SUMMARY
Rad51 is a DNA recombinase functioning in the repair of DNA double-strand breaks and the generation of genetic diversity by homologous recombination (HR). In the presence of ATP, Rad51 self-assembles into an extended polymer on single-stranded DNA to catalyze strand exchange. Inappropriate HR causes genomic instability and it is normally prevented by remodeling enzymes that antagonize the activities of Rad51 nucleoprotein filaments. In yeast, the Srs2 helicase/translocase suppresses HR by clearing Rad51 polymers from single-stranded DNA. We have examined the mechanism of disassembly of Rad51 nucleoprotein filaments by Srs2 and find that a physical interaction between Rad51 and the C-terminal region of Srs2 triggers ATP hydrolysis within the Rad51 filament, causing Rad51 to dissociate from DNA. This allosteric mechanism explains the biological specialization of Srs2 as a DNA motor protein that antagonizes HR.
INTRODUCTION
Homologous recombination (HR) is an important mechanism for the repair of DNA double-stranded breaks (DSBs), which arise normally during meiosis or as a result of exposure to DNA damaging agents (Li and Heyer, 2008). In eukaryotes, the ATP-dependent recombinase Rad51 catalyzes DNA strand exchange during HR in conjunction with pro- and anti-recombinogenic regulatory proteins that assist or inhibit Rad51’s interactions with DNA substrates (Bianco et al., 1998). Rad51 is a member of the RecA family of recombinases, which assemble into a right-handed helical filament on single-stranded DNA (ssDNA) that activates downstream steps of HR (Sung and Klein, 2006). Inherited defects in HR predispose cells to genomic instability and disease. For example, the breast cancer-associated tumor suppressor BRCA2 physically interacts with the Rad51 recombinase to regulate the assembly and stability of the Rad51 nucleoprotein filament (Pellegrini and Venkitaraman, 2004; Thorslund and West, 2007). Mutations in BRCA2 that compromise these functions are associated with an increased incidence of chromosomal rearrangements and other aberrations.
In the budding yeast Saccharomyces cerevisiae, HR is coordinated by proteins of the RAD52 epistasis group that function as positive mediators of Rad51 functions, conferring normal cellular resistance to DNA strand breaks and other types of damage (Li and Heyer, 2008; Sung and Klein, 2006; Symington, 2002). DNA DSBs are nucleolytically resected to generate a 3′ single-stranded tail of about 600 nt that serves as a template for the assembly of a pre-synaptic Rad51 filament. The Rad51 filament invades an undamaged double-stranded DNA (dsDNA) where homologous sequences can pair in a synaptic complex. Once homology is identified, the dsDNA is used as a template to extend the invading strand by DNA synthesis. This newly synthesized strand is then resolved into a continuous double-stranded DNA product by one of several pathways leading to crossovers and noncrossover products (Bianco et al., 1998; Krogh and Symington, 2004).
The nucleation of Rad51 filament growth on ssDNA provides a window of opportunity for the regulation of HR activity by both pro- and anti-recombinogenic factors. Saccharomyces cerevisiae Srs2 is a negative regulator of HR that disrupts preformed Rad51 filaments (Krejci et al., 2003; Veaute et al., 2003), and a similar activity has been recently ascribed to two mammalian RecQ-type helicases (Bugreev et al., 2007; Hu et al., 2007). The anti-recombinogenic activity of Srs2, which is attributed to the disassembly of Rad51 filaments is regulated by the recruitment of Srs2 to sites of DNA damage following checkpoint activation, possibly through phosphorylation of Srs2 by CDK-1 (Chiolo et al., 2005) and/or through the physical interaction with sumoylated PCNA (Papouli et al., 2005; Pfander et al., 2005). Srs2 physically interacts with Rad51, although the functional significance of this interaction for Srs2’s anti-recombinogenic activities has not been established (Krejci et al., 2003).
Srs2 belongs to the SF-1 superfamily of helicases/translocases and possesses a DNA unwinding activity coupled to ATP hydrolysis (Rong and Klein, 1993). These activities have been well characterized in other SF-1 family members (Lohman et al., 2008; Singleton et al., 2007). Monomers of the SF-1 enzymes Rep, PcrA, and UvrD are rapid, processive translocases on ssDNA; however, DNA unwinding activity requires oligomerization in vitro (Lohman et al., 2008). Hence, for these enzymes, DNA unwinding and ssDNA translocation activities are separable. The conserved SF-1 family motifs of Srs2 define a core architecture that is likely to closely resemble the structures of the SF-1 helicases UvrD (Lee and Yang, 2006), PcrA (Velankar et al., 1999), and Rep (Korolev et al., 1997). However, Srs2 has an additional 454 amino acids at its C-terminus (Figure 1) that mediate interactions with Rad51 (Krejci et al., 2003) and PCNA (Papouli et al., 2005; Pfander et al., 2005), and may confer specificity to the remodeling of nucleoprotein filaments. UvrD, a bacterial homolog of Srs2, similarly antagonizes the formation of RecA filaments in bacteria (Veaute et al., 2005).
Figure 1. Srs2 Translocates Unidirectionally on ssDNA.
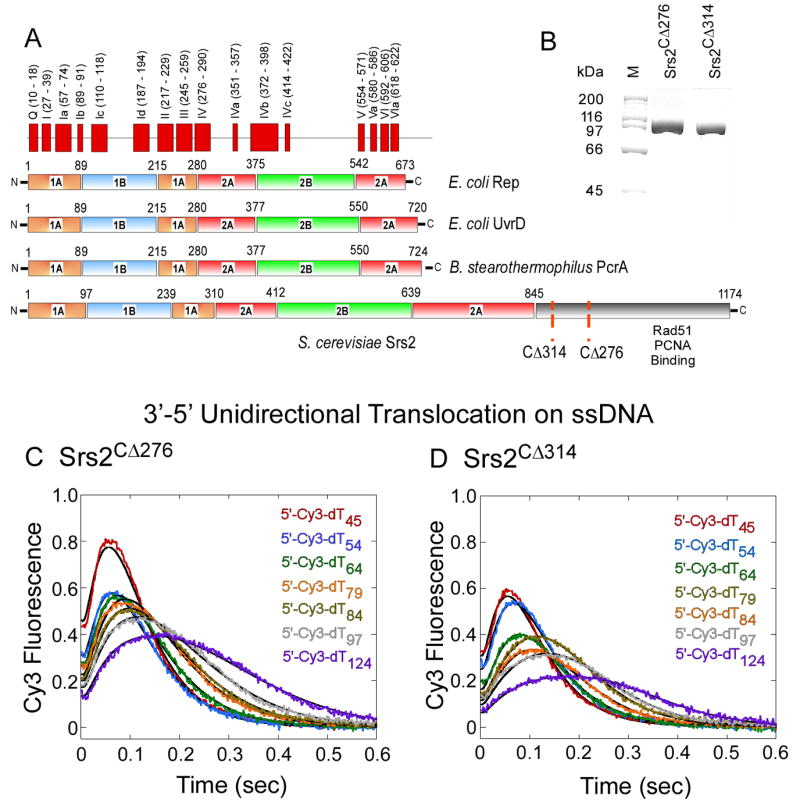
(A) Sequence alignment of Saccharomyces cerevisiae Srs2 with various bacterial SF-1 helicases. The conserved helicase motifs, 1A, 2A, 1B and 2B domains in UvrD are depicted. (B) SDS-PAGE analysis of the purified Srs2 proteins used in this study: Srs2CΔ276 and Srs2CΔ314. (C, D) Time courses monitoring Cy3 fluorescence on 5′Cy3 labeled oligonucleotides of varying lengths (25 – 124 nt) shows that Srs2CΔ276 and Srs2CΔ314 translocate unidirectionally on ssDNA.
Using real-time assays, we have monitored the dynamics of the Rad51 filament and its disassembly by Srs2. We identify features of Srs2-mediated clearing of Rad51 nucleoprotein filaments that distinguish this specialized activity from DNA translocation and unwinding activities. Our findings show that the unidirectional translocation of Srs2 on ssDNA does not by itself account for the enzymatic clearance of Rad51 filaments. Instead, the physical interaction of Srs2 with the Rad51 protein allosterically activates ATP hydrolysis within the Rad51 nucleoprotein filament to promote clearing of Rad51 from the DNA. Although the bacterial UvrD helicase can displace yeast Rad51 from DNA (Veaute et al., 2005), we show that this activity is noncatalytic and results from a binding competition with Rad51 for DNA. The anti-recombinogenic activity of Srs2 results from a protein-protein interaction that imparts biological specificity to its DNA remodeling functions.
