Abstract
During stress-induced apoptosis, the initiator caspase-9 is activated by the Apaf-1 apoptosome and must remain bound to retain significant catalytic activity. Nevertheless, in apoptotic cells the vast majority of processed caspase-9 is paradoxically observed outside the complex. We show herein that apoptosome-mediated cleavage of procaspase-9 occurs exclusively through a CARD-displacement mechanism, so that unlike the effector procaspase-3, procaspase-9 cannot be processed by the apoptosome as a typical substrate. Indeed, procaspase-9 possessed higher affinity for the apoptosome and could displace the processed caspase-9 from the complex, thereby facilitating a continuous cycle of procaspase-9 recruitment/activation, processing, and release from the complex. Owing to its rapid autocatalytic cleavage, however, procaspase-9 per se contributed little to the activation of procaspase-3. Thus, the Apaf-1 apoptosome functions as a proteolytic-based ‘molecular timer', wherein the intracellular concentration of procaspase-9 sets the overall duration of the timer, procaspase-9 autoprocessing activates the timer, and the rate at which the processed caspase-9 dissociates from the complex (and thus loses its capacity to activate procaspase-3) dictates how fast the timer ‘ticks' over.
Keywords: Apaf-1, caspase-9, apoptosome, apoptosis, molecular timer
Introduction
Stressors, such as DNA damage and growth-factor deprivation, often induce apoptosis (programmed cell death) through the activation of caspases (cysteine proteases) (Taylor et al, 2008). In these instances, proapoptotic Bcl-2 family members, such as Bax and Bak, mediate mitochondrial outer-membrane permeabilization, resulting in the release of proapoptotic factors, such as cytochrome c (Cc), from the intermembrane space into the cytosol (Chipuk and Green, 2008; Youle and Strasser, 2008). In the presence of modest levels of dATP or ATP, Cc then binds to apoptotic protease-activating factor-1 (Apaf-1)—a cytosolic adaptor protein comprised of an N-terminal caspase recruitment domain (CARD), a nucleotide binding/oligomerization domain, and a series of thirteen C-terminal WD40 repeats—and induces its oligomerization into a large heptameric complex referred to as the ‘apoptosome' (Li et al, 1997; Saleh et al, 1999; Zou et al, 1999; Cain et al, 2000; Jiang and Wang, 2000; Kim et al, 2005; Yu et al, 2005; Chandra et al, 2006). The apoptosome subsequently recruits the initiator procaspase-9 (ProC9) through its own CARD, resulting in its activation (Stennicke et al, 1999; Bratton et al, 2001).
On the basis of two prominent schools of thought, the apoptosome activates ProC9 either by facilitating its dimerization within the complex (Boatright et al, 2003; Pop et al, 2006) or by inducing a conformational change in the monomeric enzyme (Rodriguez and Lazebnik, 1999; Shiozaki et al, 2002; Chao et al, 2005), though neither model is necessarily mutually exclusive. In any event, ProC9 activity is markedly enhanced on its association with the apoptosome and undergoes subsequent autoprocessing between its large and small subunits at Asp 315 to generate the processed p35/p12 form of the enzyme (C9-p35/p12). Importantly, regardless of their processed states, caspase-9 proteins must remain bound to the complex to show significant proteolytic activity, and apoptosome-bound ProC9 or C9-p35/p12 then recruits and activates the effector procaspase-3 (ProC3) (Rodriguez and Lazebnik, 1999; Bratton et al, 2001).
A previous structural and biochemical study had suggested that the isolated CARDs in Apaf-1 and caspase-9 bind very tightly to one another (Qin et al, 1999). Intriguingly, however, on isolation of apoptosome complexes from apoptotic and Cc/dATP-activated lysates, most of the processed enzyme is generally observed outside the complex (Cain et al, 2000). Thus, it remains unclear whether a relatively small number of Apaf-1•caspase-9 apoptosome complexes, initially formed after the release of Cc, process the majority of ProC9 as a typical substrate, similar to the way the apoptosome processes ProC3 (Figure 1C, ‘CARD-static' model) or alternatively, whether ProC9 might continuously displace processed C9-p35/p12 from the complex and in turn undergo autoprocessing to complete the cycle (Figure 1C, ‘CARD-displacement' model).
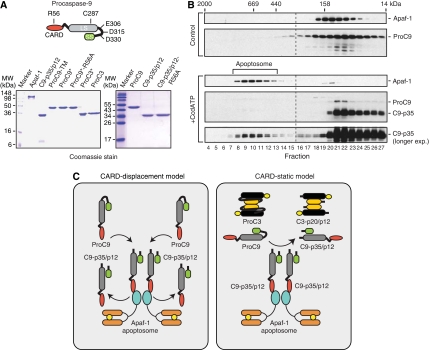
Figure 1.
Procaspase-9 autoprocessing by the Apaf-1 apoptosome. (A) A schematic of procaspase-9 is shown with its CARD, large subunit (LS), activation loop, and small subunit (SS). Crucial amino acids in the active site (C287), the CARD (R56), and the activation loop (E306, D315, and D330) are highlighted. Purified wild-type and mutant Apaf-1, caspase-9, and caspase-3 proteins (10 μg each) were subjected to SDS–PAGE and stained with Coomassie blue. (B) Recombinant Apaf-1 and ProC9 were incubated with and without Cc/dATP. The samples were subsequently fractionated by Superose-6 gel-filtration chromatography, resolved using SDS–PAGE, and immunoblotted using Apaf-1 and caspase-9 antibodies (Cain et al, 2000). (C) CARD-displacement and CARD-static models for the autoprocessing of ProC9. Details are described in the text.
