Abstract
Ubiquitin (Ub)–protein conjugates formed by purified ring-finger or U-box E3s with the E2, UbcH5, resist degradation and disassembly by 26S proteasomes. These chains contain multiple types of Ub forks in which two Ub's are linked to adjacent lysines on the proximal Ub. We tested whether cells contain factors that prevent formation of nondegradable conjugates and whether the forked chains prevent proteasomal degradation. S5a is a ubiquitin interacting motif (UIM) protein present in the cytosol and in the 26S proteasome. Addition of S5a or a GST-fusion of S5a's UIM domains to a ubiquitination reaction containing 26S proteasomes, UbcH5, an E3 (MuRF1 or CHIP), and a protein substrate, dramatically stimulated its degradation, provided S5a was present during ubiquitination. Mass spectrometry showed that S5a and GST–UIM prevented the formation of Ub forks without affecting synthesis of standard isopeptide linkages. The forked Ub chains bind poorly to 26S proteasomes unlike those synthesized with S5a present or linked to Lys63 or Lys48 chains. Thus, S5a (and presumably certain other UIM proteins) function with certain E3/E2 pairs to ensure synthesis of efficiently degraded non-forked Ub conjugates.
Keywords: nondegradable forked ubiquitin chain, proteasome, Rpn10, S5a
Introduction
Most proteins degraded by 26S proteasomes are linked to a chain of ubiquitin (Ub) molecules, in which the C-terminal carboxyl group of a Ub is coupled through an isopeptide bond to the ɛ-amino group on one of the lysines on the proximal Ub (Hershko and Ciechanover, 1998; Weissman, 2001; Glickman and Ciechanover, 2002; Pickart and Cohen, 2004; Hochstrasser, 2006). Since a Ub molecule contains seven lysines (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63), there are seven possible types of isopeptide linkages. It is generally assumed that Ub chains contain only one type of isopeptide linkage (Chau et al, 1989; Gregori et al, 1990; Wu-Baer et al, 2003), and that the nature of the Ub linkage determines the specific fate of the protein. Most proteins degraded by 26S proteasomes are believed to be linked to a homogeneous polyUb chain, in which the Ubs are coupled through isopeptide linkages to Lys48, Lys11 or Lys29 on the preceding Ub (Chau et al, 1989; Gregori et al, 1990; Johnson et al, 1995; Jin et al, 2008). PolyUb chains in which the Ubs are linked through other lysines (such as Lys63 chains) are believed to serve roles unrelated to proteasome preference (Spence et al, 1995; Deng et al, 2000; Hicke and Dunn, 2003; Pickart and Cohen, 2004). However, recent studies have called into question these widely accepted notions about proteolysis and have shown rapid degradation by purified 26S proteasomes of proteins linked to chains containing heterogeneous isopeptide linkages (Kirkpatrick et al, 2006) or to chains composed purely of Lys63 linkages (Kim et al, 2007).
Recently, we reported that the small ring-finger E3s, MuRF1 and Mdm2, and the U-box E3, CHIP, with the E2, UbcH5, form in vitro a new type of polyUb chain on substrates that does not support their rapid degradation by 26S proteasomes in contrast to homogeneous Lys48 or Lys63 chains. For example, the E3, MuRF1, forms nondegradable heterogeneous forked chains on the substrate, troponin I, with UbcH5, whereas it forms a homogenous Lys48 chain on this substrate with the E2, UbcH1, and a Lys63 chain with the dimeric E2, UbcH13/Uev1a (Supplementary Figure S1). Moreover, when the troponin I was linked by MuRF1 to a Lys48-Ub chain with UbcH1 or even to a Lys63-Ub chain with UbcH13/Uev1a, it was rapidly degraded by 26S proteasomes. The nondegradable forked chains contain all seven possible types of isopeptide linkages and forks (i.e. bifurcations) in which two Ub molecules are linked to adjacent lysine residues on the preceding Ub molecule at Lys6+Lys11, Lys27+Lys29 or Lys29+Lys33 (Kim et al, 2007). Peptides indicative of Lys27+Lys29 forks have also been observed by mass spectrometry of proteins in growing yeast (Peng et al, 2003). These chains might also contain other types of bifurcations, for example, where two Ubs are linked to nonadjacent lysines on the preceding Ub, but such forks cannot be detected yet by MALDI-TOF mass spectrometry (as the trypsin treatment would cleave at the intervening Lys or Arg residues. Supplementary Figure S1).
It is noteworthy that the ‘forked' Ub linkages were disassembled very slowly by the isopeptidases associated with the 26S proteasomes unlike the Lys48 or Lys63 linkages in the same Ub conjugates (Kim et al, 2007). One goal of these studies was to learn whether these forked heterogeneous Ub chains are responsible for the resistance to degradation of the proteins to which they are attached. For example, the resistance of the forked linkages to proteasomal isopeptidases might account for the failure of these Ub chains to support rapid degradation. Alternatively (or in addition), these anomalous chains might bind to the 26S proteasome less strongly than Lys48 or Lys63 chains, as we show here.
The formation of nondegradable Ub conjugates by the small ring-finger or U-box E3s with UbcH5 is quite surprising, as these E3s and UbcH5 function in vivo in the degradation of a variety of proteins. We therefore postulated that some additional factors are present in cells, but are missing in these purified systems, that help prevent the formation of such nondegradable chains in vivo or that facilitate their efficient degradation. These studies were undertaken to test whether any Ub-binding protein might specifically stimulate the degradation of proteins formed by these E3s with UbcH5 either by inhibiting the formation of these forked chains or by enhancing their susceptibility to 26S proteasomes. We show here that S5a/Rpn10 through its ubiquitin interacting motifs (UIM) can alter the ubiquitination process so as to prevent the formation of nondegradable forked Ub chains by UbcH5 and ring-finger or U-box E3s, and therefore can stimulate markedly the degradation of certain substrates.
In most cells, S5a/Rpn10 is primarily an abundant soluble protein, but in all cells, it also exists as a subunit of the 26S proteasome. Its functions in vivo both in the proteasome and as a free protein have long been uncertain. S5a binds Ub chains through its two C-terminal UIM domains (Hofmann and Falquet, 2001; Wang et al, 2005). It was originally discovered as a subunit of the 26S proteasome's 19S regulatory complex (PA700) and was initially proposed to be the binding site for ubiquitinated proteins (Deveraux et al, 1994; van Nocker et al, 1996a, 1996b). However, S5a/Rpn10 is not essential for viability of yeast (Lambertson et al, 1999) or for the integrity of the 19S regulatory particle (Wojcik and DeMartino, 2002). In Drosophila and yeast, ΔRpn10 mutants have major defects in the degradation of certain proteins and accumulate Ub conjugates (Rubin et al, 1997; Lambertson et al, 1999; Saeki et al, 2002; Chuang et al, 2005), especially under stressed conditions where cells generate large amounts of misfolded proteins (e.g. heat-shock), and when overall rates of proteolysis rise (Medicherla and Goldberg, 2008). It is also noteworthy that Rpn10 is essential during embryonic development of mouse (Hamazaki et al, 2007).
Here we report that S5a/Rpn10 (and perhaps related proteins) can stimulate the degradation of certain proteins by preventing the formation of nondegradable Ub chains, and that the absence of S5a/Rpn10 probably explains the failure of certain purified E3/E2 combinations to support proteasomal degradation. This requirement for S5a/Rpn10 for formation of efficiently degraded Ub conjugates may also account for the presence of free S5a/Rpn10 in the cytosol and help explain the decreased proteolysis seen in yeast or Drosophila lacking this protein.
