Abstract
Coordination of chromosome segregation and cytokinesis is crucial for efficient cell proliferation. In Bacillus subtilis, the nucleoid occlusion protein Noc protects the chromosomes by associating with the chromosome and preventing cell division in its vicinity. Using protein localization, ChAP-on-Chip and bioinformatics, we have identified a consensus Noc-binding DNA sequence (NBS), and have shown that Noc is targeted to about 70 discrete regions scattered around the chromosome, though absent from a large region around the replication terminus. Purified Noc bound specifically to an NBS in vitro. NBSs inserted near the replication terminus bound Noc–YFP and caused a delay in cell division. An autonomous plasmid carrying an NBS array recruited Noc–YFP and conferred a severe Noc-dependent inhibition of cell division. This shows that Noc is a potent inhibitor of division, but that its activity is strictly localized by the interaction with NBS sites in vivo. We propose that Noc serves not only as a spatial regulator of cell division to protect the nucleoid, but also as a timing device with an important role in the coordination of chromosome segregation and cell division.
Keywords: Bacillus subtilis, chromosome replication/segregation, coordination with cell division, Noc, specific DNA-binding sequence
Introduction
Coordination of chromosome segregation and cell division is needed in all organisms to ensure that the cells divide at the right place and time, so that bisection of the chromosome by the division apparatus occurs rarely. In bacteria, cytokinesis begins with polymerization of the tubulin homologue FtsZ into a ring structure at mid-cell. The dynamic ring (Z-ring) then serves as a scaffold for the assembly of more than 10 other proteins to form a multi-protein division machine, the divisome, which is responsible for the formation of the division septum (Errington et al, 2003; Weiss, 2004; Vicente et al, 2006; Haeusser and Levin, 2008). In rod-shaped bacteria such as Bacillus subtilis and Escherichia coli, positioning of the divisome precisely at the mid-cell is achieved through the joint action of two inhibitory factors: nucleoid occlusion and the Min system (Yu and Margolin, 1999; Rothfield et al, 2005; Barak and Wilkinson, 2007). Nucleoid occlusion prevents divisome assembly in the vicinity of the chromosome (Woldringh et al, 1990; Wu and Errington, 2004; Bernhardt and de Boer, 2005). When chromosome replication has been completed and the two daughter chromosomes have been moved towards opposite poles, this leaves spaces, in which division could occur at the mid-cell and close to the cell poles. The Min system prevents the polar potential division sites from being used, focusing the divisome to the mid-cell nucleoid-free zone (de Boer et al, 1989; Hu et al, 1999; Marston and Errington, 1999; Gregory et al, 2008; Scheffers, 2008).
Factors responsible for nucleoid occlusion have been recently identified: Noc protein in B. subtilis and SlmA in E. coli (Wu and Errington, 2004; Bernhardt and de Boer, 2005). Although the two proteins share no primary sequence homology, they exhibit similar properties. For example, both proteins are associated with the nucleoid and over-production leads to a delay in cell division; mutation of either is conditionally synthetic lethal with a min mutation, probably because of uncoordinated polymerization of FtsZ throughout the cell: an effect that can be partially rescued by over-production of FtsZ. Interestingly, SlmA is able to interact directly with FtsZ in vitro (Bernhardt and de Boer, 2005), whereas for Noc, no such interaction has as yet been shown. In Caulobacter, chromosomes adopt a more diffuse form and span the entire length of the cell. In this organism, no Min or Noc/SlmA-like protein has been identified. Instead, spatial regulation of septation requires MipZ, an FtsZ inhibitor that associates with the chromosomal origin region, and thereby couples the mid-cell localization of the FtsZ ring with the initiation of chromosome replication and segregation (Thanbichler and Shapiro, 2006). MipZ is an essential protein, emphasizing the importance of coordinating cell division with chromosome segregation.
Here, we show that the B. subtilis nucleoid occlusion protein Noc is a sequence-specific DNA-binding protein and that it has 74 binding sites on the chromosome. The Noc-binding DNA sequence (NBS) is a 14-bp long inverted repeat sequence. Autonomous plasmids carrying the NBS were able to recruit Noc onto the plasmid and inhibit cell division, showing that Noc is a potent inhibitor of cell division, but that this activity requires interaction with an NBS. Interestingly, the NBSs are distributed asymmetrically on the chromosome, being absent specifically around the replication terminus region. Recruitment of Noc into the replication terminus region by artificial introduction of NBS sites resulted in a delay in cell division. We, therefore, propose that Noc does not just serve as a spatial regulator for the site of division, but it also has a function in temporal regulation of cell division, allowing assembly of the divisome after replication traverses into the NBS-free region of the chromosome.
Results
Noc protein localizes to the nucleoid and the adjacent cell periphery
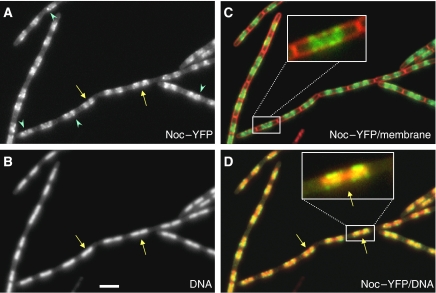
We reported earlier that Noc protein is associated with the nucleoid, based on observations of a GFP–Noc fusion (Wu and Errington, 2004). We subsequently discovered that this Noc–GFP fusion was temperature sensitive. Several new fusion constructs were then made and one of them, a Noc–YFP fusion, was found to be fully functional at temperatures >30°C. Close examination of cells expressing the Noc–YFP fusion (strain 4702) revealed a slightly different localization pattern from that of GFP–Noc, although the protein was still restricted to the general location of the nucleoid, the signal was more heterogeneous with many discrete spots evident (Figure 1 and see below). Importantly, many of the spots appeared to be associated with the cell periphery overlying the nucleoid (see inset in Figure 1C). Otherwise, the pattern of localization was similar to that reported earlier, including the frequent existence of a gap in the fluorescence pattern, relative to that of the nucleoid, near the middle of longer nucleoids (yellow arrows, see also the inset in Figure 1D). Interestingly, the localization pattern exhibited rapid but subtle changes over time, on a scale of seconds (Supplementary Figure S1), showing that the protein is very dynamic.
Figure 1.
Localization of Noc–YFP over the nucleoid and the adjacent cell periphery. Strain 4702 with a noc–yfp fusion (and without the native noc) growing exponentially in CH medium containing 0.3% xylose was examined by fluorescence microscopy. Panels show the YFP signal (A), DAPI (B), a merge of the YFP (green) and membrane (red) (C) and a merge of A and B (D) with YFP false coloured green and DAPI in red. The arrows point to central regions of the longer nucleoids, in which YFP signal is absent, and arrowheads point to spots of YFP. Scale bar, 2 μm.