In our efforts to distinguish between these two models, we have made a series of important observations using reconstituted apoptosome complexes with various caspase-9 mutants and inhibitors, such as X-linked inhibitor of apoptosis (XIAP) (Salvesen and Duckett, 2002). Collectively, our results indicate that the purpose of ProC9 autoprocessing is not to activate caspase-9, but rather to initiate a molecular timer that regulates the duration of apoptosome activity. In fact, parallels between the intrinsic proteolytic and GTPase activities of ProC9 and Ras/Rab GTPases, respectively, suggest that the basic precepts of molecular timers may be evolutionarily conserved across otherwise disparate apical signalling complexes.
Results
Procaspase-9 is processed in the Apaf-1 apoptosome but relatively little caspase-9 is retained in the complex
Caspase-9 must bind and remain bound to the Apaf-1 apoptosome to show significant catalytic activity (Rodriguez and Lazebnik, 1999). However, apoptosome complexes containing caspase-9 have been notoriously difficult to isolate from apoptotic cells, even though fully processed caspase-9 can be readily observed outside the complex (Cain et al, 2000). To recapitulate this phenomenon in vitro, we expressed and purified to homogeneity Apaf-1XL (hereafter referred to as Apaf-1) and wild-type ProC9 (Figure 1A). Apaf-1 and ProC9 were then mixed in the presence or absence of Cc and dATP (Cc/dATP) and examined by gel-filtration chromatography. In the absence of Cc/dATP, Apaf-1 and ProC9 eluted as monomeric ∼145 and ∼50 kDa proteins, respectively (Figure 1B, peak fractions 20 and 22), whereas upon addition of Cc/dATP, Apaf-1 underwent oligomerization into apoptosome complexes that contained relatively small amounts of autocatalytically processed C9-p35/p12 (Figure 1B, fractions 8–13). As in apoptotic cells, however, the vast majority of processed C9-p35/p12 was present in fractions outside the apoptosome (Figure 1B, fractions 20–27).
We speculated that at least two models might explain the complete processing of ProC9 to its C9-p35/p12 form (Figure 1C). In the CARD-displacement model, ProC9 would be recruited to the apoptosome, through CARD–CARD interactions with Apaf-1, and undergo autocatalytic processing to its C9-p35/p12 form. However, additional ProC9 would then displace C9-p35/p12, allowing for the cycle to repeat itself until all of the available ProC9 was entirely processed (Figure 1C, left panel). By contrast, in the CARD-static model, a small amount of ProC9 would be recruited to the apoptosome, but rather than being displaced, would remain tightly bound to Apaf-1 through CARD–CARD interactions. A relatively small number of apoptosome complexes would then process the vast majority of ProC9 as a typical substrate—similar to the way the apoptosome processes ProC3—irrespective of its CARD (Figure 1C, right panel).
Procaspase-9 is processed through a CARD-displacement mechanism and does not compete with procaspase-3 as a substrate for the Apaf-1 apoptosome
To distinguish between the two proposed models described above, we first expressed and purified wild-type, autocatalytically processed C9-p35/p12 and the non-cleavable procaspase-9 triple mutant (E306A–D315A–D330A; ProC9-TM) from bacteria (Figure 1A). C9-p35/p12 or ProC9-TM was then incubated in various combinations with purified Apaf-1, Cc/dATP, and either catalytically inactive procaspase-3 (C163A; ProC3*) or wild-type procaspase-3 (ProC3). When all components were present, apoptosome-bound C9-p35/p12 and ProC9-TM readily processed ProC3* to its p20 form (C3*-p20) (Figure 2A, lanes 6, 7, 13, and 14) and likewise activated ProC3, resulting in significant caspase-3-dependent cleavage of the fluorescent substrate DEVD-AMC (Figure 2B, lanes 4 and 6). Thus, as previously shown, both apoptosome-bound pro and processed forms of caspase-9 could activate procaspase-3 in vitro (Stennicke et al, 1999; Bratton et al, 2001; Srinivasula et al, 2001).
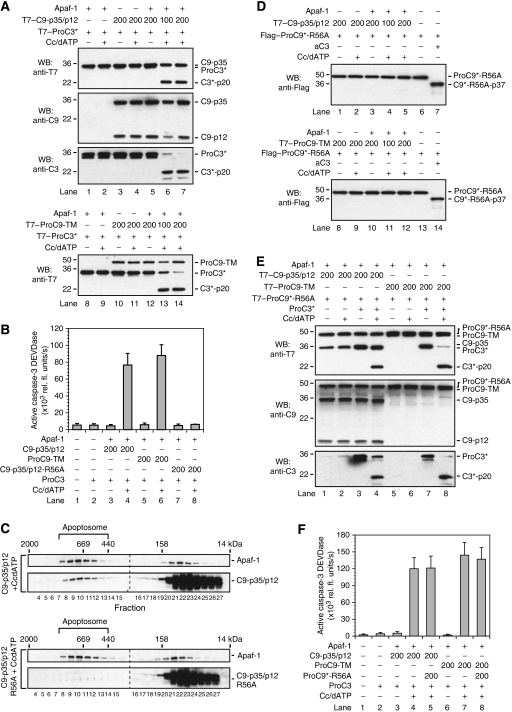
Figure 2.
