythm (Figure 5C), results similar to deletion of the entire region (Gorl et al., 2001). While these strains express similar levels of FRQ in LL, cultures with the frq5XS→D allele had significantly reduced FRQ abundance at DD8 compared to frq5XS→A (p = 0.002, unpaired t-test) and slightly reduced levels compared to WT (p = 0.086, Figure 5D). These results separate the process of FRQ degradation from periodicity and indicate that coordinated timing of phosphorylation, rather than static charge, is required to maintain rhythms.
Figure 5. Multi-site phosphorylation of PEST-1 is necessary for regulated FRQ turnover.

(A) Amino acid sequence of PEST-1 domain indicating position of 5 in vivo phosphorylation sites in red. (B) Race tube analysis of strains bearing various PEST-1 mutations. The phosphomimetic mutation (S548D) maintains a WT period; while strains bearing multiple proximal mutations in the PEST-1 domain, with all sites in (A) changed to either Ala (frq5XS→A) or Asp (frq5XS→D), abolish overt circadian rhythms. (C) Luciferase assays of strains from (B) show a low amplitude molecular rhythm in frq5XS→A. (D) Representative Western blot of frqKI and PEST-1 mutants grown in constant light (LL) or after transfer to the dark for 8h (DD8). The arrow indicates slower migrating forms of FRQ that are absent from frq5XS→A. Bottom - Densitometric analysis for 3 biological replicates (± SEM).
To test whether the C-terminus is required to promote protein stabilization, a strain was created in which the entire region encompassing all phosphorylation sites leading to short periods was deleted (frqΔC-term, Figure 6A). This strain still expresses FRQ and accumulates extra hypophosphorylated species (Figure 6B). Race tube analyses of strains expressing frqΔC-term are either arrhythmic or have severely dampened rhythms (data not shown); however, luciferase data show persistent low-amplitude short periods. Therefore, the presence of the domain stabilizes FRQ, and suggests that this stabilizing function is regulated through phase-specific phosphorylation. Finally, to test whether distal regions of FRQ have an epistatic or additive effect, the two point mutations S900A and S548A were combined intramolecularly. Early phosphorylation stabilizes FRQ, strains lacking these sites degrade faster presumably through quicker PEST-1 phosphorylation. Therefore, if later PEST-1 phosphorylation events were blocked in the context of a C-terminal mutant, the combination would be predicted to have an intermediate period, exactly what is seen in strains expressing frqS900A & S548A (Figure 6D).
Figure 6. The C-terminus of FRQ is physically required for wild-type period.

(A) Schematic of FRQ showing deleted region in frqΔC-term. This area encompasses all FRQ-stabilizing phosphorylation sites. (B) Western blot shows stable expression of FRQΔC-term in LL. Arrowhead indicates hypophosphorylated FRQ. (C) Representative luciferase traces showing short a molecular rhythm in frqΔC-term (n= number of individual measurements). (D) Mutation of distal phosphorylation sites on FRQ show an additive effect of phase-specific phosphorylation. Race tube analysis of frqS900A and the multiple mutant frqS548A & S900A.
Discussion
Circadian clocks are finely tuned cellular pacemakers found across a wide variety of phylogenetically disparate organisms. In the well-characterized fungal and animal systems, including mPER2 in mammals, Drosophila PER, and Neurospora FRQ (Edery et al., 1994; Garceau et al., 1997; Lee et al., 2001), the negative arm of the feedback loop is regulated by multi-site phosphorylation. Prior investigations of protein phosphorylation critical to the mechanism of the clock have resulted in detailed yet static descriptions of modification. Like these, the present study began with the goal of identifying all phosphorylation sites on FRQ and extended this by tracking temporal changes in phosphorylation and protein interaction.