RESULTS
Srs2 Unidirectionally Translocates on ssDNA
To obtain large quantities of Srs2 and alleviate the problem of protein aggregation, we deleted portions of the nonconserved C-terminal region and found that fragments spanning amino acids 1-860 (Srs2CΔ314 and 1-898 (Srs2CΔ276; Figure 1A) are well behaved proteins that elute from a gel filtration column as a single peak with the expected apparent molecular size for a monomer (Figures 1B & S1). Both truncated proteins exhibit similar DNA unwinding activities (data not shown), whereas the Rad51-specific functions of Srs2 are eliminated in the shorter Srs2CΔ314 construct, as shown below.
SF-1 helicases translocate with biased directionally along ssDNA in an ATP-dependent manner (Lohman et al., 2008). DNA translocation by Srs2 could assist in remodeling Rad51 nucleoprotein filaments. Translocation activity was measured on a series of single-stranded dTn oligonucleotides of different lengths (5′-Cy3-dTn; n = 45 - 124 nucleotides, Figures 1C & D) using a single turnover assay (Fischer et al., 2004; Tomko et al., 2007). The arrival of Srs2 at the 5′ end of DNA labeled with Cy3 is accompanied by an increase in Cy3 fluorescence intensity. With increasing length (n) of the DNA, the distribution of fluorescence events broadens and the average time required for Srs2 to arrive at the 5′ end increases, reflecting random binding of Srs2 to the DNA and movement in a 3′ to 5′ direction (Figure 1C & 1D). Similar experiments with the Cy3 label on the 3’ end of the DNA did not produce a change in fluorescence intensity, consistent with the unidirectional translocation of Srs2 towards the 5’ end of the DNA. Analytical ultracentrifugation experiments indicate that no more than one molecule of Srs2 is bound to each DNA under these experimental conditions (25 nM Srs2CΔ276 and 50 nM DNA; Figure S2), suggesting that a monomer of Srs2 is sufficient for translocation on ssDNA. This behavior is similar to that observed for the E. coli UvrD translocase (Fischer et al., 2004; Tomko et al., 2007). The rate of Srs2 translocation on ssDNA (approx. 300 nt s-1; Figures 1C & 1D) is significantly faster than the apparent rate of Rad51clearance by Srs2 (approx. 12 nt s-1; Figure 3). The C-terminally deleted proteins Srs2CΔ314 and Srs2CΔ276 show similar translocation activities on ssDNA (Figures 1C & 1D; Table S1) yet, as shown below, the Srs2CΔ314 mutant is selectively defective in clearing Rad51 nucleoprotein filaments.
Figure 3. Srs2 Clearing of Rad51 Filaments.
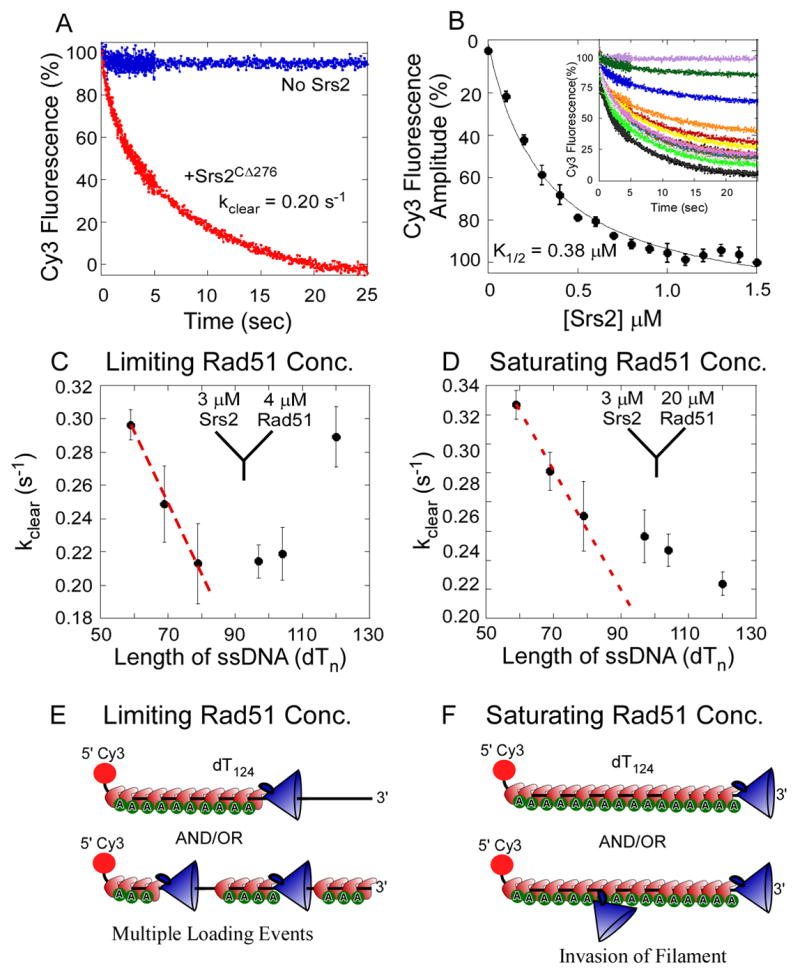
(A) In stop flow experiments, preformed Rad51 filaments were rapidly mixed with Srs2CΔ276. In the absence of Srs2, Rad51 filaments are stable (blue trace), but when challenged with Srs2 (1.5 μM post-mixing) Rad51 filaments are cleared (red trace). (B) Srs2CΔ276 concentration dependence of Rad51 clearing was measured with increasing concentrations of Srs2CΔ276 (0 - 1.5 μM post-mixing, shown as insert). The relative change in fluorescence amplitude is plotted as a function of Srs2 concentration and yields a K1/2 = 0.38 ± 0.04 μM for Rad51 filament clearing by Srs2CΔ276. (C) The Rad51 filament clearing experiment in A was repeated with 5′Cy3 ssDNA of varying lengths (dTn, n = 59-124 nt). Rad51 (2 μM post-mixing) was used in all the experiments to maintain sub-saturating Rad51-DNA filament forming conditions. The clearing rate observed for each DNA length in the presence of Srs2 (1.5 μM post-mixing) is plotted as a function of DNA length. The red dotted line is the expected trend for the clearing rate if a continuous Rad51-DNA filament is formed under these sub-saturating Rad51 concentrations. (D) The Rad51-DNA filament clearing experiment described in C was repeated with saturating concentrations of Rad51 (10 μM post-mixing). The clearing rates observed under these conditions are plotted as a function of DNA length. The red dotted line represents the expected trend for the clearing rate if Srs2 initiates clearing only at the 3′ end of the Rad51-DNA filament. Deviation from the predicted trend suggests that Srs2 can invade a Rad51 filament at multiple points and initiate clearing. (E) and (F) depict schematic representations of expected Rad51-DNA filaments formed under limiting or saturating concentrations of Rad51, and potential Srs2 loading or invading sites on the respective filaments.
Rad51 Filament Dynamics Are Coupled to ATP Turnover within the Rad51 Filament
We developed a fluorescence-based assay to monitor the kinetics of spontaneous assembly of Rad51 filaments on ssDNA and their disassembly by Srs2. Rad51 was mixed rapidly with ATP and a ssDNA (dT79 in length), labeled with Cy3 at the 3′ end or 5′ end, and the resulting increase in Cy3 fluorescence was monitored over time. The change in fluorescence upon Rad51 binding appears to be saturated faster when Cy3 is at the 5′ end compared to the 3′ end of the ssDNA (Figures 2A & 2B). Furthermore, lower concentrations of Rad51 are required to reach maximal fluorescence change with DNA labeled at the 5′ end compared to the 3′ end (Figures 2C & S3A). Differences in the responses to Rad51 monitored at the 5′ or 3′ end of the DNA are suggestive of a directional bias in filament assembly. However, the time courses of filament nucleation and growth display several phases that are indicative of additional complexities that we have not analyzed further.
Figure 2. Assembly and Disassembly of Rad51 Filaments on Single Stranded DNA.