Procaspase-9 is autoprocessed through a CARD-displacement mechanism. (A) T7-tagged C9-p35/p12 and ProC9-TM were incubated with T7-tagged ProC3* in the presence of Apaf-1 and Cc/dATP. Reactions were carried out, as described in the experimental procedures, resolved by SDS–PAGE, and immunoblotted using an anti-T7 antibody. As C9-p35/p12 and ProC3* co-migrated (top panel), the membranes in these experiments were also stripped and re-blotted for caspase-9 (anti-C9) and caspase-3 (anti-C3) using caspase-specific antibodies. (B) The initial velocities (V0) of apoptosome-bound C9-p35/p12, ProC9-TM, and C9-p35/p12-R56A on the substrate ProC3 were assessed by measuring caspase-3 DEVDase activity, as described in the Methods section. Each bar represents the mean of three separate experiments±s.e.m. (C) Recombinant wild-type C9-p35/p12 or C9-p35/p12-R56A were incubated with Apaf-1 and Cc/dATP. The samples were subsequently fractionated using Superose-6 gel-filtration chromatography, resolved by SDS–PAGE, and immunoblotted using Apaf-1 and caspase-9-specific antibodies (Cain et al, 2000). (D) T7-tagged C9-p35/p12 and ProC9-TM were incubated with Flag-tagged ProC9*-R56A in the presence of Apaf-1 and Cc/dATP. Reactions were carried out, as described in the Methods section, and immunoblotted for the processing of Flag–ProC9*-R56A using an anti-Flag antibody. In lanes 7 and 14, active caspase-3 (1 μM) was added as a positive control to confirm that ProC9*-R56A could be cleaved within its activation loop. (E, F) Apoptosome-bound T7-tagged C9-p35/p12 or ProC9-TM were incubated with T7-tagged ProC9*-R56A and either T7-tagged ProC3* or ProC3. Samples were then analysed by SDS–PAGE and immunoblotted using an anti-T7 antibody or assayed for active caspase-3 DEVDase activity. The membranes in panel (E) were also stripped and re-blotted for caspase-9 (using anti-C9) and caspase-3 (using anti-C3) using caspase-specific antibodies. Each bar represents the mean of three separate experiments±s.e.m.
Next, to determine if these apoptosome complexes could process procaspase-9 as a typical substrate, we first created and expressed a CARD-binding mutant of caspase-9 (C9-p35/p12-R56A; Figure 1A), to confirm its inability to bind to Apaf-1 (Qin et al, 1999). As expected, compared with C9-p35/p12 or ProC9-TM, C9-p35/p12-R56A failed to bind to the apoptosome, as determined through gel filtration (Figure 2C), and could not activate wild-type ProC3 following Cc/dATP-dependent activation of Apaf-1 (Figure 2B, compare lane 8 with lanes 4 and 6). Next, we incorporated an additional active-site mutation (C287A) in ProC9-R56A, so that the protease became both catalytically inactive and incapable of binding to Apaf-1 (ProC9*-R56A; Figure 1A). As before, we then carried out reconstitution experiments wherein C9-p35/p12 or ProC9-TM was incubated in various combinations with Apaf-1, Cc/dATP, and ProC9*-R56A (Figure 2D). Remarkably, neither apoptosome-bound C9-p35/p12 nor ProC9-TM could process ProC9*-R56A to its p35 fragment (Figure 2D, lanes 4, 5, 11, and 12). ProC9*-R56A was conformationally normal with a properly exposed activation loop, as addition of active caspase-3 (aC3) to the incubation mixture resulted in rapid processing of ProC9*-R56A at D330 to generate the expected p37 cleavage fragment (Figure 2D, lanes 7 and 14). Equally important, apoptosome complexes reconstituted with C9-p35/p12 or ProC9-TM readily processed ProC3* and ProC3 to their C3*-p20 and active forms, respectively, even in the presence of excess ProC9*-R56A (Figure 2E, lanes 4 and 8; Figure 2F, compare lanes 4 and 7 with lanes 5 and 8, respectively). Thus, our data strongly indicated that procaspase-9 was processed through a CARD-displacement mechanism and consequently that procaspase-9 and -3 did not compete as substrates for the apoptosome.