Noc is absent from the terminus region of the chromosome
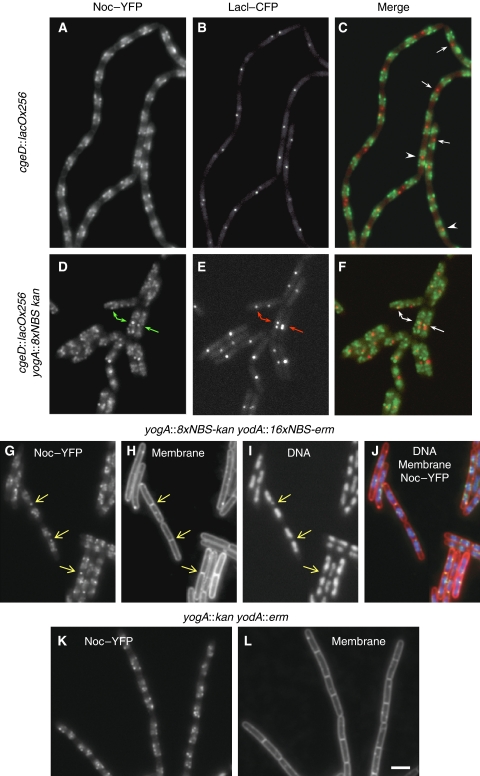
It seemed likely that the central gap in long nucleoids would represent the last replicating part of the chromosome around the replication terminus (terC) region. As we suggested earlier (Wu and Errington, 2004), absence of Noc from the terC region might allow FtsZ to begin assembling at the mid-cell before the completion of DNA replication and segregation. It would also have important implications for the timing of division and for the coordination of chromosome replication and segregation with cell division. To investigate this further, we examined the localization of the terC region using a fluorescent reporter operator system (FROS). In strain 4703, a lacO array was inserted near the terminus, about 130 kb from terC, and this was labelled with a CFP–LacI fusion. As shown in Figure 2A–C, the terC label usually appeared as a single spot or two adjacent spots close to the mid-cell. In shorter nucleoids, which we assume are only partially replicated, the TerC spots usually lie in the middle of the Noc spot clusters (arrowheads). However, in longer nucleoids, the TerC spots frequently occupied the clear space between the separated Noc clusters (arrows). Under these conditions, about 68% of the TerC spots (400 cells counted) did not coincide with Noc clusters, supporting the idea that Noc is less abundant in the terminus region than elsewhere on the chromosome.
Figure 2.
Presence and absence of Noc–YFP in the replication terminus region in the wild-type strain and in strains carrying NBS arrays near the terminus. (A–F) Dual labelling of Noc and the replication terminus region. In the otherwise wild-type strain 4703, Noc–YFP and LacI–CFP (labelling the lacO cassette near the terminus region) signals are often well separated (A–C). When an NBS array was introduced into strain 4703 at 2007 kbp, near the replication terminus, prominent spots of Noc appeared near the LacI spots (D–F). Arrows in C point to the LacI spots in cells with long nucleoids; arrowheads point to the LacI spots in cells with shorter nucleoids. (G–L) Distribution pattern of Noc–YFP and cell length difference in cells with (G–J) and without (K–L) the NBS arrays near the terminus (at 2007 and 2126 kb). Arrows in G–I point to bright spots of Noc near the mid-cell. A, D, G, K show Noc–YFP signals; B and E show the CFP–LacI spots; H and K are images of the membrane dye FM5-95 and I is the DAPI image showing the nucleoid. G is the merge of A and B; F is the merge of D and E; and J is the merge of G and I, with DNA shown in blue, membrane in red and Noc–YFP in green. Scale bar, 2 μm.
Genome-wide identification of Noc-binding sites by ChAP-on-Chip
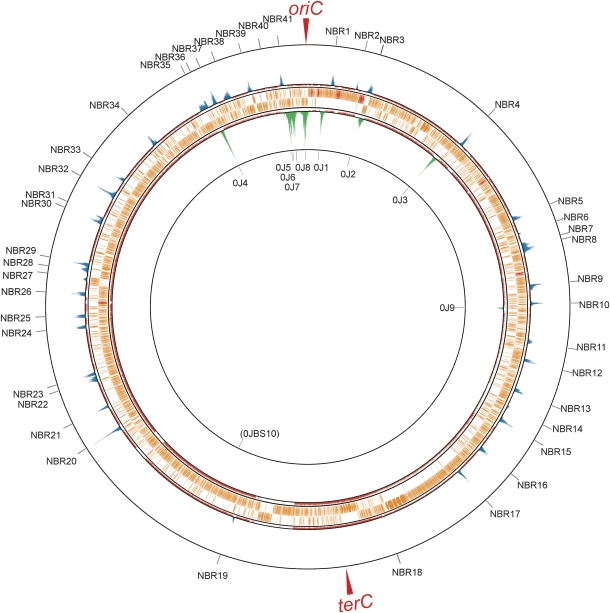
To see whether the reduced levels of Noc in the vicinity of the terC region was reflected in the specificity of its binding to DNA, we used chromatin affinity precipitation followed by microarray analysis (ChAP-on-Chip) to detect Noc binding to the chromosome in vivo. A histidine-tagged variant of Noc was constructed for this purpose and its activity was verified as described above for the YFP fusion. The results (Figure 3; Supplementary Figure S2; Supplementary Table S1) revealed a series of 41 discrete peaks (outer circles, blue peaks; labelled NBR for Noc-binding region), which were scattered around the chromosome, with the notable exception of the terC region. Thus, with the exception of two small peaks at 1873 kbp (NBR18) and 2333 kbp (NBR19) on the chromosome, the nearest peaks to terC were 411 kbp away anticlockwise (NBR17, 1606 kbp) and 760 kbp away clockwise (NBR20, 2777 kbp). An interesting feature of the peaks was that they extended over a region of 5.2–23 kbp (13.4±4.2 kbp), suggesting that Noc may bind and then spread laterally, like its relative Spo0J (Lin and Grossman, 1998; Murray et al, 2006; Breier and Grossman, 2007). The inner circle (green peaks) of Figure 3 shows the results of an experiment done earlier with cells expressing a His-tagged Spo0J protein (Ishikawa et al, 2007), which shows the expected pattern of discrete peaks located at the well-defined parS sites. The breadth of these peaks (8.3–30.7, 16.4±7.6 kbp) was generally in the same range as that of the Noc peaks, consistent with the notion that Noc, like Spo0J, spreads from preferred primary-binding sites. It was suggested that each Spo0J dimer covers about 30 bp length of DNA (Murray et al, 2006). We estimated that there are about 4500 Noc molecules per cell (data not shown), probably sufficient to cover and spread about 1–2 kbp, on average, from each binding site.
Figure 3.
Genome-wide distribution of preferred NBRs mapped by ChAP-on-Chip. Noc (outer rings) and Spo0J (inner rings)-binding signals in wild-type strains (4704 and SI002) were calculated as described in Materials and methods, and shown at their corresponding genome coordinates. Top and bottom lines indicate signal intensities of 20 and 0, respectively. Middle lines exhibit threshold values used to define the binding regions of Noc (1.5) and Spo0J (1.8). Signals above and below the threshold values are shown as blue and pink lines, respectively. ORFs (orange bars), rRNA and tRNA (red bars) are also indicated between them. The IDs of NBRs detected by our algorithm are shown at the outermost ring; 0J1–0J9 correspond to the Spo0J-binding sites reported earlier (Breier and Grossman, 2007; Murray and Errington, 2008). A minor new Spo0J-binding site was found and labelled 0JBS10. A magnified version of this data is provided in Supplementary Figure S2.