Modification of FRQ is extensive; in results qualitatively similar to recent cell-culture based studies that identified 21 phosphorylation sites on mPER2 (Vanselow et al., 2006) and up to 24 sites on Drosophila PER (Chiu et al., 2008; Kivimae et al., 2008) we also found FRQ to be highly phosphorylated with over 75 distinct confirmed sites. A caveat is that only 92% of FRQ was surveyed by tandem MS. In particular, the inability to recover residues 856-893 within the PEST-2 region, which can be phosphorylated in vitro by CK1 (Gorl et al., 2001), means that even the 75 sites identified here are an underestimate. Modifications are complex; at almost any time “FRQ” describes a heterogeneous mix of proteins with variable and distinguishable structure and surface chemistry. For example, frq undergoes temperature and circadian dependent splicing resulting in two protein isoforms; termed long- and short-FRQ (L- and S-FRQ respectively). Strains that produce S-FRQ slightly increase period while L-FRQ only strains slightly decrease period (Diernfellner et al., 2007). Here we identified several phosphorylation sites that are specific to L-FRQ. Of these, only mutation at S72→S76 increase period (Table S6), similar to S-FRQ only strains, even though both isoforms are present. In total, mutational analyses revealed that for over 60% of the confirmed phosphorylation sites, loss of single or neighboring sites have no apparent effect on the FRP. This suggests that sites may be regulated as charged domains rather than functioning individually, or contribute to other regulatory clock-mechanisms (i.e. temperature compensation, Mehra et al., 2009), although the possibility of nonfunctional “recreational” phosphorylation cannot be ruled out.
Often, putative phosphorylation targets are pursued from a candidate approach based on identifying kinase consensus targeting sites. Although precedents for this can be found within the published work in most circadian systems, an illustrative case is from our prior work (Liu et al., 2000). Deletion of a short central region of FRQ and introduction of T/S→A mutations at conserved kinase sites within this region reduces FRQ phosphorylation and increases period length. However, in our MS analysis of 15 independent samples not only were the putative sites previously characterized not seen to be phosphorylated, we failed to identify any phosphorylation events in the entire region. Absence of evidence is not evidence of absence: it may be that phosphorylation of these sites is extremely rare or destabilizes the protein to such an extent that the sites can thereafter never be seen. However, these residues lie in the midst of the FRQ-CKI interaction domain, and deletion or mutation of this region may simply reduce FRQ phosphorylation by blocking the ability of FRQ to interact with its primary kinase (He et al., 2006).
The discussion to this point has focused on what can be deduced from the static phosphorylation map, but a major aspect of our work is the description of the dynamic changes in FRQ phosphorylation and protein interaction. Indeed, one surprise was the extent and timing of phosphorylation: FRQ is extensively modified soon after synthesis, and only a few extended regions remain unmodified over its lifetime. Although the largest unmodified region is only 128 amino acids in length, the CC, FFD, FCD, and the NLS all remain unmodified, and the progress of modification of the other regions of FRQ can now be viewed in the context of our interactome data (Figure 7). FRQ is always associated with FRH perhaps explaining the absence of modifications in the FFD. Lightly phosphorylated nascent FRQ, together with FRH, rapidly associates with the WCC. This is perhaps consistent with the timing of FRQ's negative effect on WCC activity either through its phosphorylation or by promoting its clearance from the nucleus (Cha et al., 2008; Hong et al., 2008; Schafmeier et al., 2008); FRQ-WCC interactions decline thereafter, a relationship inverse to FRQ's increasing phosphorylation. FRQ recruits both CK1 and CK2 to phosphorylate and inactivate the WCC (e.g. He et al., 2006). Although we identified both subunits of CK2 in our interactome study, the small number of peptides sequenced may indicate that the interaction is transient, possibly more akin to an enzymatic relationship. Instead, we found a robust interaction between FRQ and CK1 with 55% of the predicted CK1 sites phosphorylated in vivo, and most strains mutated in these sites having period-lengthening phenotypes (Table S6). FRQ-CK1 interactions are highly regulated, increasing over 4-fold from the time of FRQ synthesis. The peak interaction between FRQ and CK1 occurs after frq expression and FRQ-WCC interaction has begun to decline. That the FRQ-WCC association decreases as the FRQ-CK1 interaction increases may suggest one role of CK1 is to modulate the FFC-WCC interaction. Late phosphorylation of FRQ has been shown to promote the accumulation the WCC (Schafmeier et al., 2006), but we found no evidence for increased association between highly phosphorylated FRQ and the WCC. While this does not rule out FRQ-dependent stabilization of the WCC, perhaps this stabilization is due to decreased interaction. However, these later interactions may be transient, as we also did not see any association between FRQ and its SCF-complex targeting protein, FWD-1.