Procaspase-9 possesses higher affinity for the apoptosome, compared with processed caspase-9, and is more active at low enzyme concentrations
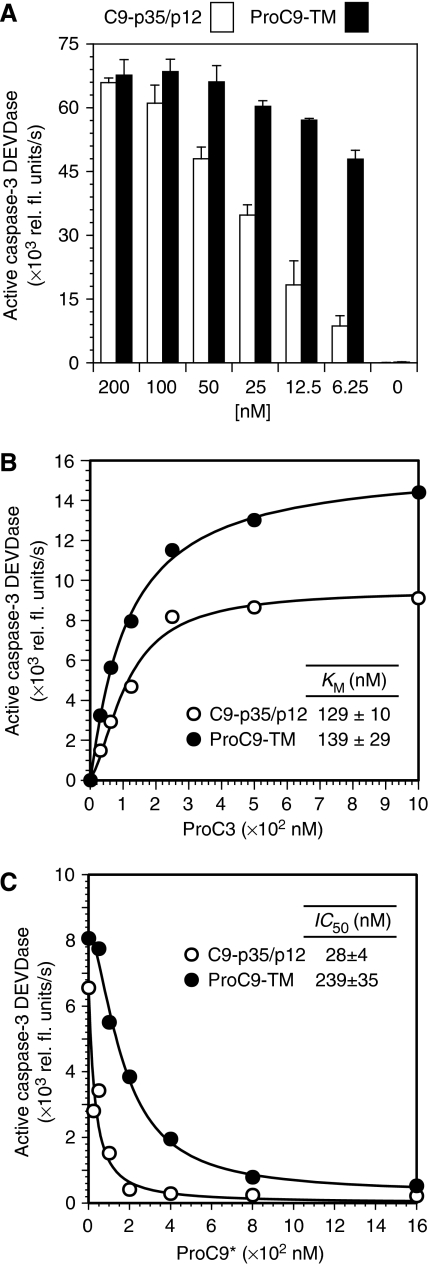
During our studies, we made an interesting observation that apoptosome-bound procaspase-9 was more active than processed caspase-9 at low enzyme concentrations. Indeed, whereas ProC9-TM and C9-p35/p12 showed similar activities at high enzyme concentrations (⩾100 nM), ProC9-TM was consistently more active at physiologically relevant concentrations (⩽25 nM) (Figure 3A). A subsequent comparison of the enzyme kinetics for ProC9-TM and C9-p35/p12 (each at 25 nM) showed that both the pro and processed forms of caspase-9 possessed identical KM values within experimental error (Figure 3B). A precise determination of their kcat values was difficult, largely because it remains unclear how many Apaf-1 and caspase-9 molecules reside within a single apoptosome complex (see discussion). Nevertheless, as the concentration of Apaf-1 remained constant in each incubation, we concluded that the Vmax for ProC9-TM was higher than the Vmax for C9-p35/p12 at low enzyme concentrations (Figure 3A, B).
Figure 3.
Autoprocessing of procaspase-9 reduces its affinity for the apoptosome. (A) Apoptosome complexes were assembled with decreasing concentrations of caspase-9 proteins (C9-p35/p12 and ProC9-TM; 200–0 nM), and the activation of ProC3 was determined by measuring DEVDase activity. Recombinant Apaf-1 and ProC3 were present in each incubation at 300 and 500 nM, respectively. Each bar represents the mean of four separate experiments±s.e.m. (B) KM values for apoptosome-bound C9-p35/p12 (25 nM) and ProC9-TM (25 nM) were determined, as described in the Methods section. The graph shown is from a single experiment; KM values in the inset represent the mean of three independent experiments±s.e.m., using at least two different protein preparations of caspase-9 and Apaf-1. (C) ProC9-TM or C9-p35/p12 were incubated with Apaf-1 and Cc/dATP, titrated with increasing concentrations of ProC9*, and finally incubated with ProC3. Apoptosome-dependent activation of ProC3 was then determined by measuring caspase-3 DEVDase activity as already described. The graph shown is from a single experiment; IC50 values in the inset represent the mean of three independent experiments±s.e.m., using at least two different protein preparations of caspase-9 and Apaf-1.
At this stage, for several reasons, we began to speculate that ProC9 might possess higher affinity for the apoptosome compared with the processed enzyme. Firstly, the Apaf-1 apoptosome is reportedly a holoenzyme (Rodriguez and Lazebnik, 1999). Secondly, ProC9 was processed through a CARD-displacement mechanism, implying that ProC9 must displace C9-p35/p12 from the apoptosome complex (Figure 2). Thirdly, Apaf-1•ProC9-TM and Apaf-1•C9-p35/p12 apoptosome complexes possessed similar KM values, but significantly different observed rates at low enzyme concentrations (Figure 3A, B). Consequently, in order to directly assess the relative affinities of pro and processed caspase-9 for the apoptosome, we established a competition assay in which catalytically inactive (C287A) procaspase-9 (ProC9*; 0–1600 nM) was used to compete with ProC9-TM (200 nM) or C9-p35/p12 (200 nM) for access to the apoptosome. Moreover, as ProC9 and ProC3 did not compete as substrates for the apoptosome (Figure 2E, F), we were able to monitor the activation of ProC3 by apoptosome complexes (DEVDase activity) as a sensitive and reproducible readout for apoptosome activity (Figure 3C). As expected, the IC50 value determined for the displacement of ProC9-TM by ProC9* was 239±35 nM compared with 28±4 nM for C9-p35/p12, indicating that ProC9 possessed an ∼10-fold higher affinity for the apoptosome compared with the processed form of the enzyme (Figure 3C).