Results
S5a/Rpn10 through its UIM domains stimulates degradation of proteins ubiquitinated by CHIP or MuRF1 with UbcH5
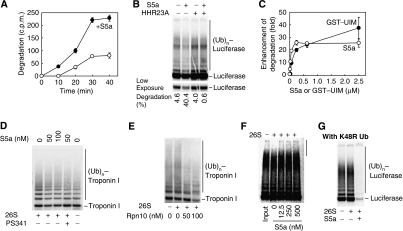
Since certain Ub-binding proteins have been reported to facilitate proteasomal degradation of Ub conjugates (Verma et al, 2004), we tested whether any known Ub-binding proteins can enhance proteasomal degradation of firefly luciferase ubiquitinated by UbcH5 and the U-box E3, CHIP. S5a was studied initially because in yeast and other cells, S5a/Rpn10 is present primarily free in the cytosol (Haracska and Udvardy, 1995; Rubin et al, 1997; Wilkinson et al, 2000), and a lack of Rpn10 in yeast and Drosophila cells leads to reduced degradation of certain proteins (Rubin et al, 1997; Lambertson et al, 1999; Saeki et al, 2002; Wojcik and DeMartino, 2002). To measure the effects of S5a on the rate of luciferase degradation, we assayed the hydrolysis of 125I–luciferase to 125I-peptide soluble in trichloroacetic acid. When S5a (50 nM) was present during the ubiquitination reaction together with 26S proteasomes (3.6 nM), luciferase (100 nM) was degraded up to threefold faster than when no S5a was added (Figure 1A). When a higher amount of S5a (500 nM) was present, luciferase was degraded even faster (Figure 1B). With increasing amounts of S5a, proteasomal degradation of luciferase increased dramatically by up to 25-fold (Figure 1C). To learn whether S5a has similar effects on proteolysis with other E3s, we assayed the degradation of troponin I by MuRF1, UbcH5 and pure 26S proteasomes (Figure 1D and Supplementary Figure S2). As found with luciferase, the presence of S5a during ubiquitination enhanced degradation of troponin I.
Figure 1.
The presence of S5a during substrate ubiquitination enhances its degradation by proteasomes. (A) S5a stimulates proteasomal degradation of luciferase. 125I–luciferase (100 nM) was denatured for 10 min at 43 °C in the presence of HSP70 (150 nM). The complex of 125I–luciferase and HSP70 was incubated with CHIP (500 nM), UbcH5 (750 nM) and purified 26S proteasomes (3.6 nM) in the presence or absence of 50 nM S5a. At different time intervals, degradation of luciferase was assayed at 37°C by measuring the appearance of TCA-soluble radioactivity with a γ-counter. (B) Addition of S5a, but not the UBL–UBA protein HHR23A stimulated Ub-dependent degradation of 125I–luciferase by 26S proteasomes. HHR23A also inhibited the enhancement of proteolysis by S5a. Purified 26S proteasomes, CHIP, UbcH5 plus S5a or HHR23A (500 nM each) were present from the start of the reaction as in (A). Ubiquitination and proteasomal degradation of 125I–luciferase were measured after 1 h using SDS–PAGE and a phosphorimager. (C) Ub-dependent degradation of 125I–luciferase increased dramatically upon addition of increasing amounts of S5a (○) or a GST fusion with the UIM domain of S5a (residues 203–329), GST–UIM (•). The reaction was carried out for 1 h and assayed as in (A), but using 125I–luciferase (250 nM), 26S proteasomes (3.6 nM), and S5a or GST–UIM were present from the outset. (D) Troponin I ubiquitinated by MuRF1 and UbcH5 in the presence of S5a (50 or 100 nM) is degraded more rapidly by 26S proteasomes. Troponin I (100 nM) was ubiquitinated by MuRF1 (500 nM) and UbcH5 (250 nM) in the presence of 26S proteasomes (3.6 nM) for 1 h at 37 °C. Degradation of troponin I was assayed by western blotting. As controls, reactions were incubated without 26S proteasomes or with 26S proteasomes preincubated with PS341 (1 μM). (E) Increasing amounts of Rpn10, the S. cerevisiae homolog of S5a, enhanced proteasomal degradation of troponin I by 26S proteasomes purified from rabbit muscle. The reaction was carried out and assayed as in (D) except that Rpn10 was used instead of human S5a. (F) Addition of S5a after the ubiquitination reaction does not enhance proteasomal degradation of ubiquitinated luciferase. Ubiquitination was carried out for 1 h at 37 °C in the absence of S5a with CHIP and UbcH5, and the Ub conjugates generated were isolated as described in the Methods. Degradation of the Ub-conjugated luciferase by 26S proteasomes (3.6 nM) was then measured at 37 °C for 1 h in the presence of increasing amounts of S5a. (G) Lys48 is not necessary for the stimulation of proteolysis by S5a. Luciferase was ubiquitinated by CHIP and UbcH5 with K48R mutant Ub in the presence of 26S proteasomes. Addition of S5a (500 nM) at the outset enhanced hydrolysis of luciferase, whereas luciferase ubiquitinated without S5a was not degraded (as was found with wild type Ub (B)).
Mammalian S5a binds to Ub chains through its two UIM domains in its C-terminal region (Hofmann and Falquet, 2001; Wang et al, 2005). We therefore tested whether the binding of these UIM-domains to the Ub-chain might also cause a stimulation of proteolysis. A fusion composed of the C-terminal half of S5a (residues 203–329) and glutathione-S-transferase (GST–UIM) markedly stimulated (up to 25-fold) proteasomal degradation of luciferase in a similar manner as S5a (Figure 1C). By contrast, GST alone had no effect (data not shown). Thus, the UIM-domains seem to be crucial for the enhancement of proteasomal degradation by S5a, and this effect of S5a does not require its N-terminal von Willebrand factor-A (vWA) homology domain (Hofmann and Falquet, 2001), which in yeast is essential for the incorporation of Rpn10 into the proteasome and for its ability to stimulate the degradation of certain proteins (Fu et al, 2001).
To test whether the yeast homolog of S5a, Rpn10, which contains only one UIM domain, can also enhance the degradation of these proteins by the Ub–proteasome pathway, we compared the proteasomal degradation of troponin I using MuRF1, UbcH5, and ATP with or without Rpn10 from Saccharomyces cerevisiae present. As shown in Figure 1E, addition of Rpn10 also increased the degradation of troponin I by rabbit-muscle 26S proteasomes. Thus, the ability to stimulate proteolysis seems to be a specific property of the UIM domain in S5a or Rpn10.
Rpn10 was proposed to enhance proteasomal degradation of certain proteins by facilitating the delivery of Ub conjugates to the proteasome (Verma et al, 2004), and the UBL–UBA proteins (e.g. Rad23) were also reported to facilitate degradation of certain substrates by such a mechanism (Chen and Madura, 2002; Medicherla et al, 2004; Verma et al, 2004). We therefore tested whether HHR23A (the human homolog of Rad23) can enhance the degradation of luciferase in a similar fashion as S5a. However, HHR23A failed to enhance proteasomal degradation of luciferase. Also the addition of HHR23A together with S5a blocked the enhancement of degradation by S5a (Figure 1B), perhaps because HHR23A bound preferentially to the Ub chains on luciferase and prevented S5a from binding to them, or because the HHR23A bound to the UIM2 domain of S5a (Hiyama et al, 1999) and prevented its activity.
To stimulate degradation, S5a must be present during the ubiquitination step
In these experiments, S5a, the ubiquitinating enzymes and proteasomes were all present simultaneously. Among the possible explanations of this large stimulation of proteolysis could be that S5a and the GST–UIM fusion act on the proteasome to enhance the binding of the Ub chain to the 26S particle or to enhance its proteolytic activity (Verma et al, 2004). However, these explanations were found to be quite unlikely as the S5a had to be present during the ubiquitination reaction to enhance proteolysis. Addition of S5a after the formation of the Ub conjugate did not enhance proteasomal degradation of 125I–luciferase. Also, S5a addition after ubiquitination inhibited degradation and deubiquitination of these conjugates by the 26S proteasome (Figure 1F). Thus, the presence of S5a during substrate ubiquitination by UbcH5 and CHIP is crucial for its enhanced degradation, and under these conditions, S5a does not seem to help in delivering the conjugates to the 26S particle, as was reported earlier with an SCF ligase by Verma et al. (Verma et al, 2004).
