Abstract
Background
The true water bugs are grouped in infraorder Nepomorpha (Insecta: Hemiptera: Heteroptera) and are of great economic importance. The phylogenetic relationships within Nepomorpha and the taxonomic hierarchies of Pleoidea and Aphelocheiroidea are uncertain. Most of the previous studies were based on morphological characters without algorithmic assessment. In the latest study, the molecular markers employed in phylogenetic analyses were partial sequences of 16S rDNA and 18S rDNA with a total length about 1 kb. Up to now, no mitochondrial genome of the true water bugs has been sequenced, which is one of the largest data sets that could be compared across animal taxa. In this study we analyzed the unresolved problems in Nepomorpha using evidence from mitochondrial genomes.
Results
Nine mitochondrial genomes of Nepomorpha and five of other hemipterans were sequenced. These mitochondrial genomes contain the commonly found 37 genes without gene rearrangements. Based on the nucleotide sequences of mt-genomes, Pleoidea is not a member of the Nepomorpha and Aphelocheiroidea should be grouped back into Naucoroidea. Phylogenetic relationships among the superfamilies of Nepomorpha were resolved robustly.
Conclusion
The mt-genome is an effective data source for resolving intraordinal phylogenetic problems at the superfamily level within Heteroptera. The mitochondrial genomes of the true water bugs are typical insect mt-genomes. Based on the nucleotide sequences of the mt-genomes, we propose the Pleoidea to be a separate heteropteran infraorder. The infraorder Nepomorpha consists of five superfamilies with the relationships (Corixoidea + ((Naucoroidea + Notonectoidea) + (Ochteroidea + Nepoidea))).
Background
The true water bugs are grouped as the infraorder Nepomorpha, one of seven infraorders within the suborder Heteroptera (Insecta: Hemiptera) [1]. This group is of tremendous economic importance because all the members, except some Corixidae, are predators [2]. Extant water bugs have been grouped into 6 [3,4] or 5 superfamilies (Notonectoidea including Pleoidea) [2]. When molecular sequence data was combined with morphological characters to analyze the phylogenetic relationships of the true water bugs for the first time, a 7-superfamily classification system was proposed, containing a newly erected superfamily Aphelocheiroidea, which had formerly belonged to Naucoroidea (Table 1) [5].
Table 1.
Previous classification systems of Nepomorpha
| Štys and Jansson 1988; Mahner 1993 | Schuh and Slater 1995 | Hebsgaard et al. 2004 | |
| Nepoidea | Belostomatidae | Belostomatidae | Belostomatidae |
| Nepidae | Nepidae | Nepidae | |
| Corixoidea | Corixidae | Corixidae | Corixidae |
| Ochteroidea | Ochteridae | Ochteridae | Ochteridae |
| Gelastocoridae | Gelastocoridae | Gelastocoridae | |
| Notonectoidea | Notonectidae | Notonectidae | Notonectidae |
| Pleidae | |||
| Helotrephidae | |||
| Pleoidea | Pleidae | Pleidae | |
| Helotrephidae | - | Helotrephidae | |
| Naucoridae | Naucoridae | Naucoridae | Naucoridae |
| Potamocoridae | Potamocoridae | ||
| Aphelocheiridae | Aphelocheiridae | ||
| Aphelocheiridae | - | - | Potamocoridae |
| Aphelocheiridae |
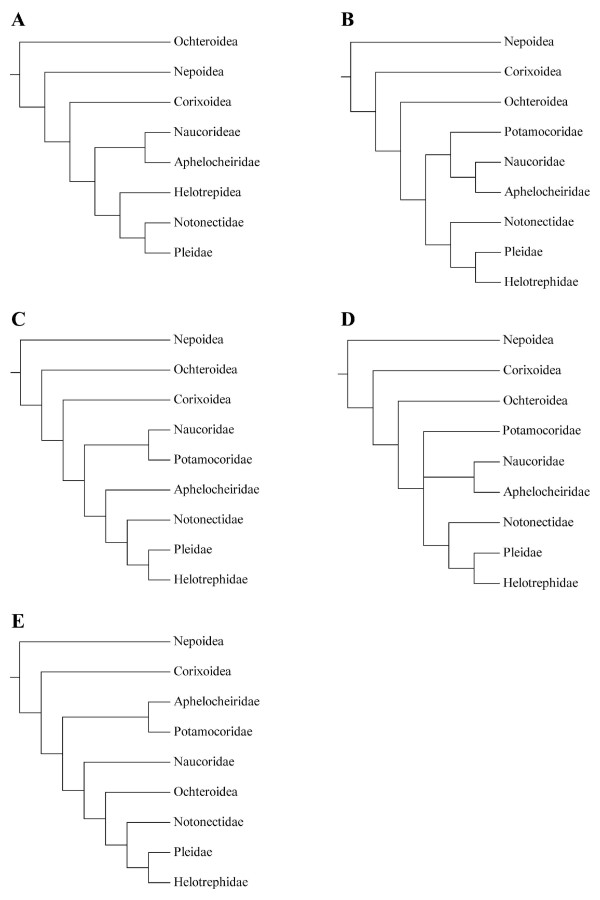
In these classification systems, the contents of Nepoidea, Corixoidea, and Ochteroidea are constant. Phylogenetic relationships proposed previously at the superfamily or family level within Nepomorpha are summarized in Figure 1. The works of China (1955) [6], Popov (1971) [7], Rieger (1976) [8], and Mahner (1993) [3] were based on morphological characters without algorithmic analysis. In their landmark work, Hebsgaard et al. (2004) reviewed these studies in detail and analyzed the phylogeny of Nepomorpha using molecular sequence data (partial sequence of 16S rDNA and 28S rDNA) combined with morphological data for the first time, raising a new superfamily Aphelocheiroidea as part of a new phylogenetic hypothesis (Figure 1E) [5].