Apoptosome activity is diminished following autoprocessing of procaspase-9, but processed caspase-9 remains primarily responsible for the activation of procaspase-3
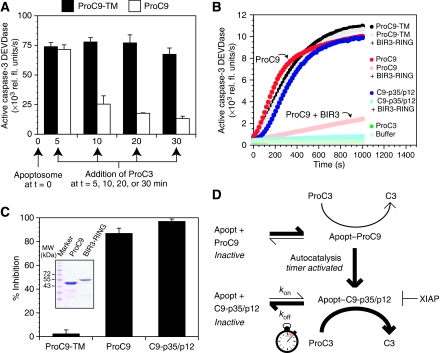
Our experiments with ProC9-TM and C9-p35/p12 strongly implied that the autoprocessing of procaspase-9 reduced its affinity for the apoptosome and consequently its activity. Nevertheless, to further confirm this model, we reconstituted apoptosome complexes with either non-cleavable ProC9-TM or wild-type ProC9 (which could undergo normal autoprocessing) and allowed the incubations to proceed for varying lengths of time (5–30 min), after which ProC3 was added to each incubation and immediately assayed for caspase-3 DEVDase activity (Figure 4A). As anticipated, non-cleavable ProC9-TM remained highly active in the apoptosome over the entire time course, whereas the activity of wild-type ProC9 was initially indistinguishable from that of ProC9-TM but rapidly and steadily declined thereafter (Figure 4A). Thus, consistent with our previous experiments with ProC9-TM and C9-p35/p12, the natural autoprocessing of wild-type ProC9 within the apoptosome resulted in a time-dependent loss of activity.
Figure 4.
Procaspase-9 undergoes rapid autoprocessing so that processed caspase-9 is primarily responsible for the activation of procaspase-3. (A) Apoptosome complexes were reconstituted with either non-cleavable ProC9-TM or wild-type ProC9 (12.5 nM) and incubated for 5–30 min, after which ProC3 (500 nM) was added to each incubation and immediately assayed for caspase-3 DEVDase activity. (B, C) ProC9-TM, ProC9, and C9-p35/p12 were incubated with Apaf-1 and Cc/dATP with and without GST–BIR3-RING of XIAP. A progression curve from one experiment is shown. Each bar represents the mean inhibition of caspase-9 activity of three independent experiments±s.e.m., in which the percent inhibition was calculated as follows: 100−[(V0BIR3-RING/V0) × 100]. Inset, purified wild-type ProC9 and GST–XIAP (BIR3-RING) were subjected to SDS–PAGE and stained with Coomassie blue. (D) Model of the Apaf-1 apoptosome as a molecular timer. Details are described in the text.
As ProC9 could either directly activate ProC3, or undergo autocatalytic processing to generate C9-p35/p12, we next sought to determine the relative contributions of ProC9 and C9-p35/p12 toward the activation of ProC3 within the apoptosome. Importantly, XIAP (and in particular its BIR3 domain) inhibits C9-p35/p12 but not ProC9 (Srinivasula et al, 2001; Bratton et al, 2002; Denault et al, 2007). We therefore carried out reconstitution experiments in which non-cleavable ProC9-TM, fully cleaved C9-p35/p12, or wild-type ProC9 were incubated with Apaf-1 and ProC3, in the presence or absence of recombinant XIAP (BIR3-RING). As expected, following the addition of Cc/dATP to the incubation mixtures, BIR3-RING had no effect on the activation of ProC3 by ProC9-TM but completely inhibited C9-p35/p12 (Figure 4B, C). In the case of wild-type ProC9, BIR3-RING largely inhibited (∼85%) the activation of ProC3 (Figure 4B, C), indicating that the rate of ProC9 autoprocessing exceeded its rate of ProC3 activation and that C9-p35/p12 was primarily responsible for the activation of ProC3.
Procaspase-9 autoprocessing activates a molecular timer that regulates apoptosome function in vitro and in cells
Collectively, to this point, our data argued strongly for a model wherein Cc/dATP induced the oligomerization of Apaf-1 into the apoptosome. ProC9 was then recruited to the complex with high affinity and either directly activated ProC3 (minor pathway) or underwent rapid autocatalytic processing to C9-p35/p12 (major pathway). This processing step alone resulted in reduced affinity of caspase-9 for the apoptosome. However, C9-p35/p12 activated ProC3 for a finite period of time until it dissociated from the complex or was displaced by additional ProC9 to reinitiate the cycle. On the basis of these data, we concluded that the Apaf-1 apoptosome was functioning as a molecular timer, in which the intracellular concentration of ProC9 set the overall duration of the timer, ProC9 autoprocessing activated the timer, and the rate at which C9-p35/p12 dissociated from the complex (koff) dictated how fast the timer ‘ticked' over (i.e. determined the time frame over which a single active C9-p35/p12 unit—be it a monomer or a dimer—activated ProC3 before dissociating from the complex) (Figure 4D).
Although the apoptosome functioned as a proteolytic-based molecular timer in vitro, we next sought to confirm that the timer likewise operated in native cell lysates and in intact cells. On the basis of our model, there were two main (and testable) predictions: firstly, if the intracellular concentration of ProC9 sets the overall duration of the timer, then the rate of caspase activation and cell death should increase commensurate with the intracellular concentration of procaspase-9. Secondly, if autoprocessing of ProC9 activates the timer, then a non-cleavable mutant of ProC9 (i.e. ProC9-TM) should disengage the timer, resulting essentially in a constitutively active apoptosome that, once formed, mediates potent killing, even at low intracellular concentrations.