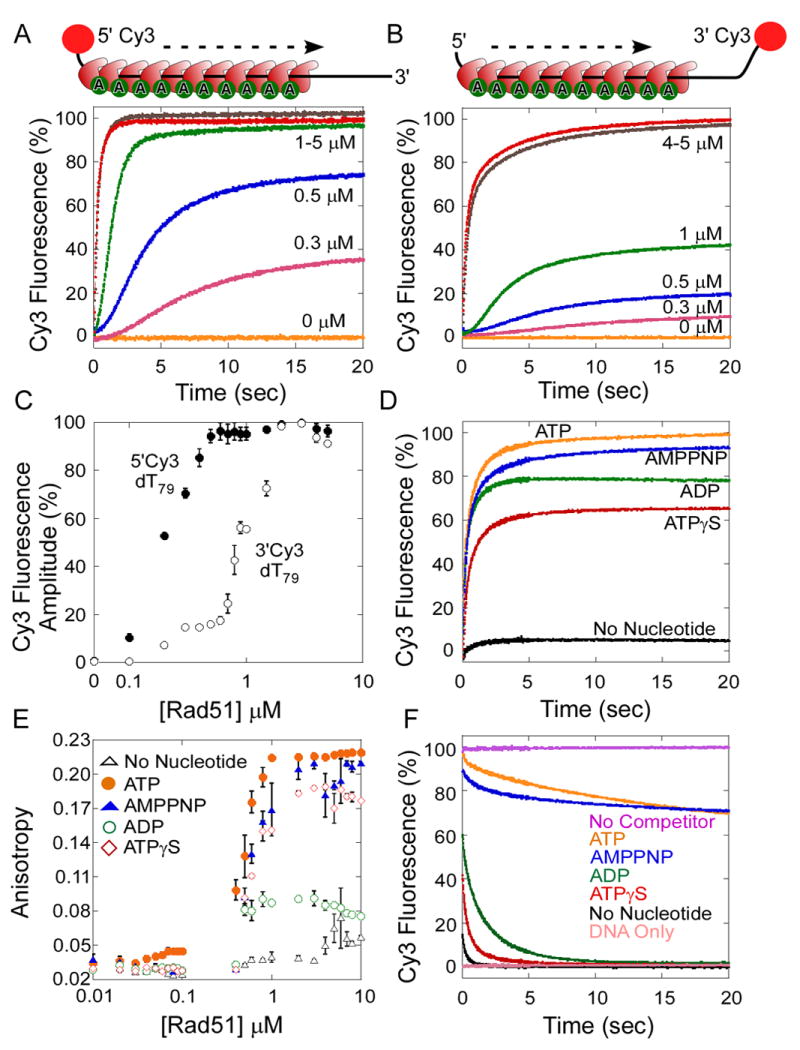
(A) Kinetics of Rad51 binding to a 5′Cy3 labeled dT79 oligonucleotide. Increasing concentrations of Rad51 (0-10 μM) were rapidly mixed with 5′Cy3-dT79 ssDNA (40 nM) and ATP-Mg2+ (5 mM) using a stopped flow instrument. The change in Cy3 fluorescence measured as a function of time reveals an increase in the Cy3 fluorescence upon Rad51 binding to the DNA. (B) Kinetics of Rad51 binding to a 3′Cy3 labeled dT79 oligonucleotide. (C) The change in fluorescence amplitude observed in A and B are plotted versus the concentration of Rad51 (log scale). Rad51 binds preferentially to the 5′ end (●) compared to the 3′ end (○) of the ssDNA. (D) Rad51 binding to ssDNA requires nucleotide. Rad51 (4 μM) was rapidly mixed with 5′Cy3-dT79 (40 nM) in the absence (black) or presence of ATP (yellow), ADP (green), AMP-PNP (blue), or ATPγS (red) (5 mM). Change in Cy3 fluorescence was measured over 25 sec post-mixing. (E) The stoichiometry of Rad51 molecules bound to a 5′-FITC labeled dT79 oligonucleotide was measured using fluorescence anisotropy. Increasing concentrations of Rad51 (0–10 μM) were mixed with 5′-FITC labeled dT79 oligonucleotide (40 nM), in the absence (△), or presence of ATP (●), ADP (○), AMP-PNP (▲), or ATPγS (◊) (2.5 mM), and the change in fluorescence anisotropy was measured. (F) Disassociation kinetics of nucleotide-bound Rad51 filaments. Preformed Rad51 filaments −Rad51 (4 μM), 5′Cy3-dT79 (40 nM), and ATP/ADP/AMP-PNP/ATPγS (5 mM), were rapidly mixed with excess unlabeled dT79 (40 μM). Rad51 filaments formed in the presence of ATP (yellow) or AMP-PNP (blue) have longer half-lives: 24.8 s and 55.0 s respectively, compared to ADP (green) or ATPγS bound molecules (red): 0.17 s or 0.09 s respectively. The filaments are stable when not challenged with competitor DNA (black).
Growth of the yeast Rad51 DNA filament is strictly dependent upon nucleotide binding to Rad51, but not on ATP hydrolysis (Bianco et al., 1998). The enhancement of Cy3 fluorescence is observed only when Rad51 and ssDNA are mixed together in the presence of ATP or its analogs AMP-PNP, ATPγS, or ADP (Figure 2D). At saturating nucleotide concentrations, the amplitude of the Cy3 fluorescence intensity change depends upon the identity of the nucleotide, with the largest changes observed in the presence of ATP. Under conditions of stoichiometric Rad51 binding (Figures 2E & S3B), one Rad51 molecule binds to 3-5 nt of ssDNA, consistent with previous reports (Zaitseva et al., 1999). In the presence of saturating ADP, the binding curve does not reach the same maximum level as seen for other nucleotides (Figure 2E), which may reflect a faster dissociation of ADP-bound Rad51 from DNA. Although ATPγS-Rad51 dissociates rapidly from the DNA (Figure 2F), it is reasonably proficient at supporting nucleoprotein filament formation, in comparison to ADP-Rad51 (Figure 2E). This difference may reflect a more favorable association rate and/or cooperative binding by ATPγS-Rad51 to DNA.
The dissociation of Rad51 nucleoprotein filaments was monitored by pre-assembling Rad51-cofactor complexes with 5′-Cy3-dT79DNA then incubating with an excess of unlabeled dT79 DNA (Figure 2F). Substantially different half-lives are observed for Rad51-DNA complexes formed with ATP (t1/2 = 24.8 ± 0.2 s), AMP-PNP (t1/2 = 55.3 ± 0.5 s), ATPγS (t1/2 = 0.1 ± 0.1 s) and ADP (t1/2 = 0.17 ± 0.1 s) using a 5’ Cy3 labeled DNA substrate. For DNA with a 3’-Cy3 label, the stabilities of Rad51-DNA complexes formed with different cofactors are ATP (t1/2 = 18.8 ± 0.3 s), AMP-PNP (t1/2 = 25.1 ± 0.3 s), ATPγS (t1/2 = 0.1 ± 0.12 s), and ADP (t1/2 = 0.17 ± 0.2 s) (data not shown). These data agree with the previous finding that ATP hydrolysis promotes Rad51 filament disassembly (Bianco et al., 1998; Chi et al., 2006). The observation that the position of the Cy3 fluorophore (5’ or 3’ end) affects the Rad51 dissociation rate suggests either that filament disassembly is faster near the 3′ end of the DNA or that the two Cy3 reporter groups respond differently to proximal Rad51 binding events.
Rad51 Filament Clearing by Srs2
To study the anti-recombinase activity of Srs2, we monitored the kinetics of ATP-bound Rad51 clearance from the 5′-Cy3-dT79 DNA (Figure 3A). Rad51-ATP filaments were assembled on the 5′-Cy3-dT79 DNA (40 nM) using excess Rad51 (4 μM) and ATP (5 mM). Filaments were mixed with Srs2CΔ276 (3 μM) to initiate clearing, and the resulting decrease in Cy3 fluorescence yielded the apparent rate of filament clearance, kclear = 0.204 ± 0.004 s-1 (Figure 3A; see Experimental Procedures). Although the decrease in 5′-Cy3 fluorescence reflects Rad51 displacement from the ssDNA, the change in Cy3 fluorescence likely occurs only after arrival of the translocating Srs2 at the 5′ end of the DNA. Under saturating Srs2 concentrations, the final fluorescence attained after the filament clearing reaction (at long times) is equivalent to the signal for Srs2 alone, indicating that there is no significant rebinding of Rad51 to the ssDNA (Figure S5).
Substoichiometric amounts of Srs2 effectively clear Rad51 filaments (Krejci et al., 2003), and we next examined filament clearing activity with increasing concentrations of Srs2 (Figure 3B). When sub-saturating concentrations of Srs2 are used, higher final Cy3 fluorescence intensity plateaus are observed that suggest inefficient Rad51 clearance and/or rebinding of Rad51 to the cleared DNA. The amplitude of the Cy3 fluorescence change, plotted as a function of Srs2 concentration, yields a K1/2 = 0.38 ± 0.002 μM Srs2CΔ276 for the filament clearing activity (Figure 3B). To test whether filament clearing is a catalytic activity of Srs2, and not the result of stoichiometric binding to DNA, we repeated the experiment using a longer dT124 substrate (Figure S4). The observed K1/2 = 0.40 ± 0.026 μM for dT124 is in good agreement with the K1/2 value for the shorter dT79 filaments. If a stoichiometric binding model were applicable, we would predict that more Srs2 would be required to block binding of Rad51 on the longer dT124 filaments. Instead, our findings are consistent with a catalytic mechanism of Rad51 filament disassembly by Srs2.