Figure 1.
Proposed phylogenetic hypotheses within Nepomorpha. A, after China (1955); B, after Popov (1971); C, after Rieger (1976); D, after Mahner (1993); E, after Hebsgaard et al. (2004).
Until now, the monophyly of each nepomorphan family, and the monophyly of superfamilies Nepoidea, Corixoidea, and Ochteroidea in Nepomorpha, have been generally accepted [3,5-8]. A close relationship between Pleidae and Helotrephidae has been supported by recent studies [3,5,7,8] and the monophyly of Aphelocheiridae + Potamocoridae was supported in the latest comprehensive study [5]. The phylogenetic relationships within Nepomorpha, however, have not reached full agreement. The unsolved problems are: 1) whether the Aphelocheiridae and Potamocoridae should be members of Naucoroidea or be raised as a separate superfamily Aphelocheiroidea; 2). whether the Pleidae and Helotrephidae should be members of Notonectoidea or be raised as a separate superfamily Pleoidea; 3) the phylogenetic relationships among the superfamilies.
The mitochondrial genome (mt-genome) is one of the largest sets of homologous genes which can be compared across animal taxa and has become an effective data source for resolving deep-level phylogenetic problems [9,10]. Within Insecta, more than one hundred mt-genomes are available now in GenBank/DDBJ/EMBL and mt-genomes have been shown to resolve intraordinal relationships, such as in Diptera [11], Hymenoptera [12], and Orthoptera [13]. There are many possible ways of using mt-genomes in phylogenetic analyses, for example by using different genes, amino acid sequences or nucleotide sequences. Using the nucleotide sequences of all available genes clearly has been shown to be the best way to extract a phylogenetic signal from mt-genomes [11,13].
Previous molecular data used for the analysis of nepomorphan relationships was about 1 kb [5] and no mt-genome data are available for any nepomorphan species. In this study, fourteen new mt-genomes were sequenced, nine of them belonging to the infraorder Nepomorpha (Table 2) (one mt-genome from our previous paper was also included in Table 2[14]). A preliminary phylogenetic framework of Nepomorpha is proposed using mt-genome data, and the relationships of Pleoidea and Aphelocheiroidea are analyzed.
Table 2.
General informatics of the taxa used in this study
| Classification | Species | Mt-genome Completeness | Size (bp) | A+T % | Accession Number |
| Archaeorrhyncha | |||||
| Fulgoroidea | |||||
| Fulgoridae | Lycorma delicatula | complete | 15410 | 76.3 | FJ456942 |
| Heteroptera | |||||
| Gerromorpha | |||||
| Hydrometroidea | |||||
| Hydrometridae | Hydrometra sp. | complete | 15416 | 78.7 | FJ456945 |
| Gerroidea | |||||
| Gerridae | Gerris sp. | complete | 15380 | 75.7 | FJ456944 |
| Leptopodomorpha | |||||
| Leptopodoidea | |||||
| Leptopodidae | Leptopus sp. | tRNA(I) – 12S | 14516 | 72.4 | FJ456946 |
| Cimicomorpha | |||||
| Reduvioidea | |||||
| Reduviidae | Valentia hoffmanni | complete | 15625 | 73.7 | FJ456952 |
| Pentatomomorpha | |||||
| Coreoidea | |||||
| Rhopalidae | Stictopleurus subviridis | complete | 15319 | 75.7 | EU826088* |
| Nepomorpha | |||||
| Ochteroidea | |||||
| Gelastocoridae | Nerthra sp. | complete | 16079 | 74.2 | FJ456943 |
| Ochteridae | Ochterus marginatus | tRNA(I) – 12S | 14609 | 72.7 | FJ456950 |
| Corixoidea | |||||
| Corixidae | Sigara septemlineata | complete | 15724 | 75.2 | FJ456941 |
| Notonectoidea | |||||
| Notonectidae | Enithares tibialis | complete | 15262 | 76.1 | FJ456949 |
| Nepoidea | |||||
| Belostomatidae | Diplonychus rusticus | tRNA(I) – 12S | 14596 | 69.8 | FJ456940 |
| Nepidae | Laccotrephes robustus | complete | 15321 | 70.6 | FJ456948 |
| Naucoroidea | |||||
| Naucoridae | Ilyocoris cimicoides | complete | 15209 | 71.0 | FJ456947 |
| Aphelocheiroidea | |||||
| Aphelocheiridae | Aphelocheirus ellipsoideus | tRNA(I) – 12S | 14574 | 74.7 | FJ456939 |
| Pleoidea | |||||
| Pleidae | Paraplea frontalis | complete | 15130 | 76.5 | FJ456951 |
* From Hua et al. (2009) [14].