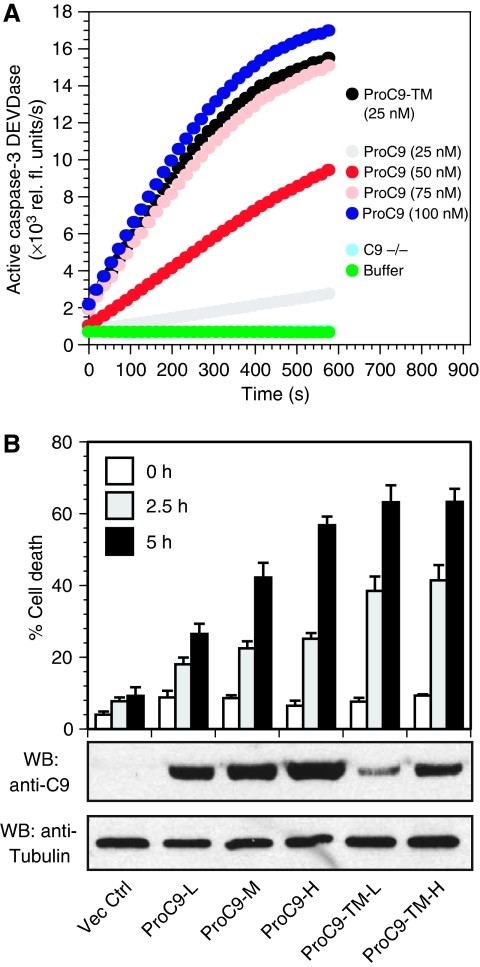
As expected, addition of wild-type ProC9 (0–100 nM) and Cc/dATP to lysates, prepared from caspase-9-deficient mouse embryonic fibroblasts (MEFs), resulted in a concentration-dependent increase in the activation of endogenous ProC3. Moreover, as observed in our pure reconstitution experiments, ProC9-TM (25 nM) showed significantly greater activity compared with wild-type ProC9 (Figure 5A, note the activity of each enzyme at 25 nM). Next, we infected caspase-9-deficient MEFs with lentiviruses engineered to express either wild-type ProC9 or ProC9-TM and selected for clones expressing either low (L), medium (M), or high (H) levels of caspase-9 proteins. The cells were then exposed to UV irradiation, and as shown in Figure 5B, cells expressing wild-type ProC9 showed a concentration-dependent increase in apoptosis. By contrast, cells expressing ProC9-TM showed profound sensitivity to UV irradiation, even at very low concentrations (Figure 5B). Thus, in summary, the Apaf-1•caspase-9 apoptosome functions as a proteolytic-based timer, both in pure reconstitution and mixed-lysate experiments, as well as in cells stimulated to undergo apoptosis.
Figure 5.
Prolonging or disengaging the proteolytic-based timer profoundly sensitizes cells to apoptosis. (A) Caspase-9-deficient lysates were reconstituted with increasing concentrations of wild-type ProC9 (0–100 nM) or non-cleavable ProC9-TM (25 nM). After the addition of Cc/dATP, apoptosome-mediated activation of endogenous ProC3 was determined by measuring caspase-3 DEVDase activity, as described in the Methods section. The results are typical of three independent experiments. (B) Caspase-9 deficient MEFs were infected with lentiviruses expressing empty vector, wild-type ProC9, or non-cleavable ProC9-TM. Stable clones, expressing varying concentrations of ProC9 or ProC9-TM (L=low; M=medium; H=high), were obtained following selection with hygromycin, and the cells were subsequently UV irradiated (8 min) on a UV illuminator and assayed for cell death by Annexin V/propidium iodide staining and flow cytometry. Each bar represents the mean of six independent experiments±s.e.m.
Discussion
The Apaf-1•procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer
Given the importance of the Apaf-1 apoptosome for stress-induced apoptosis, there has been great interest in understanding the regulation of this complex. In this study, we made several key observations regarding apoptosome function: firstly, we found unexpectedly that ProC9 was processed by the apoptosome through a CARD-displacement mechanism (Figure 1C, left panel), meaning that the apoptosome could not process ProC9 as a typical substrate, such as ProC3 (Figure 2). Secondly, based on saturation and displacement assays, we found that ProC9-TM (and by extension ProC9) bound to the apoptosome with an ∼10-fold higher affinity than C9-p35/p12, so that once processed, C9-p35/p12 dissociated from the complex or was displaced by available ProC9 (Figure 3). Thirdly, we found that the rate of wild-type ProC9 autoprocessing was rapid and resulted in a time-dependent loss in apoptosome activity. However, using a selective inhibitor of C9-p35/p12, we also found that processed caspase-9 was largely responsible for cleaving and activating ProC3 in the apoptosome (Figure 4). Thus, as already described, we proposed that the Apaf-1 apoptosome functioned as a proteolytic-based molecular timer, the first of its kind wherein the intracellular concentration of ProC9 set the overall duration of the timer, ProC9 autoprocessing activated the timer, and the rate at which C9-p35/p12 dissociated from the complex (koff) dictated how fast the timer ‘ticked' over (Figure 4D). Moreover, we found that cells, engineered to express increasing levels of wild-type ProC9, correspondingly showed increasing sensitivities to proapoptotic stimuli and that cells expressing non-cleavable ProC9-TM (which essentially disengaged the timer), were exquisitely sensitive even at very low concentrations of the enzyme (Figure 5).