Ub chains composed of Lys48 linkages are believed to be degraded preferentially by proteasomes (Chau et al, 1989; Gregori et al, 1990; Johnson et al, 1995). The Ub chains formed by UbcH5 and ring-finger or U-box E3s contain all seven possible isopeptide linkages but mainly Lys11-, Lys48- and Lys63-linkages, as well as forked linkages in which two Ub molecules are attached to the proximal Ub. These forked linkages are relatively resistant to deubiquitination by 26S proteasomes, which might account for the very slow degradation of the Ub conjugates (Kim et al, 2007). Therefore, it is possible that the presence of S5a during the ubiquitination process might enhance proteolysis by increasing the formation of Lys48 chains. We then assayed whether S5a could still enhance proteasomal degradation of luciferase ubiquitinated by CHIP and UbcH5 using a K48R Ub mutant. Although purified 26S proteasomes degraded luciferase ubiquitinated with the K48R mutant very poorly, addition of S5a during ubiquitination stimulated luciferase degradation dramatically (Figure 1G). In fact, S5a also stimulated degradation of proteins ubiquitinated with K11R, K29R or K63R Ub (Supplementary Figure S2). Thus the changes in ubiquitination that lead to greater proteolysis do not involve increased formation of Lys48 (or any other specific) linkages.
S5a blocks the formation of Lys6+Lys11, Lys27+Lys29 and Lys29+Lys33 forks, but not standard isopeptide linkages
Our finding that the presence of S5a during ubiquitination of a substrate by CHIP or MuRF1 with UbcH5 enhances degradation by proteasomes (Figure 1) strongly suggests that S5a alters the structure of the Ub chains formed on the substrate. The conjugates formed by CHIP with UbcH5 in the absence of S5a might resist proteasomal degradation because of the presence of diverse isopeptide linkages or of the forked Ub chains (Kim et al, 2007). To test if S5a might alter the nature of ubiquitin linkages formed, we analysed the isopeptide linkages in the polyUb chain formed on luciferase by CHIP and UbcH5 by mass spectrometry. The Ub–luciferase conjugates were isolated by immunoprecipitation, digested with trypsin, and subjected to nano-LC-MSMS to determine whether the different tryptic fragments that are indicative of different isopeptide Ub linkages (termed UPKs) were present (Kim et al, 2007).
Surprisingly, unlike the Ub–luciferase conjugates formed in the absence of S5a, the polyUb chain synthesized with S5a present yielded no detectable peptide in which two Ubs are linked to the lysines at Lys6 and Lys11 (UPK6/11) (Table I). Thus, S5a prevented the formation of this type of forked chain. UPK peptides indicative of other types of forks (i.e. UPK27/29 and UPK29/33) were also not found in the Ub–luciferase conjugates formed with S5a present, but were present in chains formed in its absence. Although S5a completely blocked the synthesis of Lys6+Lys11 forks, it did not affect the formation of the standard isopeptide linkages, including the abundant Lys11, Lys48 and Lys63, as well as the rarer Lys6 linkages. This selective suppression of the synthesis of forked Ub chains by S5a might account for the more rapid degradation of these Ub conjugates (see below).
Table 1.
S5a and its UIM domains prevent the formation of forked polyUb chains by CHIP and MuRF1
| Ub-peptide | CHIP | MuRF1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None | S5a | GST | GST–UIM | His-UIM1 | His-UIM2 | His-UIM1+2 | HHR23A | None | S5a | |
| Linear | ||||||||||
| UPK6 | + | + | + | + | + | + | + | + | + | + |
| UPK11 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| UPK48 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| UPK63 | +++ | +++ | + | + | + | +++ | +++ | +++ | +++ | +++ |
| Forked | ||||||||||
| UPK6/11 | + | 0 | + | 0 | + | 0 | 0 | + | + | 0 |
| UPK27/29 | + | 0 | + | 0 | + | 0 | 0 | + | + | 0 |
| UPK29/33 | + | 0 | + | 0 | + | 0 | 0 | + | + | 0 |
| PolyUb chains formed by purified UbcH5 and E3s were digested by trypsin and the quantities of UPKs were analyzed by nano-LC-MS/MS. | ||||||||||
| +, Low abundancy, <10% of total UPKs identified. | ||||||||||
| +++, High abundancy, >10% of total UPKs identified. | ||||||||||
| 0: not detected. | ||||||||||
| The pool of Ub peptides from Ub–luciferase conjugates (CHIP) or Ub-MuRF1 was identified by nano-LC-MSMS methods. S5a, fusion proteins containing S5a's UIMs or HHR23A was added to ubiquitination mixture from the outset and GST was added as a control of GST-UIM. For the reaction with GST and GST-UIM, a different batch of enzymes (E1, E2 and CHIP) from other reactions was used. | ||||||||||
This marked inhibition of the synthesis of forked chain by S5a was not restricted to ubiquitination by CHIP. Similar types of forked heterogeneous chains are formed during auto-ubiquitination of MuRF1 with UbcH5 (Kim et al, 2007). The addition of S5a to this reaction mixture also prevented the formation of Lys6+Lys11, Lys27+Lys29, and Lys29+Lys33 linkages without affecting the levels of standard isopeptide linkages (Table I). These similar findings with two quite different E3s also make it very likely that S5a might prevent fork formation by interacting with the growing Ub chain rather than with the E3.
S5a's UIM2 domain blocks formation of forked Ub chains
Since the Ub-binding C-terminal region of S5a alone could stimulate proteasomal degradation (Figure 1), we tested whether these C-terminal UIM domains could also prevent the formation of forked polyUb chains. Although GST by itself had no effect on fork formation by CHIP, the addition of the GST–UIM fusion to the ubiquitination reaction, like addition of S5a, blocked the production of Lys6+Lys11 linkages (Table I), and did not affect the synthesis of standard isopeptide linkages. Thus, the effects of S5a on fork formation, like the effects on proteolysis, are most likely because of its UIM domains, and do not require its N-terminal vWA domain. To identify the region in S5a's C-terminal half that is responsible for the inhibition of fork formation, we analyzed the types of isopeptide linkages formed on luciferase by CHIP and UbcH5 in the presence of a small portion of S5a that contains only its UIM1, UIM2 or both UIM1+UIM2 domains (fused to a His-tag). Interestingly, peptides containing only the UIM1 domain did not prevent the formation of forked Ub chains, while those containing UIM2 or UIM1+UIM2 did block the formation of Lys6+Lys11 and other forks (Table I). Thus, the UIM domain, especially UIM2, seems to be responsible for the effect of S5a.
Since HHR23A, unlike S5a, did not enhance proteasomal degradation of luciferase (Figure 1B), we analyzed the types of isopeptide linkages formed in the presence of HHR23A. Ub chains formed by CHIP and UbcH5 in the presence of HHR23A still contained Lys6+Lys11, Lys27+Lys29, and Lys29+Lys33 forks, which might explain the inability of HHR23A to stimulate proteasomal degradation of Ub conjugates formed by UbcH5.
Ub chains synthesized with S5a are susceptible to the proteasome-associated DUBs
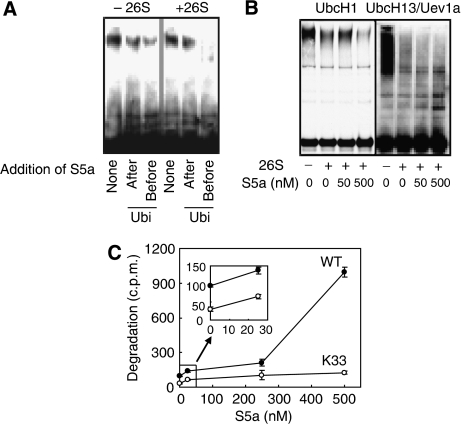
The heterogeneous forked Ub–protein conjugates and specifically the Lys6+Lys11 forks (whose content, unlike other linkages, could be assayed only semi-quantitatively) were found to be quite resistant to the isopeptidases associated with the 26S proteasome and to degradation by 26S proteasomes (Kim et al, 2007). To test whether the prevention of formation of forked Ub chains by S5a enhances the deubiquitination of the conjugates by 26S proteasomes, we isolated by immunoprecipitation the ubiquitinated luciferase formed by CHIP in the presence and absence of S5a, and then compared their rates of deubiquitination by purified 26S proteasomes. On incubation with pure proteasomes, the Ub–luciferase conjugates formed in the presence of S5a were disassembled, whereas those formed without S5a showed a minimal disassembly, as measured by their disappearance on western blotting (Figure 2A).