Results
General features of the fourteen mt-genomes of Hemiptera
Ten complete and four nearly complete mt-genomes of Hemiptera were sequenced (Table 2). Because of the polynucleotide regions, the control regions of the four incomplete mt-genomes were difficult to sequence. All mt-genomes are similar to the typical insect mt-genomes, containing the commonly found 37 genes in the same gene order as observed in Drosophila yakuba [GenBank: NC001322] [15]. No gene rearrangement or duplication has been found, indicating that the organizations of the mt-genomes in the suborder Heteroptera are more stable than those in the suborder Sternorrhyncha (Insecta: Hemiptera) [16]. Family names are used instead of the species names in following discussion for brevity since only a single representative was selected for each family.
Ten completely sequenced mt-genomes range in length from 15130 bp (Pleidae) to 16079 bp (Gelastocoridae). Lengths of the 37 coding genes of each mt-genome range from 14372 bp (Fulgoridae) to 14637 bp (Notonectidae), while the size of the control regions ranges from 608 bp (Pleidae) to 1450 bp (Gelastocoridae) (Table 3). Variance of the mt-genome size is mainly derived from the control region.
Table 3.
Statistics of the length and nucleotide composition of the genes
| Taxa | Lengths of total genes | AT% of total genes | Lengths of control region | AT% of control region |
| Fulgoridae | 14372 | 75.8 | 1043 | 83.3 |
| Hydrometridae | 14478 | 78.6 | 694 | 78.1 |
| Gerridae | 14616 | 73.1 | 781 | 66.2 |
| Leptopodidae | 14532 | 72.4 | - | - |
| Reduviidae | 14570 | 73.8 | 724 | 69.9 |
| Rhopalidae | 14563 | 75.6 | 685 | 77.1 |
| Gelastocoridae | 14631 | 75.1 | 1450 | 65.4 |
| Ochteridae | 14601 | 72.7 | - | - |
| Corixidae | 14504 | 75.4 | 772 | 76.7 |
| Notonectidae | 14637 | 76.3 | 646 | 70.4 |
| Belostomatidae | 14573 | 69.8 | - | - |
| Nepidae | 14575 | 70.9 | 751 | 65.8 |
| Naucoridae | 14611 | 71.2 | 609 | 66.0 |
| Aphelocheiridae | 14582 | 74.8 | - | - |
| Pleidae | 14559 | 76.5 | 608 | 75.0 |
Nucleotide compositions of these mt-genomes are AT-biased (Table 3). Though the control regions of four mt-genomes were not completely sequenced, this biased trend is obvious even without the control region sequence. Interestingly, in most of the completely sequenced mt-genomes, the control regions are not the most AT-rich regions (Table 3), which has been found in other insect mt-genomes [15,17]
Intergenic spacers ranging from 14 bp (Corixidae) to 23 bp (Reduviidae) were found between tRNA-Ser(UCN) and ND1 in all mt-genomes except Fulgoridae. This spacer has also been reported in other insects and some conserved motifs were identified [18-20]. In the present study, no perfectly conserved motif was found but a conserved region was identified (see additional file 1). Additionally, a unique 222 bp intergenic spacer was found between tRNA-Gly and ND3 in Hydrometridae which lacks significant BLAST similarity (megablast in nucleotide collection). The exact origin and function of such spacers are unclear, but they may be the vestiges of pseudogenes generated by the gene duplication-random loss process of rearrangement [21]
Other features of these mt-genomes, including non-traditional start codons such as TTG of CO1 in Naucoridae and GTG of ND1 in Aphelocheiridae, incomplete stop codons T or TA, and absence of the DHU arm in the secondary structure of tRNA-Ser(GCU), are found commonly in insect mt-genomes [15,18,20,22-24].