Despite its existence, one might question the purpose of such a timer, and relatedly, in the CARD-displacement model, one might wonder what would keep a small number of Apaf-1 complexes, formed in response to a relatively modest stress, from cycling through all of the available ProC9 and inducing an exaggerated apoptotic response? Considering the severe consequences of activating the apoptosome, it is critical that minor or accidental release of small quantities of Cc does not result in rapid and uncontrolled cell death. The Bcl-2 family members and intracellular nucleotides regulate the upstream release of Cc from mitochondria and it's binding to Apaf-1 (Chandra et al, 2006; Chipuk and Green, 2008). We propose that the timer provides the cell with yet another layer of regulation. As already noted, the availability of ProC9 regulates the duration of the timer, and importantly, our studies and those of others indicate that the intracellular concentration of ProC9 in cells determines their sensitivities to stress-induced apoptosis (Figure 5) (Gomyo et al, 2004; Yanamandra et al, 2004). One interesting consideration is how cells, particularly cancer cells, might combat high intracellular levels of ProC9. Several kinases that mediate growth and survival signalling, including ERK and CDK1/cyclin B1, phosphorylate ProC9 and may inhibit its recruitment to the apoptosome (Allan et al, 2003; Allan and Clarke, 2007), effectively reducing the duration of the timer. Moreover, though XIAP does not directly affect the timer per se (i.e. does not inhibit ProC9 autoprocessing or the release of C9-p35/p12 from the apoptosome), it does inhibit the activation of ProC3 and thus in essence bypasses the timer.
Stoichiometry of Apaf-1 and procaspase-9 within the apoptosome
Although not the primary focus of this study, intriguingly, the saturation and displacement assays we carried out shed some light on the issue of apoptosome stoichiometry. We routinely utilized 300 nM of Apaf-1 in our reconstitution assays, and based on the proposed seven-fold symmetry for the apoptosome, this should have resulted in ∼40 nM of active apoptosome complexes following Cc/dATP activation. However, the binding of ProC9-TM to the apoptosome, and the accompanying activation of ProC3, was saturable at ∼50–75 nM of ProC9-TM (Figure 3A). This was surprising because based on previous dogma, a minimum of 300 nM of ProC9-TM should have been required to fully occupy the apoptosome (at a 1:1 stoichiometry of Apaf-1:caspase-9) and achieve maximal activity. Instead, our data suggested that only 50–75 nM of ProC9-TM was required to saturate 40 nM of apoptosome complexes, or stated in another way, that each apoptosome complex probably contained 1–2 caspase-9 proteins. Furthermore, this interpretation was also in agreement with our displacement assays, wherein ∼25 nM of catalytically inactive ProC9* was sufficient to displace half of the C9-p35/p12 from the ∼40 nM of apoptosome complexes and correspondingly inhibit the activation of ProC3 (Figure 3C). Thus, our data suggested that Apaf-1 and caspase-9 may not be present within the apoptosome at a 1:1 stoichiometry.
It should be noted that studies using cryoelectron microscopy indicate that the apoptosome contains seven full-length Apaf-1 proteins arranged as spokes on a wheel, but caspase-9 was not visible in these relatively low-resolution (12.8–27 Å) structures (Acehan et al, 2002; Yu et al, 2005). Moreover, whereas the truncated CARDs of Apaf-1 and caspase-9 form dimers in vitro, based on the geometry of the CARD ring—comprised of seven Apaf-1 CARDs and situated directly above the central hub of the apoptosome in the fully assembled complex—it is unclear whether the apoptosome could even accommodate seven caspase-9 proteins because of steric hindrance (Acehan et al, 2002; Yu et al, 2005). Finally, the proposed stoichiometry for the apoptosome was based largely on experiments, wherein apoptosome complexes (isolated and concentrated from gel-filtration fractions) were found to contain similar levels of Apaf-1 and caspase-9 using SDS–PAGE/Coomassie blue staining (Zou et al, 1999). However, if one assumes that seven Apaf-1 (∼142 kDa) and seven caspase-9 (∼46 kDa) proteins were present within the apoptosome, then the overall size of the complex should be ∼1.3 MDa. By contrast, gel-filtration analyses of both native and recombinant apoptosome complexes suggest that these complexes may be smaller, more in the range of 700 kDa to 1 MDa (Figure 1B) (Benedict et al, 2000; Cain et al, 2001; Srinivasula et al, 2001; Chandra et al, 2006).
Owing to the inherent challenges associated with reconstituting apoptosome complexes, one might question whether our incubations simply contained fewer active apoptosome complexes than expected, each of which might have contained seven caspase-9 proteins? For this explanation to be plausible, however, ∼85% of our Apaf-1 complexes would have to have been defective. In our view, this is unlikely given that Apaf-1 was entirely monomeric before Cc/dATP activation, underwent near complete (>70%) oligomerization following activation, and did not oligomerize into the previously described large inactive Apaf-1 complexes (Cain et al, 2000). Nevertheless, additional studies will be required in future to determine the precise stoichiometry of Apaf-1 and caspase-9 in the apoptosome.