Bioinformatic identification of the likely Noc-binding site consensus sequence
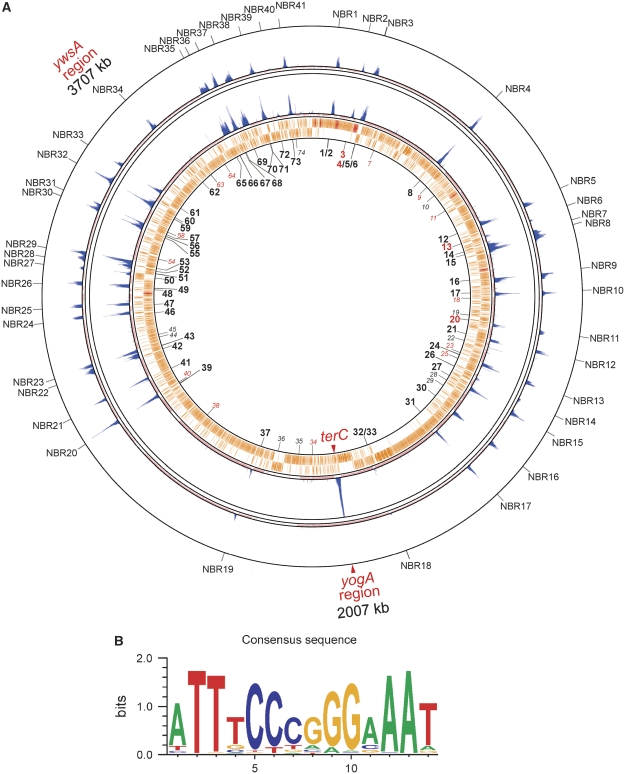
By analogy to Spo0J (Lin and Grossman, 1998), which recognizes a palindromic 16-bp sequence motif (parS), we anticipated that Noc would have a preferred DNA-binding sequence. We, therefore, used GENETYX software (GENETYX Corporation) to search for palindromic sequence motifs that were enriched in the NBRs identified by the ChAP-on-Chip experiments. The results revealed a set of closely related 14-bp palindromic sequences that might represent a preferred binding sequence for Noc (Figure 4; Supplementary Table S1). Among the 74 Noc-binding DNA sequences (NBSs) identified, 69% (51 out of 74) of them coincided with the peaks. Four of the NBSs were in the phage-related regions, which were not included in the ChAP-on-Chip analysis. No enrichment of Noc was detected in 19 of the predicted NBSs, most of which (11 out of 19) have low PMW (Position Weight Matrix) scores (<15.15; Supplementary Table S1). However, eight of the NBSs do have high PMW scores (Supplementary Table S1); these include NBS74, which overlaps with a strong Spo0J-binding region (OJ6), and NBS28, which has an identical sequence to NBS62 and NBS73. It is possible that these sequences were occupied by other proteins and, therefore, not accessible to Noc, but it is also possible that some of these sequences do not bind Noc because of variations in the flanking sequences that we have not recognized. Interestingly, the NBSs are not always located at the centre of the peaks, consistent with the above explanation that spreading/binding of Noc may be influenced by other factors present on the DNA.
Figure 4.
Altered distribution pattern of NBRs in a strain carrying an NBS insertion and deletion. (A) The binding profile of Noc in a mutant strain with a deletion at the ywsA region and an NBS insertion at the yogA region (strain 4723, inside) is shown with that of a wild-type strain, 4704 (the same data as shown in Figure 3, outside). Positions of NBSs are numbered starting from the replication origin and are indicated inside the innermost ring. NBSs with high PWM values, but without detectable Noc-binding peaks, are depicted as italic grey numbers. NBSs with low PWM values and no detectable Noc-binding peaks are numbered in red. (B) Sequence logo for all 53 of the NBSs in both directions that are located in the NBRs (106 in total).
Specific binding of purified Noc to the 14-bp consensus sequence in vitro
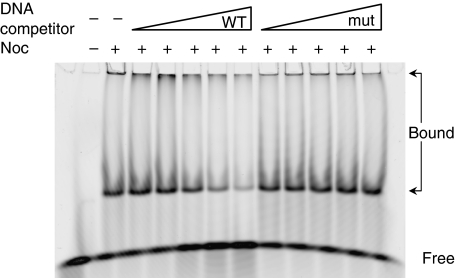
To test whether Noc is able to specially bind the 14-bp consensus in vitro, we overexpressed and purified the Noc–12xHis fusion. In parallel, we purified a mutant form of Noc (NocK164A-12xHis), which, on the basis of in vivo localization pattern, seemed not to bind to DNA (LJW and J Schneeweiss, unpublished). We first tested the NBS located in the ydbO gene at 506.3 kbp (NBS8), the enrichment of which had been confirmed by ChAP analysis (Supplementary Figure S3). The 24-bp probe comprised the 14-bp NBS consensus and 5 flanking bps on each side. Noc-probe complexes formed only when Noc-His and not when Noc(K164A)-His was used (Figure 5; Supplementary Figure S4). Furthermore, competition assays using unlabelled DNAs showed that wild-type competitor could release the labelled probe from the Noc–DNA complexes, whereas a ‘mutant' competitor, in which five bases in the consensus had been changed, did not (Figure 5). Two forms of complexes (large and small) were detected, probably because of the fact that Noc could multimerize (LJW, unpublished), and both were competed off by the specific competitor. These results confirm that Noc is a sequence-specific DNA-binding protein and that the 14-bp sequence from ydbO is one of the recognition sequences. We also tested the putative sequence located in the dhbF gene (3287 kb on the chromosome), and again found that this sequence is recognized by Noc (data not shown).
Figure 5.
Noc specifically recognizes an NBS in vitro. In the gel-shift assay, Noc–12xHis was incubated with 25 nM of a Cy5-labelled probe containing the NBS from the ydbO gene. Unlabelled competitor DNA (wild-type NBS or a mutant NBS) was present at concentrations of 0, 125, 250, 500 nM, 1 or 2 μM.
The Noc-binding site recruits Noc to the chromosome in vivo
To test whether the 14-bp putative NBS was necessary and sufficient for recruitment of Noc to specific sites on the chromosome, we first deleted part of the putative NBS (NBS 62) at 3707 kb on the chromosome and replaced it with a tetracycline-resistance (tet) gene. The results from both the ChAP-on-Chip and bioinformatics analyses indicated that this is the only putative NBS in the region (of about 300 kb) (Figures 3 and 4; Supplementary Figure S2). The site is located between the terminator of ywsB and ywsA genes; both genes of unknown function. No phenotypic effect of this mutation was evident. We then inserted eight copies of the NBS (linked to a kanamycin-resistance gene) near terC (the 2007-kbp region, between the putative terminator of yogA and the stop codon of gltB), in which predicted NBSs and ChAP-on-Chip peaks are rare. The strain (4723) also carried a noc–12xHis to enable ChAP-on-Chip analysis. Comparison of the profile obtained for this strain with that of the wild-type strain (4704, Figure 4) revealed two very specific changes. First, the peak located at 3707 kbp (ywsA region) was now absent. Second, a new peak appeared in the yogA region near terC, exactly where the array of NBS sequences had been inserted. The altered distribution pattern of Noc on the chromosome was confirmed by ChAP analysis using primer pairs from the affected regions (Supplementary Figure S3). We have also inserted the same 8xNBS array at a different location near the terminus region (1754 kbp, in between the terminators of ymfC and ymfD). Again ChAP analysis showed specific enrichment of Noc in the ymfC region, which was not detected in the unmodified strain (Supplementary Figure S3). These results strongly support the view that the consensus sequence we have identified represents a functional-binding site for Noc.
A third NBS-insertion strain we constructed (strain 4729) had two copies of the NBS inserted at 1745 kb on the chromosome (in between spoVFB and asd), again near the replication terminus. ChAP results showed an enrichment of Noc at spoVFB in this strain, but not in the otherwise wild-type strain (Supplementary Figure S3). Therefore, when the copy number of NBS at an ectopic chromosomal location was reduced to two, it was still able to recruit Noc to the site.