In considering how Srs2 initiates Rad51 filament clearance, we considered two possible scenarios in which Srs2 either initiates clearance at the 3′ end of the filament or inserts at random internal sites (Figures 3E & F). The initial rates of filament clearing were assayed on DNA of different lengths (dTn with n = 59 -124 nt) in the presence of a limiting concentration of Rad51 (2 μM post-mixing; Figure 3C) or with saturating amounts of Rad51 (10 μM, post-mixing; Figure 3D). With limiting amounts of Rad51, the longer DNA substrates (dT 87 -124) are not saturated (Figure S3B) and it is likely that multiple clusters of Rad51 polymers exist on the DNA, punctuated by gaps of exposed ssDNA (Modesti et al., 2007). A biphasic dependence of kclear on dTn is observed (Figure 3C), in which longer DNAs show faster clearance by Srs2CΔ276 when Rad51 is limiting. This is consistent with Srs2 initiating clearance of Rad51 at multiple internal sites on incompletely formed filaments (Figure 3E), such that kclear first decreases from n= 60 to 80, but then increases for n = 97 to 124 (Figure 3C). In this case, the longer DNAs (n = 97 to 124) show a faster clearance by Srs2CΔ276 presumably because Rad51 is sub-saturating. With saturating amounts of Rad51, a continual decrease in kclear is observed as a function of increasing DNA length (Figure 3D). However, since under saturating Rad51 conditions (Figure 3D) the observed rates of clearance deviate from a simple linear relationship, this may indicate imperfections in the Rad51 filaments or some limited ability of Srs2 to invade at internal sites within a Rad51 filament (Figure 3F).
A Nonconserved Region of Srs2 is Required for Rad51 Clearing Activity
The C-terminal region of Srs2 physically interacts with Rad51 (Krejci et al., 2003; Papouli et al., 2005) and with sumoylated PCNA (Pfander et al., 2005) at sites of active DNA repair. Residues 1-845 of Srs2 encompass the conserved SF-1 helicase motifs (Figure 1A), whereas the C-terminal residues 846 -1174 are not homologous with other SF-1 helicases. The Srs2CΔ276 protein spanning residues 1-898 efficiently dismantles Rad51 filaments (Figure 4A), whereas a deletion of an additional 38 C-terminal residues in the Srs2CΔ314 protein (spanning residues 1-860; Figure 1A) almost completely eliminates this Rad51-specific activity (Figure 4B). The shorter Srs2CΔ314 protein is identical to Srs2CΔ276 in all other activities examined, including DNA unwinding, DNA-dependent ATP hydrolysis, DNA binding (data not shown), and ssDNA translocation activities (Figures 1C & D). Hence, the removal of residues 861-898 from Srs2 uncouples its Rad51-dependent filament clearing activity from other DNA-dependent activities. The physical interaction of Srs2 with Rad51 may promote the loading of Srs2 on Rad51 filaments, facilitating recycling of Srs2 onto the DNA under multiple turnover conditions. These findings suggest that the C-terminal region of Srs2 has a unique and specific role in Rad51 clearing and anti-recombinogenic functions.
Figure 4. The C-terminus of Srs2 is Essential for its Physical Interaction with Rad51 and Filament Clearing Activity.
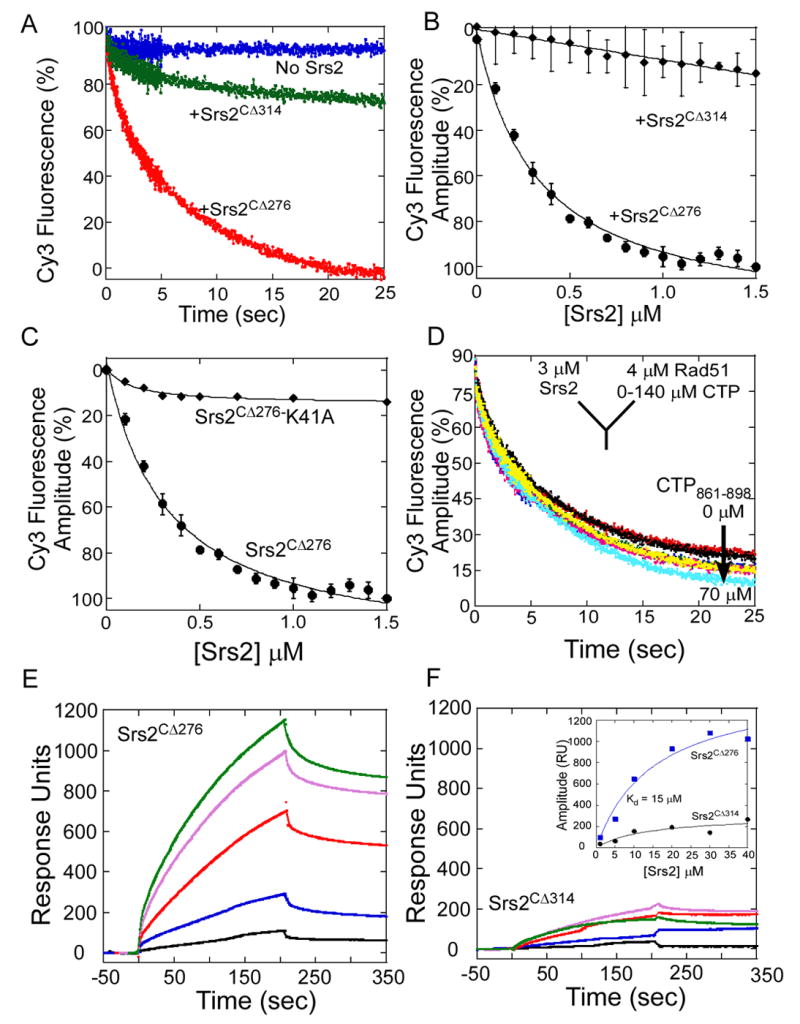
(A) Pre-formed Rad51-DNA-ATP filaments were rapidly mixed with buffer (blue trace), Srs2CΔ276 (red trace), or Srs2CΔ314 (green trace), and filament clearing monitored as a function of time. Srs2CΔ314, lacking an additional 38 residues is unable to clear a Rad51-DNA filament. (B) Srs2 concentration dependence of filament clearing shows a weak filament clearing activity for Srs2CΔ314 (◆) compared to Srs2CΔ276 (●). (C) An ATP binding/hydrolysis deficient mutant of Srs2CΔ276, Srs2CΔ276-K41A (◆), is unable to clear a preformed Rad51-DNA filament unlike the wild-type Srs2CΔ276 protein (●). (D) A synthetic peptide corresponding to the Srs2 C-terminal residues 861 – 898, is a poor competitor for Srs2CΔ276 in Rad51 filament clearing experiments. Rad51 was tethered onto a SPR chip, and increasing concentrations of (E) Srs2CΔ276, or (F) Srs2CΔ314 was passed over the chip. The insert shows the change in amplitude (response units, RU) plotted versus the concentration of Srs2, yielding a KD = 15.2 ± 5 μM for the interaction between Srs2CΔ276 and Rad51. The interaction between Srs2CΔ314 and Rad51 is very weak.
A Physical Interaction between Srs2 and Rad51 is Required for Rad51 Filament Clearing
The physical interaction of Srs2 with Rad51 was monitored on a SPR sensor chip containing immobilized Rad51. The Srs2CΔ276 protein binds to Rad51 with an apparent KD = 15 ± 5 μM, whereas the Srs2CΔ314 lacking 38 residues at its C-terminus interacts weakly with Rad51 (Figures 4E, & 4F). A synthetic peptide (CTP861-898) corresponding to the region deleted from Srs2CΔ314 did not show an interaction with Rad51 (data not shown) and high concentrations of the peptide failed to inhibit the Rad51 filament clearing activity of Srs2CΔ276 (Figure 4D). These results indicate that the peptide is missing part of the Rad51 protein binding surface or that it fails to fold properly in this context. In addition to the interaction of Srs2 with Rad51, ATP turnover by Srs2 is required for filament clearing. The ATPase defective mutant Srs2CΔ276-K41A binds to ssDNA but fails to clear Rad51 filaments (Krejci et al., 2004)(Figure 4C).