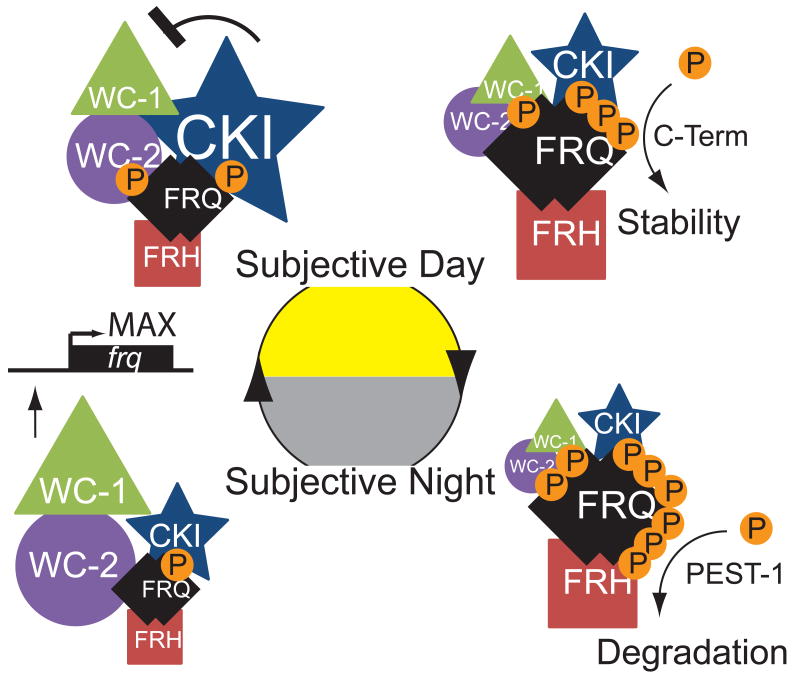
Figure 7. Model of FRQ phosphorylation and interactome.
Symbol size represents abundance of FRQ or relative steady-state interaction of FRH, WCC and CKI with FRQ. FRQ and FRH are always in a complex regardless of FRQ levels or phosphorylation. Slightly after subjective dawn, as frq mRNA levels reach a maximum, FRQ and CK1 interaction peaks, phase-lagging WCC interaction. In the subjective day as total FRQ levels increase, interaction with the WCC decreases. During this phase, the C-terminal region of FRQ is at its maximum relative phosphorylation state, helping to stabilize FRQ. During the subjective evening/night phosphorylation of the PEST-1 domain increases thereby destabilizing FRQ and interaction with the WCC is minimal. Approaching subjective dawn FRQ becomes fully phosphorylated, is degraded, and frq expression is reinitiated. New, hypophosphorylated, FRQ immediately associates with FRH and quickly increases its interaction level with the WCC to start a new cycle.
The locations and phenotypes of the original altered-function frq alleles – frq3, period 24h and frq7, period 29h - may also be informative. Both mutated residues are located within 124 amino acids of one another, within or adjacent to the single longest stretch of unmodified amino acids in FRQ. Thus, an extended region of FRQ is not phosphorylated, perhaps allowing a site for uncharacterized protein interaction or higher order structure formation in an otherwise highly modified protein. Removal of phosphorylated residues in the PEST-1 domain stabilizes FRQ, and the central part of the protein, which includes the PEST-1 and FCD, is sufficient to promote rapid degradation of a heterologous protein (Querfurth et al., 2007). One strategy to delay FRQ degradation, and therefore extend circadian period, would be to temporally regulate accessibility of this region to kinases. Interestingly, phosphorylation of the C-terminal region of FRQ phase-leads that of the PEST-1 domain, and when the C-terminal residues are mutated or the entire region deleted we observe a decrease in FRQ half-life and a shortening of period. The model of Kivimae et al. (2008), in which phosphorylation of a dPER domain causes the protein to unfold, exposing additional sites for destabilizing phosphorylation, may be applicable to FRQ, as it implies an ordered domain-specific progression of phosphorylation. Consistent with this hypothesis is the location, near the C-terminus, of another classic allele frq2, period ∼19h. This mutation may negate the stabilizing function of the C-terminus thereby leading to premature FRQ degradation.