Insertion of an NBS array near the replication terminus recruits Noc to mid-cell and leads to elongated cells
To test the functional significance of the near absence of Noc from the terminus region of the chromosome, we first examined whether insertion of an NBS array near terC affected the localization pattern of Noc, as judged by cell imaging. As described above, in otherwise wild-type cells (lacking the native noc gene), Noc–YFP fluorescent signals infrequently overlap with those of a TerC FROS label (Figure 2A–C). However, in cells containing the insertion of an NBS array (same 8xNBS array used in the above experiment) at 2007 kbp (and the deletion at 3707 kbp), about 140 kb from the TerC label, bright spots of Noc were often evident near the mid-cell. These spots were frequently adjacent to or overlapped with the TerC spots (arrows in Figure 2D–F), confirming the recruitment of Noc to the terminus region. We then inserted a second array of NBSs on the other side of terC, between the putative terminators of yodA and yodB. The extra-antibiotic-resistance marker used for the second NBS array precluded us from easily co-localizing with TerC, but as shown in Figure 2 (G–I, arrows), unusual mid-cell spots of Noc were evident in many cells of the strain with two NBS arrays in the terC region. As spots similar to these were not evident in the absence of NBS array insertions (Figure 2K and L), the results support the idea that the normal distribution of Noc on the chromosome is such that it leaves a large zone at the mid-cell available for assembly of the Z-ring in advance of the completion of chromosome replication and segregation.
If clearing Noc from the mid-cell sites was physiologically relevant, the strain with NBS arrays near terC might affect the timing and or localization of cell division. This seemed to be the case because we were able to detect a reproducible increase in cell length of the strain with terminal NBS arrays (average length 3.6 arbitrary unit) compared with the isogenic strain with no inserted arrays (3.1 arbitrary unit) (Figure 2G–L).
Binding Noc to a plasmid results in a severe block in cell division
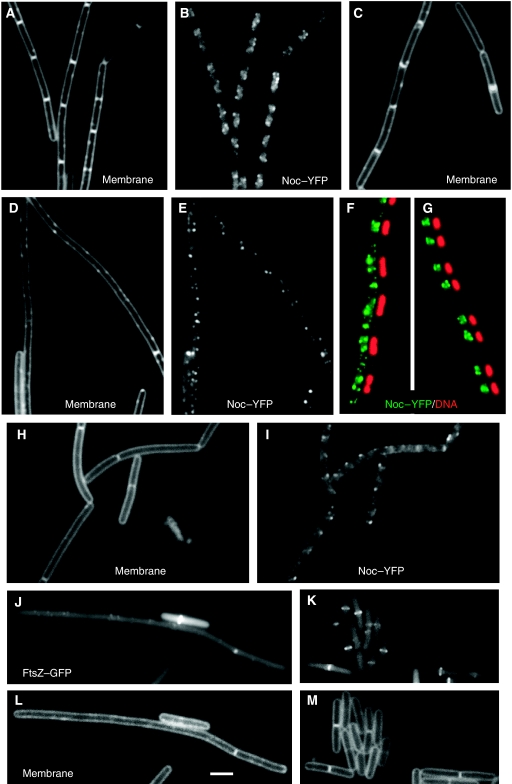
We showed earlier that overexpression of noc led to a delay in cell division and a slight increase in cell length (Wu and Errington, 2004). We wondered whether the failure to generate a more severe cell division block was due to sequestration of Noc to specific regions of the chromosome that occupy relatively defined sites along the length of the cell, thus causing a delay, at most, in the assembly of Z-rings. We have tried to isolate lethal alleles of noc, reasoning that it should be possible to impair DNA binding, but leave division inhibition activity intact. So far, it has been unsuccessful, suggesting that binding to DNA may be required in some way for inhibition of division. As an alternative way of testing whether DNA localization constrains the division inhibition activity of Noc, we tested the effects of placing an NBS on an autonomously replicating plasmid, pSpa-gfp-RBS (Bongers et al, 2005), which should be less constrained, positionally, than the chromosome. The presence of the plasmid had no detectable effect on cell length, and when Noc–YFP was expressed (as the only copy of noc and from the Pxyl promoter), it showed the normal pattern of localization (Figure 6A and B). However, when a derivative of the plasmid carrying an array of eight NBSs (pSG4929) was introduced into the cells, a filamentous phenotype was seen, indicative of a severe block in cell division (Figure 6D and E). This phenotype was accompanied by a conspicuous change in the localization of Noc, with spots now seen throughout the cells (compare Figure 6E with 6B; Supplementary Figure S5A with S5D). We assumed that the extra-nucleoidal spots of Noc were associated with the plasmid. As one way of assessing this, we treated the cells with chloramphenicol, which results in a marked condensation of the chromosome (Zimmerman, 2006). As shown in Figure 6G, Noc was almost exclusively located over the condensed nucleoids in cells containing the empty vector plasmid, whereas there were prominent additional spots of Noc in the internucleoid spaces when the plasmid carried the NBS array (Figure 6F). The filamentous phenotype was dependent on Noc, because in the absence of xylose division appeared normal (Figure 6C). Cells expressing the defective K164A mutant form of Noc also did not filament (not shown). Thus, the cell division block required both the plasmid carrying the NBS array and the functional Noc protein.
Figure 6.
Localization of Noc–YFP or FtsZ–GFP in cells harbouring multicopy plasmids. (A, B and G) Cells carrying a multicopy plasmid without an NBS (strain 4706) and grown in the presence of 0.3% xylose had normal cell length and localization pattern of Noc. (C–F) Cells harbouring a multicopy plasmid carrying an NBS array (strain 4707) grown in the presence (D–F) or absence (C) of xylose. Cells in F and G were treated with chloramphenicol. (H and I) Cells harbouring a multicopy plasmid carrying one NBS (strain 4708) grown in the presence of 0.3% xylose. (J–M) Localization of FtsZ–GFP in cells harbouring a multicopy plasmid carrying an NBS array (J and L, strain 4714) or without the array (K and M, strain 4715) grown in the presence of IPTG for the expression of noc. B, E and I show the localization of Noc–YFP; A, C, D, H, L and M are images of membranes; J and K show the localization of FtsZ–GFP; F and G are side-by-side images with Noc–YFP shown in green and DNA (DAPI) in red. Scale bar, 2 μm.
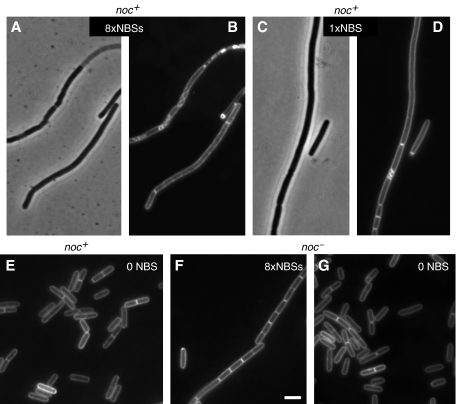
To see whether a single NBS was sufficient to affect cell division, we deleted seven copies of the NBSs from pSG4929 and introduced the resulting plasmid (pSG4930), containing only one copy of NBS, into B. subtilis. Cells again showed an altered localization pattern of Noc and were generally much longer than cells with the control plasmid, but shorter than cells with pSG4929 (Figure 6H and I). Next, we tested whether the cell division block by these multicopy plasmids required elevated levels of Noc, as the above observation was obtained under conditions in which noc–yfp was expressed from Pxyl. When the NBS array plasmid (pSG4929) was introduced into a noc null mutant strain (1282), normal-looking transformants were readily obtained (Figure 7F and G). In contrast, the wild-type strain 168 gave very few transformants, and those that were obtained grew slowly and their cells were filamentous (Figure 7A–D). This effect was seen even with a plasmid bearing a single NBS (Figure 7C and D), but not with a plasmid devoid of an NBS (Figure 7E). Therefore, even with wild-type Noc expressed at wild-type levels, the plasmid was able to compete with the chromosome and recruit sufficient Noc to severely inhibit cell division.