Srs2 Stimulates ATP Hydrolysis within the Rad51 Filament to Promote Clearing
Rad51 nucleoprotein filaments are stabilized by ATP binding (Bianco et al., 1998; Cox et al., 2005; Cox, 2007; Sung and Stratton, 1996) and ATP turnover occurs slowly within the filament (kcat = 0.004 s-1, Table S2), promoting the release of Rad51 from DNA (Figure 2F). We considered the possibility that Srs2 might stimulate ATP turnover by Rad51 in order to achieve the 30- to 50-fold stimulation of Rad51 filament disassembly by Srs2 (kclear ~ 0.2 s-1; Figure 3A). It is difficult to directly measure ATP hydrolysis by Rad51 in the presence of Srs2 because Srs2 hydrolyzes ATP rapidly (80 s-1; data not shown) in the presence of ssDNA.
To determine if ATP turnover by Rad51 limits the filament clearing activity of Srs2, we examined two mutant forms of Rad51. Rad51-K191R hydrolyzes ATP poorly (kcat = 0.001 s-1; Table S2) but is active as a recombinase in vitro (Chi et al., 2006; Sung and Stratton, 1996) and in vivo (Morgan et al., 2002). Rad51-E221D has a basal rate of ATP hydrolysis (kcat = 0.038 s-1; Table S2) that is 10-fold faster than that of wild-type Rad51. Both mutant Rad51 proteins readily form Rad51 filaments on DNA (Figure 5A) although Rad51-K191R has a 4-fold higher KD for filament formation (KD = 0.28 ± 0.005 μM, 0.24 ± 0.004 μM, and 1.21 ± 0.3 μM, for wild-type, E221D, and K191R Rad51 proteins, respectively). The Rad51-K191R protein has been reported to have normal ATP binding activity while being deficient in ATP hydrolysis (Chi et al., 2006; Sung and Stratton, 1996). However, care should be taken when interpreting results obtained with this mutant; since in comparison to wild-type Rad51, the Rad51-K191R protein shows a 90-fold higher K1/2 for the ATP dependence of DNA binding (Figure 5B; K1/2 = 1.55 ± 0.3 nM, 2.17 ± 0.6 nM, and 140 ± 10 nM for wild-type, E221D, and K191R Rad51 proteins, respectively). Thus, Rad51-K191R appears to show a defect in ATP binding, and requires higher ATP concentrations to form the Rad51-ATP-DNA filaments with this mutant. Nonetheless, with saturating amounts of ATP, the Rad51-K191R and Rad51-E221D proteins efficiently assemble into filaments on ssDNA (Figures 5A & B).
Figure 5. Rad51 Filament Disassembly is Regulated by ATP Hydrolysis Within the Rad51-DNA Filament.
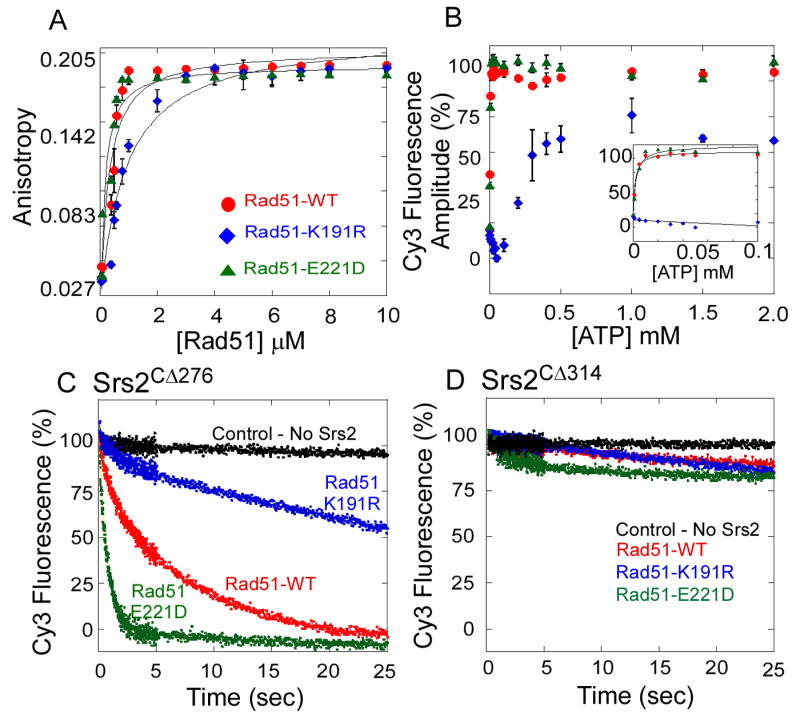
(A) Rad51-WT (●), Rad51-K191R (◆), and Rad51-E221D (▲) proteins form a filament on ssDNA measured by fluorescence anisotropy. The ATP hydrolysis deficient mutant (Rad51-K191R) binds ssDNA with lower affinity (KD = 1.21 ± 0.3 μM) compared to the wild-type Rad51 (KD = 0.28 ± 0.1 μM) or the Rad51-E221D mutant (KD = 0.21 ± 0.1 μM). (B) ATP concentration dependence of Rad51 filament formation on ssDNA measured using the stopped flow assay and monitoring changes in 5′Cy3 fluorescence on a dT79 DNA substrate. Change in fluorescence amplitude observed at each ATP concentration is plotted for wild-type Rad51 (●), Rad51-K191R (◆), and Rad51-E221D (▲). The K1/2 for ATP dependence of filament formation for wild-type Rad51 and Rad51-K191R are 1.5 ± 0.3 nM and 2.1 ± 0.6 nM respectively. The Rad51-K191R mutant (◆) does not bind to DNA at lower ATP concentrations (insert), but forms a filament at higher ATP concentrations (K1/2 = 120 ± 28 nM). (C) Srs2CΔ276 clears wild-type Rad51 (red), Rad51-K191R (blue), and Rad51-E221D (green) filaments, but (D) Srs2CΔ314 is incapable of clearing either wild-type Rad51 or mutant Rad51 filaments.
Different rates of Rad51 filament clearing by Srs2CΔ276 are observed for filaments formed by wild-type Rad51 (kclear = 0.204 ± 0.004 s-1), Rad51-E221D (kclear = 1.09 ± 0.04 s-1), and Rad51- K191R (kclear = 0.07 ± 0.01 s-1; Figure 5C). The 5- to 10-fold higher clearance rate for Rad51-E221D filaments correlates with a 10-fold higher rate of basal ATP hydrolysis by this mutant (Table S2). The clearance of Rad51-K191R filaments occurs at a significantly slower rate (Figure 5C), consistent with the slower basal rate of ATP hydrolysis by Rad51-K191R. This suggests that the rate of enzymatic clearance of Rad51 from DNA appears to be limited by ATP turnover within the Rad51 protein. However, all of these filaments require the enzymatic and Rad51-binding activities of Srs2 to disassemble the filament. The truncated Srs2CΔ314 protein, which does not interact with Rad51 (Figure 4F), is unable to clear even the destabilized filaments formed by the Rad51-E221D mutant (Figure 5D).
We next examined if Srs2 can modulate ATP turnover by Rad51 to promote filament disassembly. We took advantage of the relatively slow basal rate of ATP turnover in Rad51 bound to ssDNA by assembling filaments in the presence of α-32P-ATP before mixing with Srs2CΔ276 and a 500-fold molar excess of unlabeled ATP to lower the specific activity of radiolabeled ATP available to bind Srs2. Under these conditions, very little 32P-ADP is formed during a 30 s reaction with Srs2CΔ276 in the absence of Rad51 (Figure 6A). When Rad51 is preincubated with radiolabeled ATP and dT79 ssDNA, the addition of Srs2CΔ276 causes a concentration-dependent increase in 32P-ATP hydrolysis by Rad51 during filament clearing. Control experiments done with only Rad51, Rad51 + DNA, or Rad51 + Srs2CΔ276 (in the absence of DNA) showed no detectable ATP hydrolysis during the 30 s incubation (data not shown). A plot of the amount of 32P-ADP formed as a function of Srs2CΔ276 concentration yields a K1/2 = 0.52 ± 0.13 μM Srs2 (Figure 6A), which is comparable to the K1/2 = 0.38 ± 0.002 μM for the filament clearing reaction (Figure 3B). Similar experiments done with the mutant Rad51 nucleoprotein filaments (Figure 6C) yield K1/2 = 0.90 ± 0.08 (Rad51-K191R) and 0.34 ± 0.02 (Rad51-E221D), indicating that the Rad51-K191R mutant is more resistant to filament clearing by Srs2, whereas the Rad51-E221D mutant is cleared more rapidly than wild-type Rad51. These results suggest that ATP hydrolysis by Rad51 influences the rate of filament clearing by Srs2.
Figure 6. Srs2 Allosterically Stimulates ATP Hydrolysis within the Rad51 Filament Thereby Promoting Filament Disassembly.