Phylogenetic analyses
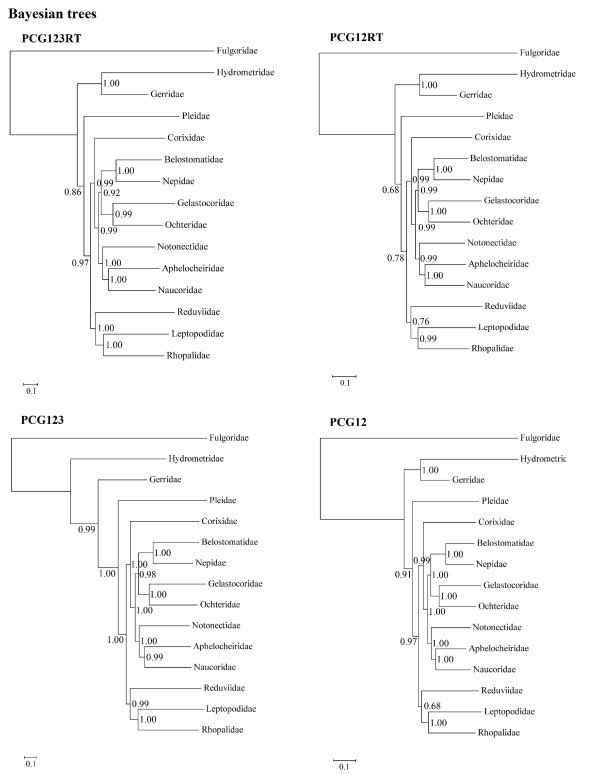
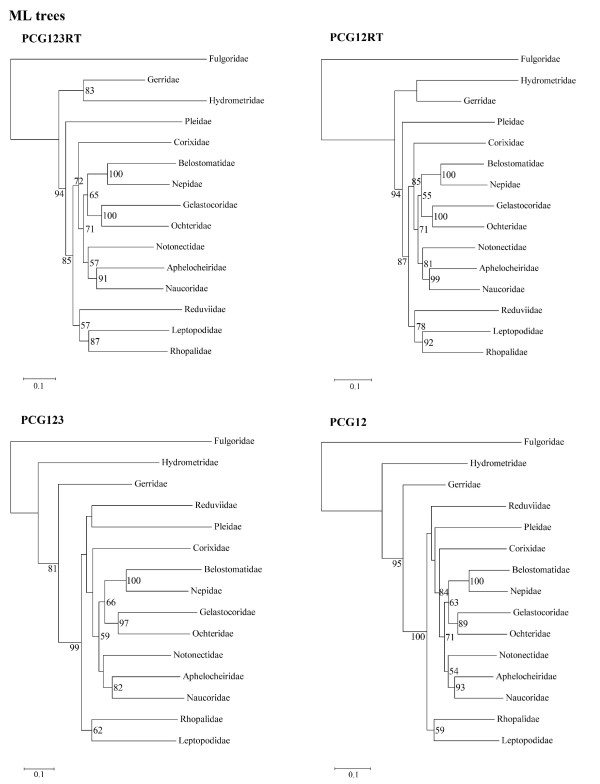
For the fifteen species, there are 14968 sites in the PCG123RT matrix (containing all three codon positions for protein coding genes (PCGs), plus the whole of the rRNA and tRNA genes), 11236 sites in the PCG12RT matrix (containing the first and the second codon positions of PCGs, plus the whole of the rRNA and tRNA genes), 11196 sites in the PCG123 matrix (containing all three codon positions of PCGs), and 7464 sites in the PCG12 matrix (containing the first and the second codon positions of PCGs). From Bayesian and ML inferences, these four matrices generated eight fully bifurcated trees with similar topology (Figure 2, Figure 3). The monophyletic Nepoidea and Ochteroidea were consistently recovered. The monophyly of the remaining nepomorphan superfamilies such as Pleoidea, Corixoidea, Notonectoidea, Naucoroidea, and Aphelocheiroidea could not be analyzed in this study as only a single representative was sampled. In all trees, the relationships within Nepomorpha were found to be constant. The Pleoidea (Pleidae) was recognized as the sister group of the clade including the infraorders Cimicomorpha (Reduviidae), Leptopodomorpha (Leptopodidae), Pentatomomorpha (Rhopalidae), and the remaining traditional Nepomorpha. Nepoidea and Ochteroidea were sister groups and this clade was found to be a sister group of Notonectoidea (Notonectidae) plus Naucoroidea (Naucoridae and Aphelocheiridae).
Figure 2.
Bayesian phylograms inferred from PCG123RT, PCG12, PCG123, and PCG12 data sets. Bayesian posterior probabilities are indicated at each node.
Figure 3.
ML phylograms inferred from PCG123RT, PCG12, PCG123, and PCG12 data sets. Bootstrap support values are indicated at each node.
Discussion
Mitochondrial genomes
The mt-genomes sequenced in this study are similar to the mt-genomes of other insects and contain very few novel features (see additional file 2 for the descriptions of these mt-genomes). It has been reported that the hemipteroid insects (including Hemiptera, Thysanoptera, Psocoptera, and Phthiraptera) have experienced increased rates of mt-genomic gene rearrangements [16,25-29]. Gene content and gene order of the mt-genomes sequenced in this study, however, are all the same as observed in Drosophila yakuba [GenBank: NC001322) [15] except for some unique intergenic spacers. In the present study, gene order provides no phylogenetic information.