Lastly, a comparison with the negative elements in other circadian systems is informative as it reveals some striking similarities. FRQ, PER, and mPER2 are all subject to multi-site phosphorylation and loss of these sites either result in period lengthening or shortening (this work, Chiu et al., 2008; Kivimae et al., 2008; Vanselow et al., 2006). In all three cases, the phosphoprotein interacts strongly with a partner (FRQ-FRH, PER-TIM, PER-CRY), and all proteins make stable associations with CK1. This casts CK1 in a role not only as a kinase acting on the negative elements, but also as a conserved central component in all clock complexes (Gallego and Virshup, 2007), serving to mediate interactions with (e.g. Yu et al., 2009) and/or phosphorylate other proteins. Indeed, it may be that, by having a time-dependent kinase-associated complex, some circadian output physiology can be regulated directly through clock-controlled PTMs, as seen with the rhythmic phosphorylation and activation of p38-like MAPK pathway in Neurospora (Vitalini et al., 2007).
While exact mechanisms whereby clock proteins act may differ slightly among circadian systems, and the sequences of the specific circadian phosphoproteins surely do, their regulation by timed phosphorylation could represent the common target of evolution in developing the circadian time delay. We think of sequence conservation as reflecting the need to preserve tertiary structured domains; however, in light of the common and extensive phosphorylation of clock proteins, an alternative view might simply be that evolution has selected for sequences that allow multiple post-translational modifications needed to facilitate opposing effects on protein stability and protein complex formation. This balance could be achieved through a dynamic cycle of clustered phosphorylations modulating the appearance and loss of charged domains. Clearly there is more to learn, but the application of high performance quantitative proteomics to enable correlation of changes in the circadian interactome with protein modifications of the components will lead the way.
Experimental Procedures
Strains and growth conditions
Strains FGSC 15070 (Δfrq∷hph+ A), FGSC 13693 (Δaga∷hph+, a), FGSC 9568 (Δmus-52∷hph+ a) were obtained from the Fungal Genetics Stock Center, Kansas City, KS. 328-4 (ras-1bd A) was used as the WT strain in this study. Transformations were performed as described (Colot et al., 2006). 541-5 (ras-1bd; frqV5H6∷bar+ a) was created by transforming pCB04 (Supplemental Data) into FGSC 9568 (Ninomiya et al., 2004) and ignite-resistant primary transformants backcrossed twice to WT. The frqKI targeting cassette, pCB05 (Supplemental Data), was introduced into 576-12 (ras-1bd; Δfrq∷hph+; Δmus-52∷hph+ a) and backcrossed to 578-3 (ras-1bd; his-3+∷Pfrq-luc+ A) to obtain homokaryon strains with and without the luciferase reporter (Gooch et al., 2008). A detailed description of the SILAC strain and growth methods can be found in the Supplemental Data. Vegetative growth cultures, race tube assays, circadian liquid culture experiments, and FRQ degradation experiments were performed as described (Garceau et al., 1997; Liu et al., 2000; Ruoff et al., 2005).
Protein preparation and Western blots
Procedures for preparation of protein lysates and Western blots were followed as described (Garceau et al., 1997). For Western blots, ≈25 μg whole-cell protein lysate was loaded per lane. α-V5 antibody (Invitrogen, Carlsbad, CA) was diluted 1:5000 for use as the primary antibody. Protein purification prior to MS analysis was performed similar to Cheng et al., 2005 using a slightly modified procedure (see Supplemental Data).
Analysis by mass spectrometry
Protein samples were analyzed by nanoscale microcapillary LC-MS/MS essentially as described (Dieguez-Acuna et al., 2005; Haas et al., 2006). For a full description, see Supplemental Data. The resulting tandem mass spectra were data-searched using the SEQUEST algorithm (Link et al., 1999), filtered to a 1% false discovery rate using the target-decoy strategy (Elias and Gygi, 2007) and reported. Phosphopeptide tandem mass spectra sequence assignments and phosphoacceptor residues were manually verified. High confidence was assigned to phosphopeptide sequences for which diagnostic, site-determining ions could be confirmed by manual inspection of their corresponding fragmentation spectra. Assignments of low confidence were reported for fragmentation spectra that confirmed both the backbone peptide sequence as well as the presence of phosphate group(s) on that sequence, but did not contain sufficient diagnostic, site-determining ions to unambiguously assign the phosphorylation locus.