Figure 2.
The presence of S5a during ubiquitination enhances deubiquitination and degradation of Ub conjugates formed only by UbcH5. (A) The ubiquitination of 125I–luciferase was carried out with or without S5a (500 nM) present for 1 h at 37 °C. After this reaction was terminated, Ub conjugates were isolated by immunoprecipitation and then incubated with 3.6 nM of purified rabbit 26S proteasomes for 1 h at 37 °C. The isopeptidase caused a decrease in the amount of high-molecular-weight conjugates (higher than 191 kDa), which were synthesized with S5a present than in the conjugates synthesized without S5a. The addition of S5a (500 nM) after ubiquitination did not promote deubiquitination by the proteasome. Ubi: Ubiquitination. (B) Addition of S5a (50 or 500 nM) during ubiquitination did not enhance the proteasomal degradation of troponin I linked to homogeneous Lys48 or Lys63 Ub chains. Ubiquitination of troponin I by MuRF1 and UbcH1 from the Lys48 chain or UbcH13/Uev1a to form the Lys63 chain were carried out as in the Figure 1D. (C) At standard concentration of S5a (500 nM), only WT Ub stimulated significantly degradation of luciferase. No significant increase in degradation with S5a was seen with the K6, K11, K27, K29, K48 and K63 single-lysine mutant Ub (data not shown). However, at low concentration of S5a (25 nM), a small but reproducible increase in Ub-dependent proteolysis was seen with K33 single lysine Ub. The reactions were carried out as in Figure 1C. Insert shows enhanced hydrolysis of 125I–luciferase with low concentrations of S5a.
S5a does not stimulate degradation of proteins linked to non-forked chains
These findings argue strongly that S5a, through its UIM domain, promotes the degradation of proteins by preventing formation of the Lys6+Lys11, Lys27+Lys29, and Lys29+Lys33 (and perhaps other) forked linkages whose presence somehow blocks degradation. If true, S5a should not stimulate the degradation of proteins linked to non-forked homogeneous Ub chains. We had earlier shown that MuRF1 with UbcH1 forms homogeneous Lys48 Ub chains on troponin I or with UbcH13/Uev1a forms Lys63 chains, and that both types of Ub conjugates are rapidly degraded by 26S proteasomes, unlike the forked Ub–troponin I conjugates formed by MuRF1 and UbcH5 (Kim et al, 2007). Therefore, by altering the E2s, we could test whether the presence of S5a during the ubiquitination reaction enhances the degradation of troponin I linked to a Lys48 chain or Lys63 chain (Figure 2B). The hydrolysis of this substrate linked to these homogeneous Ub chains was not enhanced by addition of S5a, unlike Ub–troponin I conjugates formed with UbcH5 (Figure 1D). These findings further indicate that the prevention of formation of forked chains is the primary reason for the stimulation of proteasomal degradation by S5a.
To further test whether S5a can stimulate the degradation of proteins linked to chains that are not forked but are formed by UbcH5, we assayed the effects of S5a on degradation of luciferase ubiquitinated with several mutant Ubs containing only a single lysine and therefore unable to form forked chains (Figure 2C). Although S5a consistently stimulated degradation with wild-type Ub, no significant stimulation was observed with luciferase linked to Lys6, Lys11, Lys48, or Lys63 chains through single-lysine Ub (Supplementary Figure S3). Interestingly, at low concentrations, S5a caused a small but reproducible increase in degradation of luciferase conjugated with one such mutant, the K33 single-lysine Ub (Figure 2C and Supplementary Figure S3). Nevertheless, as shown in Figure 1C, higher S5a concentrations stimulated the degradation of WT conjugates dramatically, but they caused no further increase in degradation of the conjugates linked to a K33 chain. Thus, although there might be a small capacity of S5a at low concentrations to enhance proteolysis by another mechanism (probably by helping deliver the conjugates to the 26S (Verma et al, 2004)), the large stimulation of degradation with UbcH5 correlates with its capacity to prevent formation of forked Ub-chains.
Forked Ub chains, unlike those formed with S5a present, have low affinity for the 26 proteasome
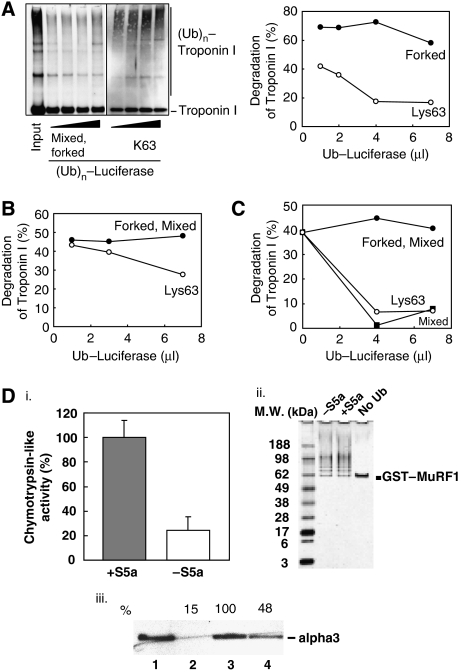
A simple explanation for the slow disassembly of the forked Ub chains by proteasomal isopeptidases (Kim et al, 2007) and for the low rate of degradation of the proteins linked to forked chains could be that these forked chains fail to bind tightly to 26S. To examine this possibility, we compared the relative affinities of the forked and non-forked Ub chains for the proteasome by testing whether polyUb conjugates made in the presence or absence of S5a could competitively inhibit the degradation of troponin I linked to homogeneous Lys63 or Lys48 chains. In control experiments, luciferase linked to a Lys63 chain (i.e. ubiquitinated by UbcH13/Uve1a and CHIP) inhibited the degradation of troponin I linked to Lys63 chains (i.e. ubiquitinated by UbcH13/Uev1a and MuRF1) and of troponin I linked to Lys48 chains (i.e. ubiquitinated by UbcH1 and MuRF1) by pure 26S proteasomes. By contrast, luciferase linked to a mixed forked Ub chain (i.e. ubiquitinated by UbcH5 and CHIP in the absence of S5a) could not inhibit the degradation of troponin I linked to a Lys63 chain (Figure 3A) or to a Lys48 Ub chain (Figure 3B). These findings argue that 26S proteasomes bind Ub–protein conjugates composed of homogeneous Lys63 and also Lys48 chains, with much higher affinities than the nondegradable forked chains. By contrast, the mixed Ub–luciferase conjugates without forks, which were formed by CHIP with UbcH5 in the presence of S5a, could inhibit the degradation of Ub–troponin I conjugates composed of Lys63 linkages (Figure 3C).
Figure 3.