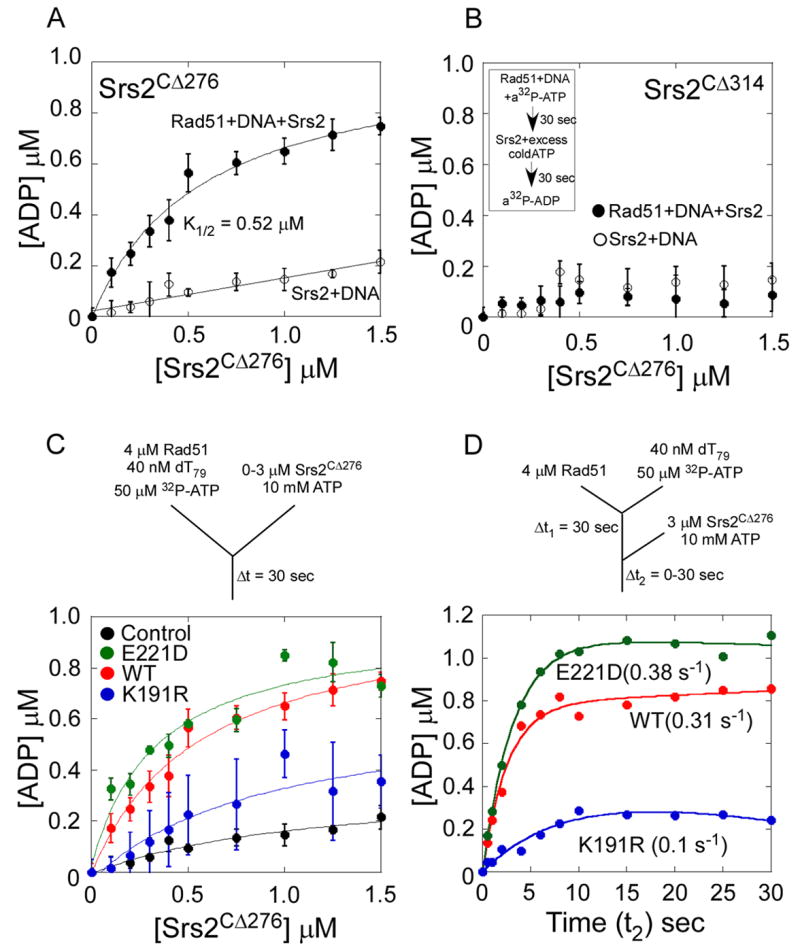
(A) Srs2CΔ276 stimulates the ATP hydrolysis within a Rad51-DNA filament. Preformed Rad51-DNA-32P-ATP filaments were mixed with increasing concentrations of Srs2CΔ276 and excess unlabeled ATP and the amount of 32P-ADP formed was measured. A stimulation of ATP hydrolysis is observed within the Rad51 filament (●), and the Srs2CΔ276 dependence of this stimulation yields a K1/2 = 0.53 ± 0.13 μM. The control experiment done in the absence of Rad51 (○) shows the background level of 32P-ATP hydrolyzed by Srs2 when a 500-fold excess of unlabeled ATP is present. (B) Srs2CΔ314 does not stimulate ATP hydrolysis within a Rad51-DNA filament. The insert shows a schematic representation of the ATPase assays described in A and B. (C) Rad51 filaments were formed on ssDNA (dT79) with either wild-type Rad51 (red), Rad51-K191R (blue), or Rad51-E221D (green) proteins in the presence of radiolabeled ATP. Srs2CΔ276 stimulates the rate of ATP hydrolysis when added to these filaments along with excess unlabeled ATP. The Srs2CΔ276 dependence of this stimulation yields a K1/2 = 0.57 ± 0.15 μM (WT), K1/2 = 0.9 ± 0.1 μM (K191R), and K1/2 = 0.34 ± 0.07 μM (E221D) respectively. The control experiment done in the absence of Rad51 (black) shows the background level of 32P-ATP hydrolyzed by Srs2 when a 500-fold excess of unlabeled ATP is present. (D) Rate of ATP hydrolysis within the various Rad51 nucleoprotein filaments during filament clearing by Srs2CΔ276 measured using a sequential mixing quenched flow setup. Rad51 (4 μM, WT-red, K191R-blue or E221D-green) was first mixed with dT79 DNA (40 nM) and 32P-ATP (50 μM) for 30 seconds and then rapidly mixed with Srs2 (3 μM) and 20 mM excess ATP for varying times (t2). The schematic for the sequential mixing experiment is shown, and the Srs2 stimulated rates of ATP hydrolysis in the wild-type and mutant Rad51 filaments are noted.
To further quantify the rate of ATP hydrolysis within the Rad51 filament during filament clearing by Srs2, we performed a sequential mixing quenched flow experiment. Modifying the strategy described above, we mixed Rad51 (4 μM) and 32P-ATP for 30 sec (t1) followed by sequential mixing with Srs2 (3 μM) and a 200 fold excess of unlabeled ATP for varying times (t2). During filament clearing by Srs2, the wild-type Rad51, Rad51-E221D and Rad51-K191R proteins hydrolyzed ATP at rates of 0.31, 0.38 and 0.1 s-1 respectively (Figure 6D).
The stimulated rates of ATP hydrolysis by these Rad51 proteins during filament clearing by Srs2 are 10-fold to 100-fold faster than the basal rates of ATP hydrolysis in the absence of Srs2 (Table S2). We note that the stimulated rates of ATP hydrolysis in Rad51 filaments (Figure 6D) are not equal to the apparent rates of filament clearance (kclear) (Figure 5C); however, this is not expected due to how the clearing rates are measured. In our filament clearing assay, the fluorescence reporter is located at the 5’ end of the ssDNA, thus a fluorescence change is detected only from the displacement of Rad51 molecules in the region proximal to the 5’ end, rather than displacement of all Rad51 molecules. In contrast, the measured ATPase activity reflects hydrolysis by all Rad51 molecules bound to DNA. Thus, the rates of ATP turnover and Rad51 filament clearance are not expected to be equivalent. Nevertheless, they show the same trends for the different Rad51 mutants.
Despite these caveats, the dose-response curves for Srs2 stimulation of Rad51 ATPase activity (K1/2 = 0.57 μM) and filament clearance (K1/2 = 0.38 μM) are indeed similar, suggesting that these activities are related mechanistically. Our results show that Srs2 stimulates the rate of ATP hydrolysis within the Rad51 filament, leading to efficient clearing of Rad51 molecules off DNA, and this activity is dependent on its physical interaction with Srs2. Whether this protein-protein interaction is solely required to engage the Rad51 filament or is an active participant in stimulating Rad51’s ATPase activity remains to be tested.
UvrD Nonspecifically Antagonizes Rad51 Filaments
The SF-1 helicase/translocase, E. coli UvrD, also can disassemble yeast Rad51 filaments (Veaute et al., 2005), although UvrD lacks the C-terminal region of Srs2 that is required for Rad51 binding interactions (Figure 1A). We therefore examined the UvrD concentration dependence for Rad51 filament clearance using the same kinetic assay (Figure 7A). The linear relationship between UvrD concentration and filament clearing activity is consistent with a non-catalytic mechanism for displacement of Rad51. Correspondingly, we do not detect binding of UvrD to Rad51 (Figure 7B), nor does UvrD stimulate ATP hydrolysis by Rad51 during Rad51 filament disassembly (Figure 7C). In addition, high concentrations of UvrD are required to destabilize Rad51 filaments in EM experiments (Figure 7D). These results suggest that UvrD displaces Rad51 by directly competing with Rad51 for DNA binding sites. In contrast, Srs2 uses a catalytic mechanism involving a physical interaction with Rad51 that stimulates ATP turnover and translocation by Srs2 to displace Rad51 from the DNA.
Figure 7. Mechanism of Clearing Rad51 Filaments by Srs2.