Figure 7.
Effect of a plasmid carrying an NBS on cell division in Noc+ cells. The cells were wild type (A–E) or noc null mutant (F, G). The plasmid contained either an array of eight copies of NBS (A, B, F), one NBS (C and D) or no NBS (E, G). A and C are phase contrast images; B, D–G are images of membrane stain. Scale bar, 2 μm.
To check whether the effect on division was due to impaired FtsZ ring formation, as shown earlier for Noc-dependent division effects, we introduced an ftsZ–gfp fusion into cells carrying the plasmid with or without the NBS array and expressing wild-type Noc from an IPTG-inducible promoter (Figure 6J–M). Again a division defect was evident (which was IPTG dependent; not shown) when the plasmid carried the NBS array (strain 4714), and this defect was accompanied by the near failure to assemble the usual bands (rings) of FtsZ at the division sites. These results show that Noc bound to DNA through one or more NBSs is an extremely potent inhibitor of division. Its activity is normally restricted topologically within the cell through association with NBSs, which cover most of the chromosome apart from the replication terminus zone.
Discussion
A specific recognition sequence (NBS) for Noc protein
A combination of ChAP-on-Chip and bioinformatics analyses allowed us to define a consensus NBS, a 14-bp inverted repeat with the sequence 5′-ATTTCCCGGGAAAT-3′. Several lines of evidence, both in vivo and in vitro, showed that this NBS is recognized specifically by Noc. Noc is closely related to the Spo0J/ParB family of proteins that are involved in plasmid and chromosome segregation. In B. subtilis, Spo0J forms foci that colocalize with oriC regions by binding to and spreading along DNA from the eight parS nucleation sites clustered around oriC (Glaser et al, 1997; Lewis and Errington, 1997; Lin and Grossman, 1998; Murray et al, 2006; Breier and Grossman, 2007). The width of the peaks of Noc binding detected by ChAP-on-Chip supports the idea that Noc spreads on DNA in a similar manner to Spo0J, and preliminary biochemical analysis suggests that Noc forms oligomers both in vitro and in vivo (LJW, unpublished).
The mechanism of division inhibition by Noc and its possible dependence on DNA binding
In E. coli, SlmA is able to bind FtsZ directly and so it probably blocks FtsZ ring assembly by depleting FtsZ from the pool of FtsZ polymers (Bernhardt and de Boer, 2005). However, it is not yet known how Noc interacts with the division machinery of B. subtilis. Our new data using a Noc–YFP fusion revealed the presence of foci of Noc associated with the cell periphery over the nucleoid. It seems likely that these foci represent proteins actively interacting with the division machinery, as the various proteins of the divisome are all directly or indirectly associated with the cell membrane. This association does not seem to require FtsZ assembly, as long filaments formed after cells were treated with FtsZ inhibitors still show peripheral Noc foci (LJW and J Schneeweiss, unpublished). Several models could explain these findings. Noc might not only target FtsZ to inhibit division, but also interact with other divisome proteins to reach the cell periphery. Second, Noc might act not on FtsZ, but on one or more yet un-identified proteins upstream of FtsZ in the assembly hierarchy of the divisome. Third, Noc might interact with non-divisome proteins at the membrane, or with the membrane itself to reach the cell periphery.
Whatever is the mechanism of inhibition of division, it now seems that this activity requires binding of Noc to DNA. The binding and spreading mechanism of Noc association with the chromosome may be ideally suited to Noc function. By analogy to Spo0J, the NBS sites presumably nucleate Noc binding, and excess Noc then spreads out along adjacent DNA. This should ensure that the chromosome is protected from guillotining by the division septum relatively independent of Noc levels in the cell. Indeed, we have noted that substantial overexpression of Noc has only a small effect on cell division (Wu and Errington, 2004). Strikingly, however, when one or more NBSs was placed on a plasmid in the presence of normal levels of Noc, a severe inhibition of division was seen. Therefore, Noc is a powerful potential inhibitor of cell division, but its activity is normally kept under tight topological control by association with chromosomal NBSs in vivo. We suggest that binding to DNA may be crucial for the inhibition of division activity of Noc. Perhaps, an array of Noc molecules lined up on DNA interacts in a multivalent manner with the polymeric proteins in the divisome.
During the B. subtilis spore development, chromosomes adopt a conformational change to form an extended structure termed as the ‘axial filament', with the two oriC regions anchored to opposite cell poles. This is followed by a division site switch, from the mid-cell to a sub-polar position. Our earlier studies have shown that the asymmetric septum is positioned such that a region about 700 kb from oriC on each chromosomal arm is enclosed in the smaller (prespore) compartment on septation (Wu and Errington, 1998). Interestingly, NBSs are less abundant in the regions about 200–700 kb from oriC on both arms of the chromosome (NBS 61 to NBS 65 on the left and NBS 6 to NBS 12 on the right). It is possible that absence of Noc in these regions allows the division septum to form asymmetrically. It would be interesting to see whether introducing NBSs to these regions would cause a block to asymmetric septation.
Positioning of NBSs on the chromosome may fine-tune the coordination of chromosome replication and cell division
Efficient cell cycle progression requires that the major cellular processes of DNA replication, segregation and cell division are well coordinated. We showed earlier that under conditions in which DNA replication has been perturbed, Noc acts as an antiguillotine device to protect the chromosome from being bisected by the division septum. The new finding that Noc is absent specifically from the replication terminus region suggests that the protein might also be a temporal regulator of cell division. Indeed, when NBSs were inserted near the terminus region, Noc was recruited to the region and this led to a delay in cell division.
The Noc-free terminus region, spanning from about 1615 to 2770 kbp on the B. subtilis chromosome, corresponds to about 28% of the chromosome. Interestingly, earlier work by Wake and co-workers using inhibitors of DNA replication on germinating spores showed that replication needs to progress through about 60–70% of the chromosome before a centrally positioned division septum can form (McGinness and Wake, 1979, 1981; Wu et al, 1995). It was noticed that the nucleoid tends to adopt a bilobed configuration at about this stage of replication, so the release of division site could be due to the appearance of a DNA-deficient space between the two chromosomal lobes. However, in the light of our new results, it now seems that the 60–70% sensitive stage corresponds roughly to the point at which the Noc-free terminus zone first comes to the mid-cell to be replicated. Thus, the positioning of the NBS and hence recruitment of Noc might serve as a timing device, allowing the division machinery to begin assembling at a defined moment late in the DNA replication cycle. The cell division septum takes several minutes to be synthesized, so that the Noc-free zone may allow the cell to anticipate the completion of replication, and close the division septum to generate new daughter cells immediately after their chromosomes are finished.
Materials and methods
Bacterial strains and plasmids
B. subtilis strains used in this study are listed in Table I, together with the plasmids used and their construction.
Table 1.