26S proteasomes had a lower affinity for luciferase linked to forked Ub chains than for luciferase linked to homogeneous Lys63 chains or non-forked chains synthesized in the presence of S5a. (A) Unlike Lys63 Ub–luciferase conjugates, mixed forked Ub–luciferase conjugates failed to inhibit proteasomal degradation of Lys63 Ub–troponin I conjugates, even at high concentrations. Troponin I linked to a Lys63-Ub chain (formed by MuRF1 and UbcH13/Uev1a as in Figure 2C) was isolated, and degradation by proteasomes was assayed with increasing amounts of luciferase linked to a mixed forked Ub chain (formed by CHIP and UbcH5 as in Figure 2A) or luciferase linked to a Lys63 Ub chain (formed by CHIP and UbcH13/Uev1a) in a similar manner. The degradation reaction was run for 1 h at 37 °C. Left panel shows the conjugates assayed with an anti-troponin I antibody. Right panel; percent degradation of Ub–troponin I was quantitated using a densitometer. (B) Unlike Lys63 Ub–luciferase conjugates, mixed forked Ub–luciferase conjugates failed to inhibit proteasomal degradation of Lys48 Ub–troponin I conjugates, even at high concentrations. The experiment was carried out as in (A), but troponin I was linked to a Lys48 Ub chain by MuRF1 and UbcH1 (formed as in Figure 2C). (C) The presence of S5a during ubiquitination of luciferase with UbcH5 and CHIP enhances affinity of the Ub conjugates for the proteasome, as shown by their ability to competitively inhibit degradation of Lys63 Ub–Troponin I conjugates. The capacity of different types of Ub–luciferase conjugates to inhibit proteasomal degradation of troponin I formed by MuRF1 and UbcH13/Uev1a was measured as in (A). Increasing amounts of luciferase linked to a forked Ub chain (formed by CHIP and UbcH5), a mixed Ub-chain lacking forks (formed by CHIP and UbcH5 with 500 nM of S5a) or a Lys63 Ub-chain (formed by CHIP and UbcH13/Uev1a), were added to reactions as in (A). The amount of Ub–troponin I conjugates remaining were measured using a densitometer. (D) Forked Ub chain has low affinity for 26S proteasomes. GST–MuRF1 bound to a glutathione-resin was incubated with E1 and UbcH5 to allow auto-ubiquitination with or without S5a present. After ubiquitination, other components of the reaction were removed by extensive washing. SDS–PAGE and silver-staining showed the purity of (Ub)n-GST–MURF1 after purification (panel ii). An equivalent amount of ubiquitinated GST–MuRF1 from each reaction was incubated with 26S proteasomes for 30 min at 4 °C. 26S proteasomes not bound to ubiquitinated GST–MuRF1 were removed by extensive washing. To measure bound particles, the chymotrypsin-like activity (suc–LLVY–amc cleavage) of 26S proteasomes bound to ubiquitinated GST–MuRF1 was measured using flourometer (panel i). The activity of 26S proteasomes bound to GST–MuRF1 ubiquitinated with S5a present was designated as 100%. To verify that the activity of 26S proteasomes shown in panel (i) represents the real amount of proteasomes, the same reactions were analyzed by western blotting (panel iii). The amount of α3 was measured by western blotting using an anti-α3 antibody (panel iii). Lane 1: input (1/10), Lane 2: No Ub, Lane 3: +S5a an Lane 4: without S5a. Amount of α3 from the reaction with Ub–MuRF1 ubiqutinated with S5a was set as 100%.
To confirm that forked Ub chains bind poorly to 26S proteasomes unlike non-forked ones made in the presence of S5a, we assayed the binding of ubiquitinated GST–MuRF1 to 26S proteasomes (Figure 3D). To facilitate the isolation of pure Ub conjugates, GST–MuRF1 was allowed to auto-ubiquitinate with or without a S5a present, while immobilized on a column, and the binding of 26S proteasomes to these immobilized conjugates was then assayed. The GST–MuRF1 ubiquitinated with UbcH5 and S5a showed a higher affinity for 26S proteasomes than GST–MuRF1 ubiquitinated without S5a (i.e. forked conjugates). This result was obtained when the quantity of proteasomes associated with Ub–MuRF1 was measured by assaying the peptidase activity or by immunoblot against the α3 subunit. These observations make it likely that the low affinity of forked Ub conjugates for the 26S proteasome is the primary reason for their low rate of degradation, and that S5a by preventing the formation of forked Ub chains by UbcH5 and ring-finger or U-box E3s enhances the capacity of the Ub conjugates to bind to the 26S proteasomes.
Discussion
S5a enhances degradation of certain Ub conjugates
It is now well established that the types of Ub chains formed on a substrate are determined by the nature of both the E2 and E3 (Kim et al, 2007). This study, however, indicates that the presence of S5a can also affect the nature of the Ub chain synthesized by U-box and ring-finger E3s, together with UbcH5 and presumably closely related E2s (e.g. Ubc4 or UbcH7). UbcH5 is probably the E2 most commonly used in in vitro assays and has been reported in vivo to catalyse the degradation of many proteins together with multiple E3s. Although with HECT-domain E3s, UbcH5 supports the formation of Lys48 or Lys63 chains, with monomeric ring-finger or U-box E3s, ubiquitination by UbcH5 seems to be a rather random process. The resulting Ub chains contain all possible isopeptide linkages and multiple types of forks, and are much more resistant to degradation by proteasomes than substrates linked to Lys48 or Lys63 chains (Kim et al, 2007). This resistance to degradation and proteasomal isopeptidases led us to postulate that these poorly degraded conjugates result from the use of purified enzymes, and that cells contain additional factors that ensure the formation of rapidly degraded Ub conjugates, but are missing when the Ub–proteasome system is reconstituted in vitro.
By preventing the ‘off-pathway' formation of nondegradable Ub conjugates, S5a seems to function like a ‘molecular chaperone' for proteolysis. Unlike any other proteasomal subunit, S5a/Rpn10 is found free in large amounts in the cytosol (van Nocker et al, 1996b; Rubin et al, 1997). Strains of yeast and Drosophila lacking S5a/Rpn10 have a reduced capacity for degradation of certain proteins (Rubin et al, 1997; Lambertson et al, 1999; Saeki et al, 2002; Chuang et al, 2005). These defects have been generally attributed to defective proteasome function in the ΔRpn10 strains, but in light of these findings, they might also result in part from the capacity of S5a/Rpn10 to prevent the formation of forked conjugates by certain E3/E2 pairs. Since S5a stimulated proteolysis only when present during the ubiquitination step, S5a must alter the conjugation process and is not simply serving either as a proteasomal receptor for polyUb chains as proposed earlier (Deveraux et al, 1994; van Nocker et al, 1996a, 1996b) or as a ‘shuttle factor' that facilitates the association of preformed conjugates with the 26S (Chen and Madura, 2002; Verma et al, 2004; Chuang et al, 2005; Richly et al, 2005). If free S5a were simply to bind to Ub chains in the cytosol, it would be expected to compete with proteasomal S5a/Rpn10 and reduce the degradation of ubiquitinated proteins (Verma et al, 2004), as was found here when S5a was added after synthesis of the conjugates. This inhibitory effect probably accounts for the early observation that addition of S5a to cell extracts reduces Ub-dependent proteolysis (Deveraux et al, 1995).
A variety of findings indicate that the ability of S5a to inhibit fork formation accounts for its capacity to dramatically stimulate the breakdown of protein ubiquitinated by UbcH5 and CHIP or MuRF1: (1) S5a had to be present during Ub conjugation both to prevent fork formation and to enhance proteasomal degradation (Figure 1F). (2) This blockage of fork formation correlated with S5a's ability to increase the disassembly of the polyUb chain by the proteasome (Figure 2A) and to increase conjugate binding to the proteasome (Figure 3). (3) S5a did not stimulate degradation when the same substrate, troponin I, was linked to homogeneous Ub chains that lack forks, that is, Lys48 or Lys63 chains formed with WT Ub by UbcH1 or UbcH13/Uev1a or Lys6, Lys11, Lys48, or Lys63 chains formed by UbcH5 with single lysine mutant Ubs (Figure 2B and C). (Although low concentrations of S5a caused a small stimulation, specifically of degradation of K33-single lysine Ub chains, this effect cannot account for the large stimulation seen with higher concentrations of S5a and wild-type Ub.) (4) Most importantly, under the conditions where they stimulated proteolysis, S5a and the fusion of GST with S5a's UIM1 and UIM2 domains prevented formation of Lys6+Lys11, Lys27+Lys29, and Lys29+Lys33 forks, but did not alter the content of standard isopeptide linkages (Table I). Together, these data strongly argue that S5a acts by preventing the formation of forked Ub chains, and no evidence was obtained for the alternative possibility that S5a enhances the capacity of 26S proteasomes to degrade or disassemble these mixed, forked conjugates.