Phylogenetic analyses
Mt-genomes provide abundant phylogenetic signal because they possess large sets of homologous genes. Multiple genes with increased sequence lengths are favorable for accurate phylogenetic analyses [30-32]. It has been shown that the best way to use mt-genomes in phylogenetic analyses is to combine all the coding genes and use nucleotide sequence data [13]. We used a data set consisting of all 37 genes (PCG123RT data set) to infer relationships within Nepomorpha. The PCG12RT, PCG123, and PCG12 data sets were analyzed to test the stability of the phylogenetic hypotheses to the inclusion of different portions of the data. Phylogenetic relationships among heteropteran infraorders are still controversial [33-36]. Taxon sampling for this study is too limited to analyze this problem in full and data for the infraorders Enicocephalomorpha and Dipsocoromorpha are not yet available. We focused on the phylogeny within infraorder Nepomorpha, especially relationships among superfamilies, using mt-genome sequences for the first time. Finally, five of the eight analyses inferred the same topology, with the remaining three tree resulting form less complete datasets (nucleotide substitution saturation analysis was also added in additional file 3) differing at a few nodes, which indicates that the mt-genome is an effective data source for resolving phylogenetic problems within Nepomorpha at the superfamily level. Because the ML and Bayesian algorithms are not sensitive to possible long-branch attraction and allow evolutionary modeling of the data [37-39], we do not think our hypotheses are artifacts.
The Problem of Pleoidea
The Pleoidea has been proposed to include Pleidae and Helotrephidae [3,5]. The mt-genome of Helotrephidae was not sequenced in this study. This family has been generally accepted as the sister group of Pleidae [3,5,7,8] except in the study of China (1955) [6].
Previous studies based on morphological characters, molecular data or combined data consistently support a monophyletic Nepomorpha [3,5-8]. In this study, however, a monophyletic Nepomorpha is supported by one analysis of the eight performed. In all previous studies, Pleoidea is the sister group of Notonectoidea [3,5,7,8] and has always been included in Notonectoidea [2]. A surprising, but strongly supported result from the mt-genome analysis was that Pleoidea is not part of Nepomorpha, but rather the sister group of the clade including Nepomorpha, Leptopodomorpha, Cimicomorpha, and Pentatomomorpha (Figure 2, Figure 3).
Based on these results we propose that the Pleoidea could be raised from a superfamily to the infraorder Plemorpha, and that the infraorder Nepomorpha consists of the remaining nepomorphans except Pleoidea. The phylogenetic position of the infraorder Plemorpha within suborder Heteroptera needs further study because mt-genome data from infraorders Enicocephalomorpha and Dipsocoromorpha are unavailable at the present time.
The Problem of Aphelocheiroidea
The recently proposed superfamily Aphelocheiroidea (including Aphelocheiridae and Potamocoridae) [5], which was considered as part of Naucoroidea by other researchers [2,3], was proposed to be the sister group of a clade consisting of Ochteroidea, Notonectoidea, and Naucoroidea [5]. In the present analysis, Aphelocheiridae and Naucoridae consistently formed a monophyletic clade with high data support (Figure 2, Figure 3). Although the mt-genome of Potamocoridae was not sequenced in this study, this family has been suggested to be the sister group of Aphelocheiridae [5], Naucoridae [8], Naucoridae + Aphelocheiridae [7], a subfamily within Naucoridae [6], or unresolved by Mahner (1993) [3]. Here we propose that the Aphelocheiroidea raised by Hebsgaard et al. (2004) [5] should be grouped within Naucoroidea again.
Phylogeny of "Nepomorpha"
The traditional infraorder Nepomorpha is not monophyletic and it should only contain Corixoidea, Nepoidea, Ochteroidea, Notonectoidea, and Naucoroidea. The superfamilies Nepoidea (including Nepidae and Belostomatidae) and Ochteroidea (including Ochteridae and Gelastocoridae) are monophyletic and sister-groups in all trees with high support, as is generally accepted [3,5-8]. Nepoidea has been considered as the most basal branch of Nepomorpha in previous studies [3,5,7,8]. The position of Ochteroidea varied between previous researchers [3,5-8]. Because of the novel position inferred for Pleoidea, the sister-group relationship between Pleoidea and Notonectoidea which has been consistently proposed [3,5-8] was not supported and Notonectoidea was inferred to be the sister group of Naucoroidea.
The placement of Corixoidea is very different from previous hypotheses. Traditionally, this superfamily was placed as the sister group of a clade composed of Naucoroidea, Ochteroidea, and Notonectoidea [3,5,7] or the sister group of a clade composed of Naucoroidea and Notonectoidea [6,8]. In our results, Corixoidea is always the most basal clade within Nepomorpha, with Naucoroidea, Notonectoidea, Ochteroidea, and Nepoidea forming a monophyletic group.
