EMBO J 28, 1867–1877 (2009); published online 08 July 2009
Polyubiquitin (polyUb) is a diverse signal in terms of both chain length and linkage type (Ikeda and Dikic, 2008). Chains can be formed through covalent conjugation of ubiquitin (Ub) to any of the seven lysine residues on the preceding Ub (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, or Lys63), and in some instances two Ub molecules can simultaneously modify two lysine residues on a single Ub resulting in branched chains (Kim et al, 2007). Ubiquitin chain-length and linkage type, along with affinity for proteasome receptors and ease of deubiquitination, all contribute towards setting substrate hierarchy. Hence, polyUb-binding proteins influence the specificity and efficiency of intra-cellular proteolysis. One such auxiliary factor, S5a (or Rpn10), partakes in shuttling polyUb conjugates, limiting the access of competing substrate carriers, and in anchoring Ub chains at the proteasome (Deveraux et al, 1994; Glickman et al, 1998; Matiuhin et al, 2008). It now seems that S5a/Rpn10 also functions in upstream events by blocking the synthesis of low-priority forked chains and promoting the formation of unbranched chains with high affinity for the proteasome (Kim et al, 2009).
In an attempt to understand why some conjugation reactions generate polyubiquitinated substrates that are poorly degraded, and how Ub-binding proteins support proteosomal degradation, Kim et al have added purified S5a to a coupled ubiquitination–degradation assay. The presence of S5a in the reaction containing a substrate (luciferase), ubiquitination enzymes (an E2, UbcH5, and the U-box E3 ligase CHIP) and 26S proteasome significantly increased the rate of luciferase degradation. S5a had a similar effect on the degradation of another substrate (troponin I) ubiquitinated by UbcH5 and another E3 ubiquitin ligase (MuRF1). The yeast homologue of S5a, Rpn10, enhanced the proteasomal degradation of troponin I to the same extent as S5a.
In order to understand the source of proteolysis enhancement, S5a was then added to the reaction mix after the formation of ubiquitin conjugates. This move actually inhibited the degradation of the luciferase substrate, indicating that S5a is involved in early steps of targeting proteins for degradation. The use of mutated ubiquitin restricted in the linkages it can form (K48R, K11R, K29R, or K63R) did not alter the effect of S5a on degradation, indicating that degradation is not achieved by increasing the levels of any specific chain linkage. Nevertheless, mass spectrometry analysis of conjugates formed by CHIP with UbcH5 showed a decrease in forked linkages when S5a was added to the ubiquitination reaction. Forked chains are those in which two Ub molecules are linked to two adjacent lysines on the preceding Ub molecule (in essence, rather than forming elongated chains, some polyUbs might exist as a branched bush; see Figure 1A). It is important to note that because of technical limitations only simultaneous modification on adjacent lysines (K6/11, K27/29, or 29/33) was assayed in the accompanying study. The effect of S5a on the formation of forked chains was also detected during ubiquitination by the MuRF1 and UbcH5 pair of enzymes. Hydrolysis of substrates linked to homogenous non-forked Ub chains was not enhanced by addition of S5a and the authors therefore conclude that S5a interacts with the growing polyUb chain to prevent fork formation.
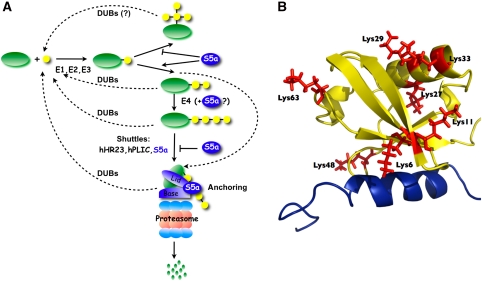
Figure 1.