The presence of forks in the conjugates formed by UbcH5 seems responsible for their relative resistance to proteasomal isopeptidases, and this resistance might contribute to the inability of proteasomes to rapidly degrade these ubiquitinated proteins. However, probably a more important factor in their resistance to proteolysis is that the forked Ub conjugates have little or no affinity for the 26S particle, since they could not competitively inhibit proteasomal degradation of Lys63- or Lys48-linked Ub conjugates. By contrast, the non-forked, mixed conjugates formed with S5a present competitively inhibit the degradation of these Lys63 or Lys48 linked substrates. Also our assay of conjugate binding to the 26S clearly showed that forked ubiquitin chains have a lower affinity than non-forked ubiquitin chains (Figure 3). Thus, preventing fork formation clearly enhanced conjugate binding to the 26S complex.
An important, initially surprising finding was that the presence of Lys6, Lys11 and Lys63 linkages in the Ub chains did not prevent the dramatic (up to 25-fold) stimulation of proteasomal degradation by high S5a concentrations. Thus, S5a enhanced proteasomal degradation of luciferase and troponin I ubiquitinated by UbcH5 and suppressed fork formation without significantly altering the content of these various isopeptide linkages. It is widely stated that only Ub chains composed of Lys48, Lys29 or Lys 11 linkages can target proteins to the 26S proteasome (Finley et al, 1994; Johnson et al, 1995; Pickart and Cohen, 2004; Jin et al, 2008) and that other linkages serve distinct functions in vivo. However, we observed that troponin I linked to Lys63 chains is degraded rapidly by purified mammalian 26S proteasomes, in fact even more rapidly than Lys48 chains (Kim et al, 2007) and that such chains bind strongly to these particles (Figure 3, and (Hofmann and Pickart, 2001)). Also, cyclin B1 ubiquitinated by APC degraded rapidly, although it contains several types of linkages or might lack Lys48 linkages (Kirkpatrick et al, 2006). Furthermore, the observation that the proteasome-associated Ub ligase, Hul5, synthesizes Lys63 chains on Ub conjugates and enhances their degradation, also suggests a preference of the 26S for Lys63 chains (Crosas et al, 2006). Thus, in vivo, there probably are cytosolic proteins that bind selectively to Lys63 chains and prevent their rapid hydrolysis.
Our earlier findings (Kim et al, 2007) suggested that the slow cleavage of forked linkages by proteasomal isopeptidases, especially the Lys6+Lys11 forks (which are the most reliably quantitated forks) might account for the resistance to proteasomal degradation. In this study, we showed that S5a prevents the formation of all three types of forked linkages (Kim et al, 2007). It is also possible that these E3s with UbcH5 might link two Ub chains to more distant Lys residues on the proximal Ub, but such bifurcated structures (unlike those where two Ub chains are linked to adjacent lysine residues) do not withstand tryptic digestion and therefore cannot be detected by the current approaches. If such distant forks are formed, they would also be likely to retard proteasomal degradation.
Although S5a/Rpn10 seems to be essential during mouse development (Hamazaki et al, 2007), cells lacking S5a are viable (Lambertson et al, 1999; Wojcik and DeMartino, 2002), contain functional 26S proteasomes (Verma et al, 2004), and show defects in the degradation of only a limited groups of proteins (Rubin et al, 1997; Lambertson et al, 1999; Saeki et al, 2002), perhaps because these proteins are ubiquitinated by ring-finger or U-box E3s with UbcH5. The limited functional defect in the ΔRpn10 cells under normal conditions might indicate that other Ub-binding proteins (especially other UIM proteins) can compensate for the lack of free S5a. Although the UBA protein, HHR23A (the human homolog of Rad23), did not suppress fork formation (Table I) and did not stimulate proteolysis, under the condition used here, in contrast to S5a or the GST–UIMS5a fusion, Rad23, at lower concentrations, was reported to enhance proteasomal degradation by delivering Ub conjugates to the 26S proteasome (Verma et al, 2004). In this study, we also noted that S5a, at low concentrations, could slightly increase proteolysis by a mechanism not involving the suppression of fork formation (Figure 2C and Supplementary Figure S3), since it occurred with non-forked K33 single-lysine Ubs. Thus, free S5a might stimulate Ub-dependent proteolysis by multiple mechanisms (see below and (Matiuhin et al, 2008)) in addition to the large stimulation by preventing fork formation by certain E3/E2 pairs. The crucial role of S5a in ensuring formation of degradable Ub conjugates probably accounts for the presence of free S5a in the cytosol. In related experiments (Kim and Goldberg, unpublished observations), we have shown that in many mammalian cells, most S5a is free in the cytosol, as was found earlier in Drosophila and yeast (Haracska and Udvardy, 1995; Rubin et al, 1997; Wilkinson et al, 2000). Surprisingly, however, in HeLa cells, most S5a is in the proteasome, and little is free as reported earlier (Hendil et al, 2002). Possibly in these cells and in S5a-deficient lines, fork formation is more frequent than in other cells. Alternatively, other cell Ub-binding proteins might function in these cells in place of S5a to prevent fork formation.
Probable mechanism of the inhibition of fork formation
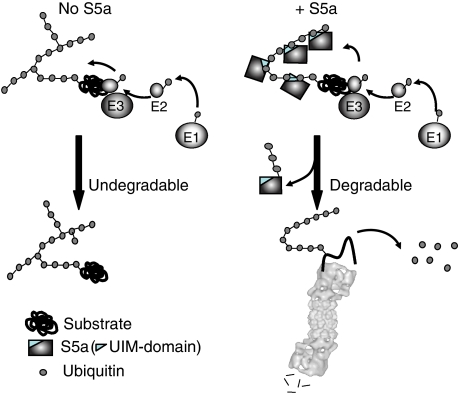
Most likely, S5a prevents the synthesis of forked chains by binding to the growing polyUb chain through its UIM domains and sterically blocking the linkage to a proximal Ub of additional Ub molecules released from UbcH5 (Figure 4). An analysis of S5a's structure by NMR has indicated that its two UIM domains can bind simultaneously to distant Ub moieties (Wang et al, 2005). However, it is unclear exactly which portions of S5a might shield the terminal ubiquitin from multiple ubiquitinations. The UIM domains fused to GST can prevent the formation of forked chain in similar fashion as S5a. The strongest evidence that S5a bound to growing Ub chains shields the lysines from the highly reactive Ub is that the S5a molecule itself also gets ubiquitinated (Uchiki et al, 2009). Under our typical experimental conditions, S5a and the GST–UIM fusion were found to be extensively ubiquitinated by CHIP or MuRF1 with UbcH5 during the ubiquitination of the substrate or auto-ubiquitination of the E3s. In yeast, Rpn10 (Crosas et al, 2006) and in mammals, S5a (Uchiki et al, 2009) are rapidly degraded presumably because of their continual ubiquitination. In addition, other proteins that contain UIM domains tend to be ubiquitinated in vivo (Klapisz et al, 2002; Oldham et al, 2002; Polo et al, 2002; Hicke and Dunn, 2003; Timsit et al, 2005), and after ubiquitination, these proteins have been reported to show reduced affinity for polyUb chains (Miller et al, 2004). In a related study, we have obtained strong evidence that ubiquitination of the UIM protein is a consequence of its binding to the growing Ub chain and that S5a (unlike a typical E3 substrate) is ubiquitinated by a very wide variety of E3s with UbcH5 (Uchiki et al, 2009).
Figure 4.