Furthermore, Lycorma delicatula (Insecta: Hemiptera: Archaeorrhyncha) was removed from the data sets and the phylogeny was re-analyzed with the same methods. Same position of Pleoidea and phylogenetic hypotheses within Nepomorpha could be drawn (see additional file 4). Finally, the infraorder Nepomorpha should consist of five superfamilies with the phylogenetic hypothesis of (Corixoidea + ((Naucoroidea + Notonectoidea) + (Ochteroidea + Nepoidea))).
Conclusion
Although previous studies based on morphological characters alone or combined with DNA sequence no longer than 1 kb confirmed the monophyly of Nepomorpha, the phylogenetic inference with the evidence from mitochondrial genomes in this study supports the raise of a separate infraorder Plemorpha which belonged to Nepomorpha before. The well-resolved nepomorphan phylogenetic relationships at superfamily level allow a better understanding of evolutionary patterns within this group and provide a robust framework for comparative studies of nepomorphans. The present study demonstrates the great effectiveness of mitochondrial genome for inferring phylogenetic relationships at superfamily level. Furthermore, this study also suggests the need of using multiple genes for future phylogenetic analyses of highly debated phylogenies.
Methods
According to the seven-superfamily system of Nepomorpha proposed by Hebsgaard et al. (2004) [5], the representatives of each superfamily were selected (Table 2). Representatives of the infraorders Gerromorpha, Leptopodomorpha, Cimicomorpha and Pentatomomorpha were also included (Table 2). A representative of the suborder Archaeorrhyncha (Insecta: Hemiptera), Lycorma delicatula (White), was chosen to root the trees.
A single individual of each species was preserved in 95% ethanol at -20°C and total genomic DNA was extracted using the method based on CTAB [40]. PCRs were performed with TaKaRa LA PCR Kit Ver.2.1 following the manufacturer's recommendations. The primers are listed in additional file 5. PCR products were electrophoresed in 0.7% agarose gel, purified, and then both strands were sequenced with primer walking by Beijing Sunbiotech Co. Ltd.
The complete sequences of each gene were used for phylogenetic analysis (excluding stop codons of the PCGs). All PCGs were aligned based on amino acid sequence alignments in MEGA version 4.0 [41]. The rRNAs and the tRNAs were aligned with CLUSTAL X version 1.83 [42] under the default settings. Ambiguously aligned regions of PCGs and rRNA genes were carefully adjusted by hand. Transfer RNA alignments were corrected according to secondary structure. The aligned sequences were concatenated as four matrices used in phylogenetic analyses: 1) The PCG123RT matrix, including all three codon positions of PCGs, rRNA genes, and tRNA genes; 2) the PCG12RT matrix, including the first and the second codon positions of PCGs, rRNA genes, and tRNA genes; 3) the PCG123 matrix, including all the three codon positions of PCGs; 4) the PCG12 matrix, including the first and the second codon positions of PCGs.
MrBayes Version 3.1.1 [43] and a PHYML online web server [44] were employed to reconstruct the phylogenetic trees under the GTR model. In Bayesian inference, two simultaneous runs of 3,000,000 generations were conducted for each matrix. Trees inferred prior to stationarity were discarded as burn-in, and the remaining trees were used to construct a 50% majority-rule consensus tree. In ML analysis, the parameters were estimated during analysis and the node support values were assessed by bootstrap resampling (BP) [45] calculated using 100 replicates.
Abbreviations
CO1, CO2, and CO3: were the abbreviations of cytochrome c oxidase subunit I, II, and III genes; CytB: cytochrome b gene; ATP6 and ATP8: ATP synthase F0 subunit 6 and 8 genes; ND1, ND2, ND3, ND4, ND4L, ND5, ND6: NADH dehydrogenase subunit 1–6 and 4L genes. Transfer RNA genes were labeled according to the IUPAC-IUB single letter code for the specified amino acid; in cases where there was more than one tRNA for a particular amino acid, they were distinguished by their anticodons. PCG: protein coding gene.
Authors' contributions
JH designed the experiments, carried out the data analyses and drafted the manuscript. ML, PD, and YC participated in the experiments and helped prepare the additional files. QX helped draft the manuscript. WB directed this study and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Alignments of the conserved sequence blocks of the intergenic spacers between tRNA-S(TGA) and ND1. The data provided represent the alignments of the conserved sequence blocks of the intergenic spacers between tRNA-S(TGA) and ND1. The alignments were generated by plotting the identities to a standard as a dot, and a gap as a dash.
General information of the mt-genomes in this study. The data provided represent the putative secondary structure of tRNAs, codon usage in each mt-genome, and analyses of nucleotide compositions of each mt-genome.
Nucleotide substitution saturation analysis. The data provided represent the nucleotide substitution saturation analysis of different portions of datasets.
Bayesian and ML phylograms inferred from the data sets without Lycorma delicatula (Insecta: Hemiptera: Archaeorrhyncha). The data provided represent Bayesian and ML phylograms inferred from the data sets without Lycorma delicatula (Insecta: Hemiptera: Archaeorrhyncha).