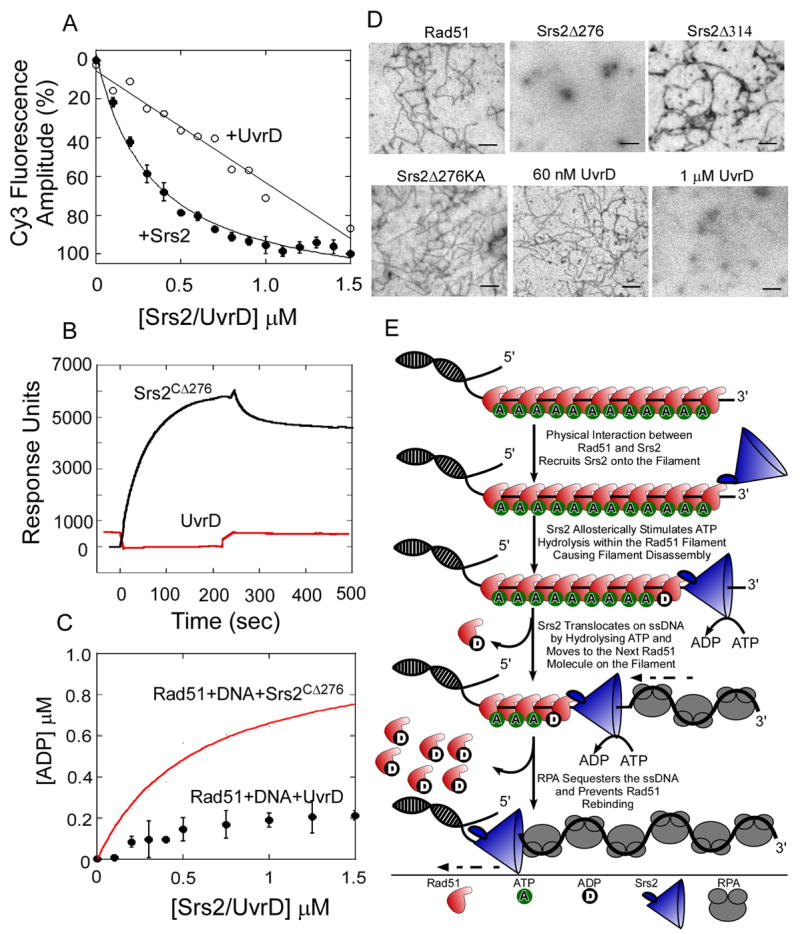
(A) In stop flow experiments, preformed Rad51 filaments were rapidly mixed with increasing concentrations of Srs2CΔ276 (●) or UvrD (○). The relative change in fluorescence amplitude is plotted as a function of Srs2/UvrD concentration. (B) Rad51 was tethered onto a SPR chip and 7 μM UvrD (red) or Srs2CΔ276 (black) was passed over the chip. The change in response units (RU) is plotted versus time. Srs2CΔ276 binds to Rad51 but no interaction between Rad51 and UvrD is observed. (C) UvrD does not stimulate the rate of ATP hydrolysis within a Rad51-DNA filament (●) unlike Srs2CΔ276 (red trace). (D) EM analysis of Rad51 filaments formed on M13 mp18(+) ssDNA and challenged with RPA, along with 60 nM Srs2CΔ276 , Srs2CΔ314 , Srs2 Srs2CΔ276 KA, UvrD, or 1 μM UvrD. 60 nM Srs2CΔ276 or 1 μM UvrD are capable of clearing filaments, whereas Rad51 filaments are intact when challenged with 60 nM Srs2CΔ314, Srs2CΔ276 KA, or UvrD. (E) The physical interaction between Srs2 and Rad51 is required for Srs2 to gain access onto the Rad51 nucleoprotein filament. ATP hydrolysis within the Rad51-DNA filament, allosterically modulated by Srs2, promotes filament disassembly, and the ssDNA translocation activity of Srs2 enables processive 3′ to 5′ clearing of the filament. The free DNA is sequestered by RPA and prevents reformation of the Rad51 filament.
DISCUSSION
Srs2 functions as an anti-recombinase by catalyzing the displacement of the Rad51 recombinase from presynaptic filaments (Krejci et al., 2003; Veaute et al., 2003). It was reasonable to assume that the ATP-dependent motor activity of Srs2 on ssDNA would be sufficient to displace Rad51 from the DNA (Singleton et al., 2007). However, our findings indicate that the Rad51 filament is resistant to clearing by Srs2 when ATP hydrolysis is impaired by mutation of Rad51 (Figure 6C). This suggests that Rad51 is an active participant in the clearing of presynaptic filaments by Srs2. We show that Srs2 stimulates ATP hydrolysis within the Rad51 filament, causing the Rad51 molecules to disassociate from the DNA. A specific protein-protein interaction between the C-terminal region of Srs2 and Rad51 is required for the clearing of presynaptic Rad51 filaments as well as the stimulation of ATP hydrolysis by Srs2. We hypothesize that once Srs2 loads onto the filament through a physical interaction with Rad51, it clears Rad51 filaments by allosterically activating ATP hydrolysis within Rad51 (Figure 7E). In this mechanism of remodeling the Rad51 presynaptic filament, the ATP-dependent ssDNA translocation activity of Srs2 may simply position Srs2 for binding to each successive Rad51 subunit and promote its dissociation from DNA.
Crystal structures of Rad51 and bacterial RecA-type homologs indicate that ATP binds at the interface between subunits of the protein polymer (Conway et al., 2004; Qian et al., 2006; Zhang et al., 2008). The physical interaction of Srs2 with Rad51 may alter packing interactions between Rad51 monomers within the filament, configuring the Rad51 active site for turnover of ATP. The energy required for this conformational change could be contributed by the binding interactions between Srs2 and Rad51, and/or by the mechanical motion of Srs2 on ssDNA. Once a Rad51 molecule is displaced from DNA, Srs2 can translocate along the ssDNA to the next Rad51 subunit in the filament to continue the disassembly process.
Although Srs2 binds to ssDNA with high affinity (KD = 32 ± 4 nM; data not shown), it is not known how Srs2 loads onto DNA coated with Rad51. The physical interaction of Srs2 with Rad51 might assist loading onto the filament by triggering Rad51 release and providing access to a ssDNA binding site. Once Srs2 is loaded on the DNA, the ssDNA would become available for translocation and processive clearing of Rad51. In addition, the capacity of Srs2 to invade Rad51 filaments at internal sites (Figures 3C, 3D & S4) may increase the efficiency of clearing longer Rad51 filaments several kilobases in length to prevent recombination.
The C-terminal region of Srs2 can be envisioned as a control node that integrates the motor activity of Srs2 with ATP turnover in the Rad51 filament. Srs2 functions in the recovery from checkpoint mediated cell cycle arrest, and several sites of cell cycle-dependent phosphorylation are also located within the C-terminal region of Srs2 (Chiolo et al., 2005). These posttranslational modifications of Srs2 could potentially regulate interactions with Rad51 filaments, enhancing the spatiotemporal control of anti-recombinogenic activity.
EXPERIMENTAL PROCEDURES
Oligonucleotides and Proteins
Unlabeled, Cy3-labeled, and fluorescein isothiocyanate-labeled DNA oligonucleotides were synthesized using an ABI 394 DNA synthesizer and gel purified before annealing. The C-terminal deletion constructs of Srs2, Srs2CΔ276 and Srs2CΔ314, were overexpressed in E. coli and purified as described (Krejci et al., 2003) with minor modifications (supplemental data). The active site mutant Srs2CΔ276-K41A was generated using site-directed mutagenesis (Quickchange, Stratagene Inc.), and purified identically. The yRad51 expression plasmid was a kind gift from Dr. Patrick Sung (Yale University) and used to generate mutants of yRad51 by site directed mutagenesis. Both WT and mutant Rad51 proteins were purified as described (Conway et al., 2004). UvrD was purified as described (Runyon et al., 1993) and RPA was a kind gift from Dr. Peter Burgers (Washington University). The Srs2 C-terminal peptide (CTP, residues 861-898) was synthesized using solid-phase synthesis and purified by HPLC, dried and resuspended in 50 % acetonitrile.
Rad51 Filament Assembly and Dissociation Kinetics
Stopped flow fluorescence experiments were carried out in a SX2000 instrument (Applied Photophysics, UK). The mixing schemes for the various experiments are diagrammed in the respective figures. Equal volumes from both syringes were mixed together at 25 °C, and the change in Cy3 fluorescence was monitored using a 530 nm long pass filter after excitation at 515 nm. All stopped flow experiments were carried out in SF-Buffer (50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, and 50 mM NaCl).