The ubiquitin-binding protein, S5a/Rpn10, trails ubiquitin conjugates along their trajectory. (A) A general scheme of the ubiquitin–proteasome pathway updated with new results described by Kim et al (2009) in this issue. Ubiquitination enzymes (E1, E2, and E3) conjugate the carboxy terminus of ubiquitin (yellow) to a lysine residue on a substrate (green) selected for degradation. Subsequent conjugation might be sequential, leading to extended polymeric ubiquitin (lower arrow), or simultaneous at multiple lysines on a single ubiquitin link forming branched chains (upward arrow). The presence of S5a (blue) during conjugation promotes extended chains over branched ones (Kim et al, 2009). Polyubiquitin-binding proteins, among them S5a, shuttle elongated chains to the proteasome (Elsasser et al, 2004; Grabbe and Dikic, 2009; Verma et al, 2004). However, S5a also imposes a threshold on substrate delivery by competing with other polyUb shuttles for proteasome binding (Matiuhin et al, 2008). In some cases (Kim et al, 2009), S5a might aid E3s in transfering the conjugates directly to the proteasome, bypassing the downstream steps. At the proteasome, S5a partakes in anchoring the substrate while it is processed and unfolded for proteolysis (Deveraux et al, 1994; Glickman et al, 1998). At any number of junctions, deubiquitinating enzymes shave or trim polyubiquitin chains, thereby reversing conjugation and enforcing quality control. (B) Structure of S5a UIM in complex with Ub based on published NMR structure (pdb 1YX6 (Walters et al, 2002), generated with Pymol). Hydrophobic residues on the UIM region of S5a (blue ribbon) interact with a patch of hydrophobic residues on Ub (yellow backbone), exposing most of the seven lysines on the far side of Ub. With the possible exception of Lys6 (and to a lesser extent Lys48), access to most lysines is not shielded on anchoring of a single Ub to S5a; therefore, S5a might restrict build-up of forked chains in another manner by co-ordinating access of components of the ubiquination machinery.
What hurdle do forked Ub chains pose on protein degradation? Part of the effect of S5a seems to stem from the difficulty in processing (or deubiquitinating) forked chains. In an elegant set of experiments, the authors go on to show that forked Ub chains bind poorly to 26S, in contrast to non-forked ones. In a competition assay, luciferase linked to mixed forked Ub chains (ubiquitinated in the absence of S5a) was unable to inhibit the proteasomal degradation of troponin I linked to Lys63 or Lys48 chains, whereas luciferase linked to Lys63 chains or mixed non-forked chains (ubiquitinated in the presence of S5a) caused a significant decrease in the degradation of troponin I bound to homogeneous chains. Furthermore, in a binding assay, MuRF1 autoubiquitinated in the presence of S5a showed higher affinity for purified 26S than MuRF1 ubiquitinated in the absence of S5a. Poor anchoring to the proteasome (by S5a or another receptor; Figure 1A) might be the cause for the apparent stability and slow rate of processing or proteolysis of these conjugates.
The accompanying manuscript by Kim et al (2009) opens up a porthole to an exciting new layer of complexity in directing the ubiquitination process. Ubiquitin chain length and linkage type, along with the affinity for proteasome receptors and ease of deubiquitination, all contribute towards setting substrate hierarchy (Figure 1A). To these selection processes, one can now add a new checkpoint: at early stages of ubiquitination S5a limits the formation of forked chains, in which chains are extended at more than one lysine on a given Ub. Yet, as with many new observations, some amount of caution should be exercised when considering the general implications. So far branched chains have been identified mostly in vitro and their formation is strongly depended on the E2 used in the ubiquitination reaction. Among all possible branched modifications, only three forks at adjacent lysines on a single ubiquitin (K6/11, K27/29, and K29/33) have been documented. An accurate quantification of branched ubiquitin relative to total Ub-in-chains is yet to be carried out, although the assumption is that the ratio of forked modifications over extended chains is low (Kim et al, 2009). Furthermore, S5a UIM binds Ub through a patch of hydrophobic residues (Walters et al, 2002), leaving most lysine residues exposed for unrestricted conjugation to subsequent Ub (Figure 1B). This implies an intricate mechanism for S5a to block synthesis of forked chains, involving interactions between multiple components of the synthesis machinery, perhaps by getting polyubiquitinated itself, thus deflecting the imprecise ubiquitination of the substrate (Kim et al, 2009).
Nonconformity in ubiquitin polymerization—e.g. arborization in contrast to extension—might lead to accumulation of highly stable ubiquitin conjugates that are poorly recognized and slow to be removed. As previously shown in vivo, expression of Rpn10 or its UIM domain alone was able to reverse the accumulation of poorly turned-over polyUb conjugates under an induced stress condition (Matiuhin et al, 2008). The corrective properties of S5a in directing proper chain formation and prioritizing proteasome-bound substrates might be useful for enhancing protein turnover or overcoming stress conditions associated with malfunctions in the ubiquitin–proteasome system.
Footnotes
The authors declare that they have no conflict of interest.
References
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M (1994) A 26S subunit that binds ubiquitin conjugates. J Biol Chem 269: 7059–7061 [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem 279: 26817–26822 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9/Signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Grabbe C, Dikic I (2009) Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem Rev 109: 1481–1494 [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin–protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL (2009) S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J 28: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell 32: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin–proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41: 1767–1777 [DOI] [PubMed] [Google Scholar]