B. subtilis strains and plasmids
| Strain/plasmid | Relevant genotypea | Construction, source or referenceb |
|---|---|---|
| B. subtilis | ||
| 168ED | trpC2 | Laboratory stock |
| AT62 | cgeD∷pAT12(cat lacOx256) veg∷pAT27(erm Pveg-gfpF64L S65T-lacI) | Teleman et al (1998) |
| KPL682 | phe trp thrC∷Ppen-lacIÄ11-cfp(W7) mls | Lemon and Grossman (2000) |
| 1282 | trpC2 Δnoc∷tet | Wu and Errington, 2004 |
| 1283 | trpC2 Δnoc∷spc | Wu and Errington, 2004 |
| 2020 | trpC2 amyE∷(spc Pxyl-gfpmut1-ftsZ) | J Sievers (unpublished) |
| 4701 | trpC2 amyE∷(spc Pxyl-noc-yfpmut1) | pSG4925 → 168 (Sp) |
| 4702 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut1) | 4701 DNA → 1282 (Sp) |
| 4703 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut1) cgeD∷pAT12(cat lacOx256) | AT62 DNA → 4702 (Cm) |
| 4704 | trpC2 noc∷pSG4927(‘noc-12xhis) | pSG4927 → 168 (EL) |
| 4705 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut1) cgeD∷pAT12(cat lacOx256) thrC∷Ppen-lacIÄ11-cfp(W7) mls | KPL682 → 4703 (EL) |
| 4706 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut2) pPspaS-gfp-RBS | pPspaS-gfp-RBS → 4702 (Em) |
| 4707 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut2) pSG4929(PspaS8xNBS(ydbO) erm) | pSG4929 → 4702 (Em) |
| 4708 | trpC2 Δnoc∷tet amyE∷(spc Pxyl-noc-yfpmut2) pSG4930(PspaS1xNBS(ydbO) erm) | pSG4930 → 4702 (Em) |
| 4712 | trpC2 noc∷pSG4934 (kan Pspac-'noc) | pSG4934 → 168 (km) |
| 4713 | trpC2 noc∷pSG4934 (kan Pspac-'noc) ΔamyE∷(spc Pxyl-gfpmut1-ftsZ) | 2020 DNA → 4712 (Sp) |
| 4714 | trpC2 noc∷pSG4934 (kan Pspac-'noc) ΔamyE∷(spc Pxyl-gfpmut1-ftsZ) pSG4929(PspaS 8xNBS(ydbO) erm) | pSG4929 → 4713 (Em) |
| 4715 | trpC2 noc∷pSG4934 (kan Pspac-'noc) ΔamyE∷(spc Pxyl-gfpmut1-ftsZ) pPspaS-gfp-RBS | pPspaS-gfp-RBS → 4713 (Em) |
| 4716 | trpC2 ΔywsB(NBS)∷tet | See Materials and methods |
| 4717 | trpC2 ywsB∷tet | See Materials and methods |
| 4718 | trpC2 yogA∷(8xNBS(ydbO) kan) | See Materials and methods |
| 4720 | trpC2 yogA∷ kan | See Materials and methods |
| 4721 | trpC2 yodA∷(16xNBS(ydbO) erm) | See Materials and methods |
| 4722 | trpC2 yodA∷erm | See Materials and methods |
| 4723 | trpC2 noc∷pSG4927(‘noc-12xhis) ΔywsB(NBS)∷tet yogA∷(8xydbO kan) | 4716 DNA → 4704 (Te), then 4718 DNA → resulting strain (Km) |
| 4727 | trpC2 noc∷pSG4927(‘noc-12xhis) ywsB∷tet yogA∷kan | 4717 DNA → 4704 (Te), then 4720 DNA → resulting strain (Km) |
| 4729 | trpC2 noc∷pSG4927(‘noc-12xhis) ΔywsB(NBS)∷tet spoVFB∷pSG4935 (kan ‘spoVFB 2xNBS(ydbO)) | 4716 DNA → 4704 (Te), then pSG4935 → resulting strain (Km) |
| Plasmids | ||
| pUK19 | bla kan | B Haldenwang (unpublished) |
| pSG840 | bla erm | Laboratory stock |
| pSG441 | bla aph-A3 lacI pspac | Laboratory stock |
| pMUTinHis | bla erm lacI Pspac-12xhis | Ishikawa et al (2006) |
| pET16B | bla lacI Pspac-10xhis | Novagen |
| pSG5472 | bla amyE' spc Pxyl-yfpmut1 ‘amyE | A Formstone (unpublished) |
| pSG1154 | bla amyE' spc Pxyl-gfpmut1 ‘amyE | Lewis and Marston (1999) |
| pSpa-gfp-RBS | PspaS-gfp erm | J-W Veening (unpublished) |
| pSG4924 | bla amyE' spc Pxyl-yfpmut1 ‘amyE | yfp (PCR from pSG5472, EcoRI+SpeI) into pSG1154 (EcoRI+SpeI) |
| pSG4926 | bla amyE' spc Pxyl-noc yfpmut1 ‘amyE | noc+RBS (PCR, AvrII+SalI) into pSG4924 (AvrII+XhoI) |
| pSG4927 | bla erm lacI Pspac- noc-12xhis | ‘noc (PCR, EcoRI+XhoI) into pMUTinHis (EcoRI+XhoI). |
| pSG4928 | bla erm lacI Pspac-noc(K164A)-12xhis | Site-directed mutagenesis from pSG4927 |
| pSG4929 | PspaS 8xNBS(ydbO) erm | See Materials and methods |
| pSG4930 | PspaS 1xNBS(ydbO) erm | See Materials and methods |
| pSG4931 | pET16B-noc-12xhis | See Materials and methods |
| pSG4932 | pET16B-noc(K164A)-12xhis | See Materials and methods |
| pSG4934 | bla aph-A3 lacI Pspac-noc' | noc (PCR, EcoRV+BgiII) into pSG441 (SmaI+BgiII) |
| pSG4935 | bla kan ‘spoVFB 2xNBS(ydbO) | ‘spoVFB (PCR, SacI+XhoI) into pUK19-2xNBS (SacI+XhoI) |
| a'X or X', the 5′ end or the 3′ end of the gene X has been truncated. Resistance gene abbreviations as follows: bla, ampicillin; cat, chloramphenicol; erm and mls, erythromycin; kan, kanamycin; spc, spectinomycin; tet, tetracycline. | ||
| bFor strains constructed by transformation, the source of the DNA used in the transformation is given first, with restriction enzymes, where used. The recipient strain is indicated after the arrow, with selected marker in parentheses: Cm, chloramphenicol; Em and EL, erythromycin; Km, kanamycin; Sp, spectinomycin; Te, tetracycline. | ||
General methods
B. subtilis cells were made competent for transformation with DNA either by the method of Kunst and Rapoport (Kunst and Rapoport, 1995) or by the method of Anagnostopoulos and Spizizen (Anagnostopoulos and Spizizen, 1961) as modified by Jenkinson (Jenkinson, 1983). DNA manipulations and E. coli transformations were carried out using standard methods (Sambrook et al, 1989). Solid medium used for growing B. subtilis was nutrient agar (Oxoid) and liquid media were PAB (Oxoid Antibiotic Medium no 3), CH medium or LB. Chloramphenicol (5 μg ml−1), kanamycin (5 μg ml−1), tetracycline (12 μg ml−1), erythromycin (1 μg ml−1) and lincomycin (25 μg ml−1) were added as required. Media used for growing E. coli were LB (Sambrook et al, 1989) and nutrient agar supplemented with ampicillin (100 μg ml−1), erythromycin (100 μg ml−1) or chloramphenicol (20 μg ml−1) as required. Transformants of B. subtilis harbouring plasmids were initially selected on plates containing erythromycin at 2 μg ml−1, followed by screening on plates containing 5 μg ml−1 of erythromycin.