Primers used in PCR. The data provided represent primers used in amplification of mt-genomes.
Acknowledgments
Acknowledgements
We thank Profs. Shuzhi Ren, and Guoqing Liu, Nankai University, for identifying the species of Sigara septemlineata and Enithares tibialis. Thanks two reviewers for reviewing the manuscript and providing very helpful advices. Thanks review 2 for his very kind annotation of the manuscript to help us to revise it. This project was supported by the Natural Science Foundation of China (No. 30725005 and No. J0630963), and by the Ministry of Education of China (No. 20050055027).
Contributor Information
Jimeng Hua, Email: nkhuajimeng@163.com.
Ming Li, Email: tnliming@yahoo.com.cn.
Pengzhi Dong, Email: pengzhiryn@163.com.
Ying Cui, Email: cql8179270@126.com.
Qiang Xie, Email: qiangxie@nankai.edu.cn.
Wenjun Bu, Email: wenjunbu@nankai.edu.cn.
References
- Štys P, Kerzhner I. The rank and nomenclature of higher taxa in recent Heteroptera. Acta Ent Bohemoslov. 1975;72:65–79. [Google Scholar]
- Schuh RT, Slater JA. True bugs of the world (Hemiptera: Heteroptera) classification and natural history. Cornell University Press, Ithaca, New York; 1995. [Google Scholar]
- Mahner M. Systema cryptoceratum phylogeneticum (Insecta, Heteroptera) Zoologica. 1993;143 [Google Scholar]
- Štys P, Jansson A. Check-list of recent family-group and genus-group names of Nepomorpha (Heteroptera) of the world. Acta Entomologica Fennica. 1988;50:1–44. [Google Scholar]
- Hebsgaard MB, Andersen NM, Damgaard J. Phylogeny of the true water bugs (Nepomorpha: Hemiptera-Heteroptera) based on 16S and 28S rDNA and morphology. Systematic Entomology. 2004;29:488–508. doi: 10.1111/j.0307-6970.2004.00254.x. [DOI] [Google Scholar]
- China WE. The evolution of the water bugs. Symposium on organic evolution; India Bulletin of the National Institute of Science. 1955. pp. 91–103.
- Popov YA. Trudy Paleontological Institute Academy of Science. Vol. 129. USSR: Nauk; 1971. Historical development of the hemipterous infraorder Nepomorpha; pp. 1–228. [Google Scholar]
- Rieger C. Skelett und Muskulatur des Kopfes und Prothorax von Ochterus marginatus Latreille. Zoomorphologie. 1976;83:109–191. doi: 10.1007/BF00993483. [DOI] [Google Scholar]
- Cameron SL, Beckenbach AT, Dowton M, Whiting MF. Evidence from mitochondrial genomics on interordinal relationships in insects. Arthropod Systematics & Phylogeny. 2006;64:27–34. [Google Scholar]
- Dellaporta SL, Xu A, Sagasser S, Jakob W, Moreno MA, Buss LW, Schierwater B. Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci USA. 2006;103:8751–8756. doi: 10.1073/pnas.0602076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL, Lambkin CL, Barker SC, Whiting MF. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Systematic Entomology. 2007;32:40–59. doi: 10.1111/j.1365-3113.2006.00355.x. [DOI] [Google Scholar]
- Cameron SL, Dowton M, Castro LR, Ruberu K, Whiting MF, Austin AD, Diement K, Stevens J. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome. 2008;51:800–808. doi: 10.1139/G08-066. [DOI] [PubMed] [Google Scholar]
- Fenn JD, Song H, Cameron SL, Whiting MF. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol Phylogenet Evol. 2008;49:59–68. doi: 10.1016/j.ympev.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Hua J, Dong P, Li M, Cui Y, Zhu W, Xie Q, Bu W. The analysis of mitochondrial genome of Stictopleurus subviridis Hsiao (Insecta: Hemiptera-Heteroptera: Rhopalidae) Acta Zootaxonomica Sinica. 2009;34:1–9. [Google Scholar]
- Clary DO, Wolstenholme DR. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Thao ML, Baumann L, Baumann P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha) BMC Evol Biol. 2004;4:25. doi: 10.1186/1471-2148-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-X, Hewitt GM. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochemical Systematics and Ecology. 1997;25:99–120. doi: 10.1016/S0305-1978(96)00042-7. [DOI] [Google Scholar]
- Cameron SL, Whiting MF. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 2008;408:112–123. doi: 10.1016/j.gene.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Hua J, Li M, Dong P, Cui Y, Xie Q, Bu W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera) BMC Genomics. 2008;9:610. doi: 10.1186/1471-2164-9-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 2008;25:2499–2509. doi: 10.1093/molbev/msn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda) BMC Genomics. 2006;7:182. doi: 10.1186/1471-2164-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL, Whiting MF. Mitochondrial genomic comparisons of the subterranean termites from the Genus Reticulitermes (Insecta: Isoptera: Rhinotermitidae) Genome. 2007;50:188–202. doi: 10.1139/G06-148. [DOI] [PubMed] [Google Scholar]