Single strand DNA (Cy3-dT) (oligodeoxythymidylate (dT)) labeled at the 3’ or 5’ terminus with Cy3 (40 nM) and 5 mM ATP were rapidly mixed with increasing concentrations of Rad51 (0 –20 μM). Two control experiments were conducted to normalize the experimental time courses. The ssDNA only control was obtained by rapidly mixing ssDNA with buffer. The filament control was obtained by rapidly mixing Rad51:DNA filament with buffer. Each control yielded a constant fluorescence signal that was used to determine the percent change in Cy3 fluorescence for each experimental time course using equation (1).
| (1) |
The effect of nucleotides on Rad51 DNA filament assembly was measured by rapidly mixing a solution of Cy3-dT79 (40 nM) and 5 mM nucleotide (ATP, ADP, AMP-PNP, or ATPγS) with Rad51 (4 μM). The fluorescence signal measured in the presence of ATP was considered maximal (100%) relative to the ssDNA only control. The dissociation rate of Rad51 from ssDNA was measured by incubating Rad51 (4 μM), Cy3-dT79 (40 nM), and ATP (5 mM) for 1 min then rapidly mixed the solution with excess unlabeled dT79 competitor DNA (40 μM). The apparent dissociation rates for the various time courses were determined by fitting the data to a single exponential (2), where A and k are the amplitude and the rate constant of the fluorescence change corresponding to spontaneous filament disassembly:
| (2) |
Kinetics of Rad51 Filament Clearing by Srs2
Rad51-(Cy3-dT)-ATP filaments were assembled by preincubating Rad51 (4 μM), 5′ or 3′ Cy3-dT79 (40 nM), and ATP (5 mM) followed by rapidly mixing with increasing concentrations of Srs2 (0 – 3 μM). Two control experiments were conducted to normalize the experimental time courses as above. In addition to the filament control a Srs2 disassembled filament control was conducted by preparing a solution of ATP (5 mM), Cy3-dT79 (40 nM), and Srs2 (3 μM) then rapidly mixing it with buffer. Each control yielded a constant fluorescence signal (see figure S5) that was used to determine the percent change in Cy3 fluorescence for each filament clearing time course using equation (3).
| (3) |
The filament clearing time courses were analyzed by fitting to a single exponential (1) to obtain an apparent filament clearing rate constant (k = kclear) and amplitude change (A). The amplitudes were then plotted as a function of Srs2 concentration and the resulting plot fit to a hyperbola to obtain K1/2 values.
We were also able to fit the data reasonably well to a double exponential equation, capturing a rapid but small amplitude component at early time points that is independent of Srs2 concentration and a slower component that consists of the majority of the amplitude change and is dependent on Srs2 concentration. Similar K1/2 values, within error, were obtained with either analysis and the apparent kclear obtained from the single exponential fit was similar to the slower component in the two exponential fits. At this time we do not know what the fast phase is attributed to, but it appears to have little effect on the determined K1/2 values and thus the conclusions we have drawn from the data.
The clearing rates for Rad51-K191R and Rad51-E221D were measured similarly by pre-forming Rad51-(Cy3-dT))-ATP filaments with Rad51-K191R or Rad51-E221D (4 μM), Cy3-dT79 (40 nM), and ATP (5 mM) and rapidly mixed with Srs2 (3 μM). To study the effects of saturating or limiting concentrations of Rad51 on filament clearance by Srs2, Rad51 filaments were pre-formed with Cy3-dT79 (40 nM), ATP (5 mM), and either 4 μM or 20 μM Rad51, then rapidly mixed with Srs2 (3 μM). The apparent filament clearing rates for the various traces were measured by fitting the data to equation (1).
Srs2 ssDNA Translocation Kinetics
Srs2 ssDNA translocation experiments were performed under single round conditions (no rebinding of Srs2 to ssDNA) using a fluorescence stopped-flow assay to monitor the arrival of Srs2 at the 5’-end of the DNA using 5’-Cy3-(dT)L substrates of various lengths (L) as described (Fischer et al., 2004). For ssDNA translocation assays, Srs2CΔ276 or Srs2CΔ314 (25 nM, final) was pre-incubated with excess ssDNA (50 nM, final) in one syringe and reactions initiated by rapid mixing with buffer containing ATP (0.5 mM, final) and heparin (4 mg/ml, final). Heparin functions as a trap to sequester the Srs2 molecules as they dissociate from the DNA ends after a single round of translocation. All concentrations are denoted as the final concentrations, post-mixing. Analysis of the resulting time courses is discussed in supplemental experimental procedures.
SPR Analyses of Rad51-Srs2 Interactions
0.2 mg/ml of Rad51 or BSA (final volume of 100 μl in 10 mM sodium acetate, pH 4.0) was coupled to a sensor chip derivatized with 0.05 M N-hydroxysuccinimide and N-ethyl-N’-(dimethylaminopropyl)carbodiimide, and equilibrated in BC-Buffer (50 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, pH 8.0, 10 % glycerol, 150 mM NaCl, 5 mM β-mercaptoethanol, and 0.01 % NP-40). Increasing concentrations of Srs2 (0 – 40 μM; or 7 μM UvrD) in 100 μl reaction volume were passed over the chip. The increase in response units (RU) was measured over 200 seconds during the binding reaction, followed by washing with BC buffer and monitoring disassociation for an additional 200 seconds. The chip was regenerated in between runs with a mixture of 10 μl 0.2 %SDS and 10 mM NaOH. The change in RU at ΔT = 0 sec was subtracted from RU at ΔT = 200 sec to obtain the change in amplitude. The RU amplitude change was plotted against Srs2 concentration and fit to a hyperbola to obtain a KD for the interaction.
ATPase Assays
To analyze the K1/2 for ATP hydrolysis within the Rad51 filament, 50 μM of α32P-ATP was mixed with 2 μM Rad51 and 40 nM dT79 DNA in 10 μl reactions in ATPase buffer (120 mM Tris-Cl, pH7.5, 50 mM NaCl, 10 mM MgCl2, 0.5 mM βME, and 100 μg/ml BSA) and incubated for 30 seconds at 25 °C. Different concentrations of Srs2 (0 – 1.5 μM) was added along with 25 mM unlabeled ATP-Mg2+. The reaction was incubated for an additional 30 seconds, and stopped by adding 10 μl of 0.5 M EDTA. ADP formation was monitored by spotting 1 μl of the reaction on a TLC plate, dried, and resolved with 0.6 M phosphate buffer (pH 3.4).
The rate of ATP hydrolysis during filament clearing was measured on a KinTek Quenched Flow System using a sequential mixing setup. To form the nucleoprotein filaments, equal volumes (18 μl each) of 4 μM Rad51, 40 nM dT79 DNA and 50 μM 32P-ATP in the SF-Buffer were rapidly mixed for 30 sec (t1) during the first mixing. This reaction was then sequentially mixed for varying times (t2 = 0 – 30 sec) with 3 μM Srs2CΔ276. The reaction was quenched with 70 μl quench solution (1M HCl), neutralized with 27 μl neutralization buffer (2.5 M NaOH, 0.5 M EDTA and 0.5 M Tris). ADP formation was monitored by spotting 1 μl of the reaction on a TLC plate, dried, and resolved with 0.6 M phosphate buffer (pH 3.4).
Electron Microscopy
EM analyses of Rad51 filaments were performed as described (Krejci et al., 2003). Srs2 or UvrD (60 nM or 1 μM) were mixed with RPA (350 nM) then added to Rad51 filaments assembled on M13mp18(+) ssDNA (2.4 μM Rad51; 7.5 μM nucleotides in M13 DNA) in EM buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10 % glycerol and 0.5 mM EDTA) and incubated at 37 °C for 3 minutes. 3 μl of the reaction was spotted onto freshly glow discharged carbon-coated copper grids for 2 min. The grids were rinsed two times in R buffer and stained with 0.75% uranyl formate (Electron Microscopy Sciences, Hatfield, PA) for 30 sec. Excess liquid was wicked away and the grids were allowed to air dry. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA). Images were acquired at 40,000x or 60,000x magnification.
Supplementary Material
Acknowledgments
We thank Patrick Sung for the kind gift of the yRad51 protein overexpression plasmid, Jurek Majka, and Bill Nolan for assistance with experiments, and Wandy Beatty for the EM analysis. This work was supported by research grants to TE (R01GM059902 and NCI-PO1CA092584), TML (GM045948), and LK (WT076476).
ABBREVIATIONS
- HR
Homologous Recombination
- DSBs
Double Stranded Breaks
- ssDNA
Single Stranded DNA
- dsDNA
Double Stranded DNA
- nt
nucleotide
- bp
base pair
- aa
amino acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J Biol Chem. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Modesti M, Ristic D, van der Heijden T, Dekker C, van Mameren J, Peterman EJ, Wuite GJ, Kanaar R, Wyman C. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Morgan EA, Shah N, Symington LS. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol Cell Biol. 2002;22:6336–6343. doi: 10.1128/MCB.22.18.6336-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Qian X, He Y, Ma X, Fodje MN, Grochulski P, Luo Y. Calcium stiffens archaeal Rad51 recombinase from Methanococcus voltae for homologous recombination. J Biol Chem. 2006;281:39380–39387. doi: 10.1074/jbc.M607785200. [DOI] [PubMed] [Google Scholar]
- Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- Runyon GT, Wong I, Lohman TM. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen C, Fu X, Gu S, Mao Y, Xie Y, Huang Y, Li Y. A meta-analysis of DNA repair gene XPC polymorphisms and cancer risk. J Hum Genet. 2008;53:18–33. doi: 10.1007/s10038-007-0215-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