Fluorescence microscopy
Cells containing gfp/yfp/cfp fusion were all grown at 30°C. Cells were stained and viewed on agarose (1.2%) slides and images were obtained as described earlier (Wu and Errington, 2004). When FM5-95 was used for staining the membranes, 70 μl of the culture was mixed with 1 μl of the FM5-95 (Molecular Probe) solution (200 μg ml–1) in an Eppendorf. The gfp–ftsZ fusion was expressed ectopically from an inducible Pxyl promoter integrated at amyE with 0.08% inducer (xylose) in the presence of the wild-type untagged copy of FtsZ. The noc–yfp fusion was expressed ectopically from an inducible Pxyl promoter integrated at amyE with 0.2–0.5% xylose. Timelapse microscopy was performed using a Yokogawa Spinning Disc Confocal System, with a 491-nm laser and coupled to a Coolsnap HQ2 Camera, and the images were collected using 0.3 s exposure time.
ChAP-on-Chip analysis
ChAP-on-chip analysis was performed as described earlier(Ishikawa et al, 2007), using affinity-purified Noc-complexes from each His-tagged B. subtilis strain. Noc-binding signals were analysed and visualized by a software package, in sillico Molecular Cloning Array Edition (imc_ae, in sillico biology, inc.), as the values that divided signal intensities of DNA in the affinity-purified fraction (ChAP DNA) by those of DNA isolated from the whole cell extract fraction before the purification (control DNA), as described earlier (Cho et al, 2008), with the following modifications. To remove abnormal low signals from those of the control DNA, the lowest 10% signals were removed from the control DNA data before the division. The two highest signals in every 100 probe along the genome were eliminated to remove abnormal high peaks after the division. Note that it was confirmed by imc_ae software that the overall peak patterns were not changed by these procedures. Analysis of Spo0J was also carried out in the same way, based on earlier published data (Ishikawa et al, 2007). Raw data (CEL format) from the ChAP-on-chip experiments described here have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/microarray-as/ae/) under accession number E-MEXP-2133.
Data analysis
Protein-binding peaks were automatically detected as following. The signals with higher values than threshold, which were determined as ⩾1.4 for Noc and ⩾3.0 for Spo0J depending on their background levels, were concatenated when the distance of neighbouring signals was less than 400 bp, and the regions containing ⩾50 signals were defined as protein-binding regions. Signals on ribosomal RNA, which make signals higher than background level because of their high copy number on genome, were removed from the result.
The PWM values for NBSs were created from data set of all NBSs involved NBRs using web-based programs: WebLog 3 for creating sequence logo (Crooks et al, 2004) and Virtual Footprint Version 3.0 for search of NBSs with high PWM scores on the B. subtilis genome (Munch et al, 2005).
Plasmid construction
We initially attempted to construct NBS arrays consisting of NBSs from three different locations (in the ydbO, ykoW and dhbF genes, respectively) in the vector pUK19 by annealing complimentary primer pairs and several rounds of restriction enzyme digestion and ligation. However, the first construct obtained consisted of only one NBS, in which the ydbO fragment containing the NBS (digested with EcoRV and SalI) had been inserted between SmaI and SalI in pUK19. The plasmid was named pUK19-1xNBS and was used to amplify the copy number of the NBS. To do this, the NBS(ydbO) fragment was purified from pUK19-1xNBS (after digestion with EcoRI and SalI) and ligated to pUK19-1xNBS digested with EcoRI and XhoI to duplicate the NBS(ydbO) fragment. The process was repeated a few times until the number of NBS reached eight copies, giving pUK19-2xNBS, pUK19-4xNBS and pUK19-8xNBS, respectively.
The 8xNBS from pUK19-8xNBS was also subcloned into pSG840 using SphI and XhoI (for the insert) or SalI (for pSG840) to generate pSG840-8xNBS. The pSG840-16xNBS, carrying 16xNBS, was constructed in several steps: first, the 8xNBS fragment (obtained by polymerase chain reaction (PCR) from pUK19-8xNBS using primers pUK19-F and pUK19-R(KpnI)) was digested with XhoI and then ligated to the SalI-digested pUK19-8xNBS. The ligation product was then digested with XhoI and SphI, and the 16xNBS fragment was gel purified before being ligated to SphI and SalI-digested pSG840.
To introduce an NBS array into the B. subtilis high copy plasmid pSpa-gfp-RBS, the 8xNBS(ydbO) fragment was isolated from pSG840-8xNBS after restriction enzyme digestion with HindIII and XbaI, then ligated to the vector (obtained by PCR using primers Pspaspn-R(XbaI) and GFP-F(XhoI)) digested with the same enzymes. The resulting plasmid, in which the 8xNBS array had replaced gfp in pSpa-gfp-RBS, was named pSG4929. For pSG4930, that carried only one copy of NBS, pSG4929 was digested with SmaI and re-ligated to excise seven copies of the NBS.
The pSG4935 was constructed to allow introduction of NBSs at the spoVFB locus (at 1744.6 kb) on the B. subtilis chromosome by single cross-over integration. The 460-bp ‘spoVFB fragment (including the stop codon, but without the N-terminus coding portion) was amplified from the chromosomal DNA of the B. subtilis wild-type strain 168ED using primers spoVFB-F(SacI) and spoVFB-R(XhoI), digested with SacI and XhoI and then inserted into pUK19-2xNBS between the SacI and XhoI sites.
The pSG4931 and pSG4932 were constructed for overexpression of the wild-type and mutant noc–12xhis fusions in E. coli; noc–12xhis and noc(K164A)-12xhis were obtained by PCR from chromosomal DNAs of B. subtilis strains 4704 and 4705 (carrying the 12xhis fusion), respectively, using primers yyaA-F(BspHI) and pmutinHis-R(BamHI). The PCR products were then digested with BspHI and BamHI, and cloned into pET16B between the NcoI and BamHI sites.
Construction of B. subtilis strains containing NBS deletion/insertions
The NBS located at 3706 kb on the B. subtilis chromosome is just downstream of the terminator of ywsB and upstream of the coding sequence of ywsA. To delete this NBS, DNA fragments (about 1.8 kb long) from upstream and downstream of the NBS were amplified from the wild-type strain 168 by PCR, digested with HindIII and NotI, respectively, then ligated to the tetracycline-resistance gene excised from pBEST309 using the same enzymes. The ligation mixture was transformed into the wild-type B. subtilis strain 168 directly with the selection for tetracycline resistance. The ‘downstream' fragment contained only the last 5 bps of the NBS, and so insertion of the tet gene resulted in deletion of most of the NBS. A control construct was constructed in the same way, except that the ‘downstream' fragment contained the whole of the NBS and, therefore, the NBS would be retained after insertion of tet. Several transformants for each construct were examined by PCR and sequencing to confirm the insertional modifications, and one of the NBS deletion strains was designated 4716 and one of the control constructs was designated 4717.