Proposed mechanism on how S5a prevents the formation of forked polyUb chains. Without S5a, ring-finger and U-box Ub ligases with UbcH5 form non-degradable Ub conjugates containing forked polyUb chains, because of the reaction of a lysine on a proximal Ub with a highly reactive Ub released from UbcH5. The resulting Ub conjugate is poorly degraded by proteasomes because of the presence of forks in the polyUb chain which reduces binding to the 26S proteasome. S5a binds to the growing polyUb chain and blocks the available lysines except those on the terminal Ub and then shields the chain so as to prevent the formation of more than one isopeptide linkage on one Ub moiety. During this process, the bound S5a is itself ubiquitinated.
Perhaps the strongest evidence that this ubiquitination of S5a is linked to its ability to prevent fork formation (Figure 4) is that S5a gets ubiquitinated by CHIP or MuRF1 with UbcH5, but not when these E3s function with UbcH1 or Ubc13/Uev1, which attach Ub moieties in a precise manner yielding homogenous linkages. By contrast, with UbcH5, ubiquitination is a rather nonspecific process in which the highly reactive Ub, released from UbcH5, reacts with any Lys on the proximal Ub and on occasion reacts with two Lys residues on the same Ub, but not if S5a is present to bind to the growing chain and to shield the proximal Ubs. Under these conditions, the Lys residues on S5a might instead become ubiquitinated. Thus, this tendency of S5a to be ubiquitinated might be an accidental consequence of its shielding function (i.e. a consequence of its residing in a dangerous neighborhood), rather than being an essential mechanism for the protection against fork formation.
Because the capacities of S5a to prevent fork formation and to stimulate proteolysis are also seen with GST–UIMS5a, and because cells lacking S5a show relatively minor defects in degradation, this new ‘chaperoning' effect is probably not a unique property of S5a. Possibly, some other UIM family members and perhaps other types of Ub-binding proteins serve similar roles in blocking fork formation. There is growing evidence in yeast that the UBA proteins, Rad23 and Dsk2, promote degradation by facilitating the delivery of certain Ub-conjugated proteins to the proteasome (Wilkinson et al, 2001; Chen and Madura, 2002; Medicherla et al, 2004; Verma et al, 2004; Richly et al, 2005), and can function in concert with S5a in promoting degradation (Lambertson et al, 1999; Verma et al, 2004). Recently, extraproteasomal Rpn10 has been reported to regulate Dsk2's access to proteasomes in yeast (Matiuhin et al, 2008). Such a role for Rpn10 seems independent of that described here and does not seem specific to certain E3/E2 pairs. Since HHR23A did not prevent fork formation and failed to stimulate proteolysis under the present conditions, there seem to be multiple mechanisms by which a Ub-binding protein might stimulate proteasomal degradation.
These findings indicate that S5a (and probably some other Ub-binding proteins) serve an important chaperone-like function for two major families of Ub ligases, the U-box and ring-finger E3s, when they function with UbcH5. Other E3 families, even those containing a ring-finger subunit, such as the dimeric E3, Brca1/Bard1 (Nishikawa et al, 2004), probably are much less prone to form forks since they transfer the Ub moiety in a stereo-specific manner to form homogenous chains, as do the HECT-domain E3s, E6AP and Nedd4 (Kim et al, 2007). Thus, the formation of a forked polyUb chain and the requirement for a chaperone-like cofactor, like S5a, to prevent fork formation clearly depend on the nature of the E3/E2's ligation mechanism.
Possible biological significance of S5a's preventing fork formation
The in vivo significance of fork formation and the resultant slowing of proteasomal degradation are uncertain. It is even possible that this tendency to form forked chains and its inhibition by S5a/Rpn10 (or a related protein) are regulated processes that serve to control the rate of proteolysis under certain conditions. The levels of S5a seems to be regulated in mammals, because the amount of free (nonproteasomal) S5a varies between different cell types (Kim and Goldberg, unpublished observations), the expression of different S5a isoforms changes during mouse development (Kikukawa et al, 2002), and S5a expression can be activated by c-Src as an antiapoptotic response (Gus et al, 2007). However, we believe it is more likely that fork formation is simply an untoward (‘off-pathway') consequence of the catalytic mechanism of UbcH5 functioning with U-box or ring-finger E3s. S5a thus seems to function as a molecular chaperone that prevents the synthesis of nondegradable forked Ub conjugates and enhances the efficiency of proteasomal degradation. Therefore, a failure of S5a/Rpn10 (or related proteins) to block fork formation might possibly contribute to the increased levels of Ub conjugates reported in Rpn10-deficient strains. It is noteworthy that Ub conjugates that are resistant to efficient proteasomal degradation and deubiquitination are also found in a variety of late-onset diseases, especially neurodegenerative diseases (Sherman and Goldberg, 2001; Ciechanover and Brundin, 2003) where they accumulate as intracellular inclusions. One possible explanation of their unusual stability could be the presence of forked chains. It is noteworthy that after heat shock or exposure to oxygen radicals, S5a/Rpn10 and Ubc4/5 seem essential for the rapid degradation of the damaged proteins (Medicherla and Goldberg, 2008). Unfortunately, reliable analytic methods are not yet available to quantify the actual abundance of forks in the highly heterogeneous pools of Ub chains in normal and stressed cells and to verify the importance of S5a (and other proteins) in protecting against accumulation of such non-degradable Ub conjugates.
Materials and methods
Ubiquitination
Ubiquitination of 125I–luciferase by CHIP and auto-ubiquitination of MuRF1 were assayed as described earlier (Kim et al, 2007). 125I–luciferase (100 nM) was heat-treated to 43 °C for 10 min in the presence of Hsp70 (150 nM) and then cooled on ice for 5 min. Hsp70–luciferase complex was added to the ubiquitination mixture containing 6His-E1 (50 nM), UbcH5 (750 nM), 6His-CHIP (500 nM), and ubiquitin (59 μM) in a buffer composed of 20 mM Tris–HCl (pH7.6), 20 mM KCl, 5 mM MgCl2, 2 mM ATP, and 1 mM DTT. The actual enzyme concentrations used in each individual experiment are indicated in the captions. If required, indicated amount of S5a, GST–UIM (containing residues 203–329 of S5a) or HHR23A purified from E. coli was added to the reaction. After ubiquitination of 125I–luciferase (for 1 h at 37°C), the ubiquitinated proteins were resolved on SDS–PAGE and then measured using a phosphorimager (BioRad). Ub–MuRF1 conjugates were resolved on SDS–PAGE and detected by Coomassie blue staining. Troponin I was ubiquitinated as described earlier (Kim et al, 2007) and Ub–troponin I conjugates were detected by western blotting using an anti-troponin I antibody. The quantity of each band was calculated by Quantity One software (BioRad).
Mass spectrometry
To identify the nature of the isopeptide linkages in the polyUb chains, Ub conjugates were treated exhaustively with trypsin, and the fragments were analyzed by mass spectrometry (Kim et al, 2007). Ub–luciferase conjugates were isolated with an anti-luciferase antibody and then resolved on SDS–PAGE. The auto-ubiquitinated MuRF1 was resolved on SDS–PAGE without immunoprecipitation. The bands of Ub conjugates larger than 100 kDa were excised from the SDS–PAGE gel and were digested with sequencing-grade trypsin (Promega) as described (Shevchenko et al, 1996). Digested samples were loaded onto a fused silica microcapillary C18 column (Magic, Michrom BioResources, Auburn, CA) prepared in-house (75 μm inner diameter, 10 cm in length). An Agilent 1100 high-pressure liquid chromatography (HPLC) system (Agilent Technologies, Palo Alto, CA) was used to deliver a gradient across a flow splitter to the column for over 40 min. The column eluant was directed into an LCQ–Deca electrospray ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA), and the eluted peptides were dynamically selected for fragmentation by the operating software. The acquired MS/MS data was analyzed with the non-redundant mouse database from NCBI, using SEQUEST database search tool for peptide identification. Modifications were permitted to allow for the detection of the following (mass shift shown in Daltons): oxidized methionine (+16) and ubiquitinated lysine (+114).