SILAC quantification was performed using a highly in-house-modified version of the XPRESS algorithm (http://tools.proteomecenter.org, Han et al., 2001), in which SEQUEST results are assigned a heavy-to-light (H/L) ratio value based on integration of precursor ion chromatograms extracted to high mass precision (+/- 2.5 part-per-million). After reporting, SILAC ratios were handled as described in Supplemental Data.
Supplementary Material
Acknowledgments
We would like to thank Luis F. Larrondo for kindly providing strains, methods, and advice for the measurement of rhythms via luciferase. NIGMS T32GM008704 supported C.L.B, P20 RR018787 supported A.N.K and S.A.G, and J.J.L and J.C.D. (RO1s GM34985 and GM08336 and P01 GM068087) supported the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Hughes C, Stewart MH, Doble B, Bhatia M, Lajoie GA. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics. 2008;7:1587–1597. doi: 10.1074/mcp.M800113-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner M, Schafmeier T. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev. 2006;20:1061–1074. doi: 10.1101/gad.1410406. [DOI] [PubMed] [Google Scholar]
- Cha J, Chang SS, Huang G, Cheng P, Liu Y. Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J. 2008;27:3246–3255. doi: 10.1038/emboj.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieguez-Acuna FJ, Gerber SA, Kodama S, Elias JE, Beausoleil SA, Faustman D, Gygi SP. Characterization of mouse spleen cells by subtractive proteomics. Mol Cell Proteomics. 2005;4:1459–1470. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 2007;581:5759–5764. doi: 10.1016/j.febslet.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Proteins in the Neurospora circadian clockworks. J Biol Chem. 2006;281:28489–28493. doi: 10.1074/jbc.R600018200. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 2001;20:7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cha J, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, Dunlap JC. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, Liu Y. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21:3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimae S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs EG. Nobel Lecture. Protein phosphorylation and cellular regulation I. Biosci Rep. 1993;13:127–142. doi: 10.1007/BF01149958. [DOI] [PubMed] [Google Scholar]
- Larrondo LF, Colot HV, Baker CL, Loros JJ, Dunlap JC. Fungal Functional Genomics: Tunable Knockout-Knockin-expression and tagging strategies. Eukaryot Cell. 2009 doi: 10.1128/EC.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci U S A. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC. A role for Casein Kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009 doi: 10.1016/j.cell.2009.03.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M, Mazzotta G, Chen Z, Roenneberg T. The right place at the right time: regulation of daily timing by phosphorylation. Genes Dev. 2006;20:2629–2623. doi: 10.1101/gad.1479706. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Perkins DD, Radford A, Sachs MS. The Neurospora Compendium - Chromosomal Loci. Academic Press; 2001. [Google Scholar]
- Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- Pregueiro AM, Liu Q, Baker CL, Dunlap JC, Loros JJ. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner A, Heise F, Lauinger L, Neiss A, Tataroglu O, Brunner M, Schafmeier T. Posttranslational regulation of Neurospora circadian clock by CK1a-dependent phosphorylation. Cold Spring Harb Symp Quant Biol. 2007;72:177–183. doi: 10.1101/sqb.2007.72.025. [DOI] [PubMed] [Google Scholar]
- Ruoff P, Loros JJ, Dunlap JC. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2005;102:17681–17686. doi: 10.1073/pnas.0505137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T, Diernfellner A, Schafer A, Dintsis O, Neiss A, Brunner M. Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes Dev. 2008;22:3397–3402. doi: 10.1101/gad.507408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T, Kaldi K, Diernfellner A, Mohr C, Brunner M. Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes Dev. 2006;20:297–306. doi: 10.1101/gad.360906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalini MW, de Paula RM, Goldsmith CS, Jones CA, Borkovich KA, Bell-Pedersen D. Circadian rhythmicity mediated by temporal regulation of the activity of p38 MAPK. Proc Natl Acad Sci U S A. 2007;104:18223–18228. doi: 10.1073/pnas.0704900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a non-catalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009 doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.