Insertion of NBS arrays at 2007 and 2126 kb on the chromosome was done using the same method. The insertion point at the 2007-kb position was between the stop codon of yogA and the putative terminator of gltB. DNA fragments (about 2 kb long) from upstream and downstream of the insertion point were amplified from the genomic DNA of strain 168 by PCR, digested with NdeI and SalI, respectively, then ligated to the 8xydbO-kan cassette excised from pUK19-8xNBS using NdeI and XhoI, or the kan cassette from pUK19. The insertion point at the 2126-kb position was between the putative terminators of yogA and yogB. The DNA fragments from upstream and downstream of the insertion point were digested with EcoRI and SalI, respectively. The 16xydbO-erm cassette and the erm cassette were excised from pSG840-16xNBS and pSG840, respectively, using the same enzymes. Several transformants for each construct were examined by PCR and sequencing to confirm the modifications. The 8xydbO-kan-insertion strain at 2007 kb was desinated 4718 and the kan-insertion strain (control) 4719. The 16xydbO-erm-insertion strain at 2126 kb was desinated 4721 and the erm-insertion strain (control) 4722.
Protein expression and purification
E. coli strains BL21(DE3)/pLys harbouring plasmids pSG4931 or pSG4932 were grown in LB at 37°C to an OD600 of 0.5, at which point IPTG was added to a final concentration of 3 mM to induce protein expression. After 3 h at 30°C, cells were harvested and resuspended in CelLytic-B Plus (Sigma) to lyse the cells. Benzonase was omitted from the lysis buffer, but NaCl was included at a final concentration of 400 mM. The lysate was incubated at room temperature by shaking for 15 min, then sonicated on ice (10 s at level 4 using a Sonics Vibracell, three rounds) to break the DNA. To precipitate DNA, 30% streptomycin sulphate (in wash buffer) was added to the lysate (to a final concentration of 2.2%), and the mixture was incubated at 4°C by shaking for 30 min, then centrifuged first at 3273 g and then at 17 000 g at 4°C for 15 min each. The supernatant was then used for purification using HIS-Select Spin Columns (Sigma). Briefly, the cleared lysate was passed through the column (pre-equilibrated with wash buffer) and the column was washed six times with wash buffer. The Noc–12xHis fusion protein was eluted with elution buffer. Buffer exchange was performed using PD-10 desalting columns (Amersham) with storage buffer, and samples were concentrated using an Ultrafree-10 centrifugal filter device (Millipore). The fusion proteins were estimated to be >98% pure as judged by SDS–PAGE. Wash buffer contained 500 mM NaCl, 50 mM sodium phosphate (pH 8), 15 mM imidazole, 10% glycerol and EDTA-free protease inhibitor (Roche). Elution buffer contained 500 mM NaCl, 50 mM sodium phosphate (pH 8), 250 mM imidazole, 10% glycerol and EDTA-free protease inhibitor (Roche). Storage buffer contained 300 mM NaCl, 30 mM HEPES-KOH (pH 7.4), 1 mM DTT and 10% glycerol.
Electrophoretic mobility-shift assay
DNA fragments used for the assay were obtained by annealing pairs of complimentary oligonucleotides. The cy5-ydbO-F (AAAAAGTTTCCCGGGCAATAATTT) and cy5-ydbO–R (AAATTATTGCCCGGGAAACTTTTT) contained the NBS from ydbO and had been labelled at their 5′-termini with a Cy5 fluorophore (Sigma-Genosys). The unlabelled competitor DNAs contained either the wild-type NBS site (sequence same as above) or a mutant site [ydbOmut-F (AAAAAGTATCATGGCCTATAATTT) and its compliment ydbOmut–R (AAATTATAGGCCATGATACTTTTT)]. Binding reactions (18 μl) were performed in 20 mM HEPES (PH8.0), 1 mM DTT, 200 mM KoAc, 5 mM MgCl2, 100 μg ml–1 BSA, 10% glycerol, 0.025% nonidetP-40, 0.09 μg ul–1 of Poly (dI:dC), 790 ng of protein and 25 nM of labelled DNA probe. Where unlabelled 24 bp competitor DNA was included, its concentration ranged from 125, 250, 500, 1000 and 2000 nM. The reactions were assembled on ice and then incubated for 25 min at room temperature before 16.8 ul was loaded onto a pre-run 5% acrylamyde gel (in 0.5 × TBE) and electrophoresed at 100 V for 1.5 h at 4°C. Gels were imaged directly using a Typhoon Trio imager and bands were analysed using ImageQuant TL software (V2005).
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
Work in the Errington laboratory was supported by a grant from the Biotechnology and Biological Sciences Research Council, and that in the Ogasawara laboratory by a KAKENHI grant-in-aid for scientific research in the Priority Area ‘Systems Genomics' from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Jan-Willem Veening, Alex Formstone and Bill Haldenwang for providing vectors.
References
- Anagnostopoulos C, Spizizen J (1961) Requirements for transformation in Bacillus subtilis. J Bacteriol 81: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak I, Wilkinson AJ (2007) Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol Rev 31: 311–326 [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers RS, Veening JW, Van Wieringen M, Kuipers OP, Kleerebezem M (2005) Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl Environ Microbiol 71: 8818–8824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Grossman AD (2007) Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol 64: 703–718 [DOI] [PubMed] [Google Scholar]
- Cho E, Ogasawara N, Ishikawa S (2008) The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet Syst 83: 111–125 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ (2003) Cytokinesis in bacteria. Microbiol Mol Biol Rev 67: 52–65, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J (1997) Dynamic, mitotic-like behaviour of a bacterial protein required for accurate chromosome partitioning. Genes Dev 11: 1160–1168 [DOI] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K (2008) Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev 22: 3475–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Levin PA (2008) The great divide: coordinating cell cycle events during bacterial growth and division. Curr Opin Microbiol 11: 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA 96: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N (2006) A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol Microbiol 60: 1364–1380 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y, Kurokawa K, Oshima T, Ogasawara N (2007) Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res 14: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF (1983) Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol 129: 1945–1958 [DOI] [PubMed] [Google Scholar]
- Kunst F, Rapoport G (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol 177: 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD (2000) Movement of replicating DNA through a stationary replisome. Mol Cell 6: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Errington J (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol 25: 945–954 [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227: 101–109 [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Grossman AD (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685 [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol 33: 84–96 [DOI] [PubMed] [Google Scholar]
- McGinness T, Wake RG (1979) Division septation in the absence of chromosome termination in Bacillus subtilis. J Mol Biol 134: 251–264 [DOI] [PubMed] [Google Scholar]
- McGinness T, Wake RG (1981) A fixed amount of chromosome replication needed for premature division septation in Bacillus subtilis. J Mol Biol 146: 173–177 [DOI] [PubMed] [Google Scholar]
- Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D (2005) Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21: 4187–4189 [DOI] [PubMed] [Google Scholar]
- Murray H, Errington J (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84 [DOI] [PubMed] [Google Scholar]
- Murray H, Ferreira H, Errington J (2006) The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol 61: 1352–1361 [DOI] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL (2005) Spatial control of bacterial division-site placement. Nat Rev Microbiol 3: 959–968 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Scheffers DJ (2008) The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett 582: 2601–2608 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC-H, Grossman AD, Losick R (1998) Chromosome arrangement within a bacterium. Curr Biol 8: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J (2006) Septum enlightenment: assembly of bacterial division proteins. J Bacteriol 188: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS (2004) Bacterial cell division and the septal ring. Mol Microbiol 54: 588–597 [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N (1990) Role of the nucleoid in the toporegulation of division. Res Microbiol 141: 39–49 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (1998) Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol 27: 777–786 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Franks AH, Wake RG (1995) Replication through the terminus region of the Bacillus subtilis chromosome is not essential for the formation of a division septum that partitions the DNA. J Bacteriol 177: 5711–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W (1999) FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Zimmerman SB (2006) Shape and compaction of Escherichia coli nucleoids. J Struct Biol 156: 255–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File