Deubiquitination of Ub–luciferase conjugates
125I–luciferase was ubiquitinated with or without S5a present. The reactions were terminated by adding EDTA (final concentration 10 mM) and then purified with an anti-luciferase antibody and protein A/G resin. The resin was washed with 1 × conjugation buffer thrice. 26S proteasomes (3.6 nM) was added and then incubated at 37 °C for 1 h. Ub–luciferase resolved on SDS–PAGE, and then the remaining quantity of large conjugates was measured using a phosphorimager.
Proteasomal degradation
125I–luciferase was ubiquitinated in the presence of 3.6 nM of rabbit-muscle 26S proteasomes in a final volume of 50 μl with or without S5a, and wild-type or a single lysine Ub mutant. After 1 h, the reaction was stopped by adding 200 μl of 7% TCA, and the acid-soluble radioactivity was measured using a gamma counter. The same reaction was resolved by SDS–PAGE, and then analyzed using the phosphoimager. Degradation of troponin I was assayed as described earlier (Kim et al, 2007) and measured by western blotting using an anti-troponin I antibody.
Competition of Ub chains for a proteasome
Troponin I was ubiquitinated by resin-bound MuRF1 with either UbcH1 or UbcH13/Uev1a. The supernatant of reaction was used to prepare Ub–troponin I conjugates. Ub–luciferase conjugates were prepared using CHIP and UbcH5 with or without S5a or by CHIP and UbcH13/Uev1a. After the reaction, CHIP, E1 and Hsp70 were removed using their His6-tag. Purified 26S proteasomes (6 nM) and ATP (2 mM) were added to measure the degradation of Ub–troponin I conjugates. To test the competition for the proteasome, Ub–luciferase conjugates linked to a different type of Ub chain were added to the reaction. After 1 h at 37°C, remaining Ub–troponin I was detected by western blotting.
Affinity of ubiquitinated GST–MuRF1 for 26S proteasomes
GST–MuRF1 bound to glutathione-resin was incubated with E1, UbcH5 and Ub in the presence of ATP. S5a was added to designated reactions. After 3 h at 37°C, other components than GST–MuRF1 was washed off by extensive washing with washing buffer 1 (50 mM Tris (pH7.5), 100 mM NaCl, 40% glycerol and 1 mM DTT) three times. 20 μl of 25% suspension of resin in washing buffer was incubated with 2 μl of purified 26S proteasomes and 20 μl of binding buffer (40 mM Tris (pH 7.5), 80 mM NaCl, 20 mM MgCl2, 2 mM ATP, 0.05% Triton X-100 and 2 mg/ml BSA) at 4 °C for 30 min with rotation. Unbound proteasomes were removed by washing thrice using with 500 μl of washing buffer 2 (20 mM Tris (pH 7.5), 40 mM NaCl, 10 mM MgCl2, 1 mM ATP and 0.025% Triton X-100). The amount of bound 26S proteasomes was assayed by measuring the particle's chymotrypsin-like activity using the fluorogenic substrate suc–LLVY–amc and also by western blotting using an anti-α3 antibody. Similar results were obtained with each method.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
The authors are grateful to Mary Dethavong for valuable assistance in the preparation of this paper. We thank John O'Bryan for helping us with S5a-constructs for S5a and UIM-proteins, Kylie Walters for helping us with UIM constructs for UIMs of S5a and Daniel Finley for helpful discussions. This work was supported by grants from the NIGMS, the High Q Foundation and the Fund for Innovation from Elan Corporation to ALG, from the Korean Ministry of Education, Science and Technology, FRP08 A1-032 of the 21C Frontier Functional Proteomics Program to KPK and from the NIH to SPG.
References
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Chen L, Madura K (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol 22: 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K (2005) Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40: 427–446 [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M (1994) A 26S proteasome subunit that binds ubiquitin conjugates. J Biol Chem 269: 7059–7061 [PubMed] [Google Scholar]
- Deveraux Q, van Nocker S, Mahaffey D, Vierstra R, Rechsteiner M (1995) Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J Biol Chem 270: 29660–29663 [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol 14: 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD (2001) Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. J 20: 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Gregori L, Poosch MS, Cousins G, Chau V (1990) A uniform isopeptide-linked multiubiquitin chain is sufficient to target substrate for degradation in ubiquitin-mediated proteolysis. J Biol Chem 265: 8354–8357 [PubMed] [Google Scholar]
- Gus Y, Karni R, Levitzki A (2007) Subunit S5a of the 26S proteasome is regulated by antiapoptotic signals. J 274: 2815–2831 [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Sasaki K, Kawahara H, Hisanaga S, Tanaka K, Murata S (2007) Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol 27: 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Udvardy A (1995) Cloning and sequencing a non-ATPase subunit of the regulatory complex of the Drosophila 26S protease. Eur J Biochem 231: 720–725 [DOI] [PubMed] [Google Scholar]
- Hendil KB, Hartmann-Petersen R, Tanaka K (2002) 26 S proteasomes function as stable entities. J Mol Biol 315: 627–636 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem 274: 28019–28025 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26: 347–350 [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem 276: 27936–27943 [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kikukawa Y, Shimada M, Suzuki N, Tanaka K, Yokosawa H, Kawahara H (2002) The 26S proteasome Rpn10 gene encoding splicing isoforms: evolutional conservation of the genomic organization in vertebrates. Biol Chem 383: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2(s) and ubiquitin-protein ligases (E3(s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8: 700–710 [DOI] [PubMed] [Google Scholar]
- Klapisz E, Sorokina I, Lemeer S, Pijnenburg M, Verkleij AJ, van Bergen en Henegouwen PM (2002) A ubiquitin-interacting motif (UIM) is essential for Eps15 and Eps15R ubiquitination. J Biol Chem 277: 30746–30753 [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K (1999) Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153: 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell 32: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL (2008) Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol 182: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Kostova Z, Schaefer A, Wolf DH (2004) A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep 5: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SL, Malotky E, O'Bryan JP (2004) Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J Biol Chem 279: 33528–33537 [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T (2004) Mass spectrometric and mutational analyses reveal lys-6-linked polyubiquitin chains catalyzed by brca1-bard1 ubiquitin ligase. J Biol Chem 279: 3916–3924 [DOI] [PubMed] [Google Scholar]
- Oldham CE, Mohney RP, Miller SL, Hanes RN, O'Bryan JP (2002) The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr Biol 12: 1112–1116 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE (2004) Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 5: 177–187 [DOI] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP (2002) A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S (2005) A series of ubiquitin binding factors connects cdc48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120: 73–84 [DOI] [PubMed] [Google Scholar]
- Rubin DM, van Nocker S, Glickman M, Coux O, Wefes I, Sadis S, Fu H, Goldberg A, Vierstra R, Finley D (1997) ATPase and ubiquitin-binding proteins of the yeast proteasome. Mol Biol Rep 24: 17–26 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Saitoh A, Toh-e A, Yokosawa H (2002) Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin- dependent proteolysis. Biochem Biophys Res Commun 293: 986–992 [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29: 15–32 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Miller SL, Mohney RP, O'Bryan JP (2005) The U-box ligase carboxyl-terminus of Hsc 70-interacting protein ubiquitylates Epsin. Biochem Biophys Res Commun 328: 550–559 [DOI] [PubMed] [Google Scholar]
- Uchiki T, Kim HT, Zhai B, Gygi SP, Johnston JA, O'Bryan JP, Goldberg AL (2009) The UIM protein, S5A, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J Biol Chem (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Deveraux Q, Rechsteiner M, Vierstra RD (1996a) Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci USA 93: 856–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD (1996b) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol 16: 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ (2005) Structure of s5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol 348: 727–739 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Ferrell K, Penney M, Wallace M, Dubiel W, Gordon C (2000) Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J Biol Chem 275: 15182–15192 [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol 3: 939–943 [DOI] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN (2002) Analysis of Drosophila 26 S proteasome using RNA interference. J Biol Chem 277: 6188–6197 [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lagrazon K, Yuan W, Baer R (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem 278: 34743–34746 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3