- Carapelli A, Vannini L, Nardi F, Boore JL, Beani L, Dallai R, Frati F. The mitochondrial genome of the entomophagous endoparasite Xenos vesparum (Insecta: Strepsiptera) Gene. 2006;376:248–259. doi: 10.1016/j.gene.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Salvato P, Simonato M, Battisti A, Negrisolo E. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae) BMC Genomics. 2008;9:331. doi: 10.1186/1471-2164-9-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacin C, Shao R, Cameron S, Barker SC. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta) Insect Mol Biol. 2006;15:63–68. doi: 10.1111/j.1365-2583.2005.00608.x. [DOI] [PubMed] [Google Scholar]
- Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- Shao R, Campbell NJ, Barker SC. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera) Mol Biol Evol. 2001;18:858–865. doi: 10.1093/oxfordjournals.molbev.a003867. [DOI] [PubMed] [Google Scholar]
- Shao R, Campbell NJ, Schmidt ER, Barker SC. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol. 2001;18:1828–1832. doi: 10.1093/oxfordjournals.molbev.a003970. [DOI] [PubMed] [Google Scholar]
- Cameron SL, Johnson KP, Whiting MF. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. J Mol Evol. 2007;65:589–604. doi: 10.1007/s00239-007-9042-8. [DOI] [PubMed] [Google Scholar]
- Mattern MY. Molecular phylogeny of the Gasterosteidae: the importance of using multiple genes. Mol Phylogenet Evol. 2004;30:366–377. doi: 10.1016/S1055-7903(03)00190-8. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 2000;17:437–455. doi: 10.1006/mpev.2000.0839. [DOI] [PubMed] [Google Scholar]
- Wortley AH, Rudall PJ, Harris DJ, Scotland RW. How much data are needed to resolve a difficult phylogeny? Case study in Lamiales. Syst Biol. 2005;54:697–709. doi: 10.1080/10635150500221028. [DOI] [PubMed] [Google Scholar]
- Schuh RT. Review of R. H. Cobben, Evolutionary trends in Heteroptera. Part II. Mouthpart -structures and feeding strategies. Systematic Zoology. 1979;28:653–656. doi: 10.2307/2412577. [DOI] [Google Scholar]
- Schuh RT. The influence of cladistics on heteropteran classification. Annual Review of Entomology. 1986;31:67–93. doi: 10.1146/annurev.en.31.010186.000435. [DOI] [Google Scholar]
- Wheeler WC, Schuh RT, Bang R. Cladistic relationships among higher groups of Heteroptera: congruence between morphological and molecular data sets. Entomol Scand. 1993;24:121–137. [Google Scholar]
- Xie Q, Tian Y, Zheng L, Bu W. 18S rRNA hyper-elongation and the phylogeny of Euhemiptera (Insecta: Hemiptera) Mol Phylogenet Evol. 2008;47:463–471. doi: 10.1016/j.ympev.2008.01.024. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Scally M, Madsen O, Stanhope MJ, de Jong WW, Catzeflis FM, Springer MS, Douzery EJ. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol Biol Evol. 2002;19:1656–1671. doi: 10.1093/oxfordjournals.molbev.a003989. [DOI] [PubMed] [Google Scholar]
- Holder M, Lewis PO. Phylogeny estimation: traditional and bayesian approaches. Nature Reviews Genetics. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 2006;63:826–841. doi: 10.1007/s00239-005-0293-y. [DOI] [PubMed] [Google Scholar]
- Reineke A, Karlovsky P, Zebitz CP. Preparation and purification of DNA from insects for AFLP analysis. Insect Mol Biol. 1998;7:95–99. doi: 10.1046/j.1365-2583.1998.71048.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments of the conserved sequence blocks of the intergenic spacers between tRNA-S(TGA) and ND1. The data provided represent the alignments of the conserved sequence blocks of the intergenic spacers between tRNA-S(TGA) and ND1. The alignments were generated by plotting the identities to a standard as a dot, and a gap as a dash.
General information of the mt-genomes in this study. The data provided represent the putative secondary structure of tRNAs, codon usage in each mt-genome, and analyses of nucleotide compositions of each mt-genome.
Nucleotide substitution saturation analysis. The data provided represent the nucleotide substitution saturation analysis of different portions of datasets.
Bayesian and ML phylograms inferred from the data sets without Lycorma delicatula (Insecta: Hemiptera: Archaeorrhyncha). The data provided represent Bayesian and ML phylograms inferred from the data sets without Lycorma delicatula (Insecta: Hemiptera: Archaeorrhyncha).
Primers used in PCR. The data provided represent primers used in amplification of mt-genomes.