Are molecular timers functionally conserved across apical signalling complexes?
Although we were unable to identify other examples of proteolytic-based molecular timers in the literature, we suggest that the basic tenets of molecular timers are present in other disparate signalling complexes. For example, members of the Ras/Rab GTPase superfamily cycle between their active GTP-bound and inactive GDP-bound states. Therefore, the intrinsic GTPase activity of a Ras/Rab protein activates its molecular timer and the rate at which the protein dissociates from its effectors constitutes the timer. Importantly, a number of Ras/Rab regulators have also been identified, including the guanine nucleotide exchange factor proteins (GEFs) and GTPase-activating proteins (GAPs), which activate Ras/Rab proteins and hasten their inactivation, respectively (Siderovski and Willard, 2005).
Although structurally and mechanistically distinct from GAPs and GEFs, there are factors that have arguably similar roles in the context of the apoptosome. Indeed, GAPs enhance the GTPase activity of Ras/Rab proteins, and despite the fact that oligomerized Apaf-1 is required to activate caspase-9 through binding, ironically, it may also increase the intrinsic autoprocessing activity of ProC9, leading to the dissociation of C9-p35/p12 from the complex. In addition, analogous to GEFs, which maintain Ras/Rab proteins in their activated states, the tumour suppressor PHAPI, together with cellular apoptosis susceptibility protein (CAS) and Hsp70, accelerates nucleotide exchange and facilitates the appropriate oligomerization of Apaf-1 into functional apoptosome complexes (Hill et al, 2004; Schafer et al, 2006; Kim et al, 2008). Finally, in comparing our gel-filtration results, following reconstitution of the recombinant apoptosome, with those previously isolated from Cc/dATP-activated native cell lysates, it seems that in the latter case processed caspase-9 may be retained in the apoptosome at higher concentrations (Figure 1B) (Cain et al, 1999, 2000). Importantly, as we have shown, the timer remains highly functional in lysates and in intact MEFs from caspase-9-deficient cells (Figure 5). It is however intriguing to speculate that factors might exist in other cells, which sustain the association of C9-p35/p12 with the apoptosome and in effect delay the timer, resulting in prolonged activity of the complex. Thus, in future studies, it will be important to determine if other proteins, which reportedly associate with the apoptosome (Schafer and Kornbluth, 2006), inhibit or activate this caspase-activating complex through regulation of its molecular timer.
Materials and methods
Reagents
Cc (Sigma, catalogue number C3131); dATP (Sigma, catalogue number D-4788); DEVD-AMC (MP Biomedicals); T7-Tag monoclonal antibody (Novagen, catalogue number 69522-3); Anti-Flag M2 monoclonal antibody (Sigma, catalogue number F 3165).
Cloning
ProC9 was cloned into BamHI/XhoI sites of pET21b vector (Novagen) and various mutants—R56A, C287A, R56A–C287A, and E306A–D315A–D330A—were generated by site-directed mutagenesis. Similarly, ProC3 was cloned into pET21b and an active-site mutant (C163A) was generated by site-directed mutagenesis. The BIR3-RING fragment of XIAP was cloned into pGEX-4T-1 (Pharmacia) at EcoRI/NotI sites.
Protein expression and purification
Apaf-1XL and ProC3 were expressed in Hi-five cells (Invitrogen) using a baculoviral expression system (Chandra et al, 2006). All recombinant caspase-9 proteins, ProC3 (C163A), and fully mature active C3 were expressed in E. coli strain BL21(DE3)pLysS (Novagen) and purified using an FPLC coupled to a Ni2+–NTA column (Qiagen). The proteins were then dialyzed and further purified to homogeneity by anion-exchange chromatography (Mono-Q, Amersham Biosciences). Finally, the concentration of each caspase was determined using the Bradford assay, and for each active enzyme, by performing active-site titrations with zVAD-fmk (Stennicke and Salvesen, 2000).
Gel-filtration analysis
Apoptosome complexes were reconstituted using Apaf-1 (1 μM), ProC9 (1 μM), with or without Cc (10 μM), dATP (2 mM), and MgCl2 (2 mM) [i.e. Cc/dATP] in a final volume of 200 μl and incubated at 25°C for 45 min. The samples were subjected to Superose-6 gel-filtration chromatography, and fractions (500 μl) were then immunoblotted using rabbit polyclonal antibodies against Apaf-1 and caspase-9 (Cain et al, 2000).
Apoptosome-reconstitution and caspase-activity assays
Recombinant apoptosome complexes were reconstituted using Apaf-1 (300 nM or 43 nM of heptameric apoptosome complexes), T7-tagged caspase-9 (C9-p35/p12 or ProC9-TM; 100–200 nM), and Cc/dATP in the presence of T7-tagged ProC3* (500 nM) and/or Flag-tagged ProC9*-R56A (500 nM) in a final volume of 25 μl. Samples were incubated at 37°C for 30 min, separated on a 15% SDS–PAGE, and immunoblotted using T7 and Flag antibodies. For fluorescence-based caspase-9-activity assays, the apoptosome was assembled using Apaf-1 (300 nM or 43 nM of heptameric apoptosome complexes), caspase-9 (C9-p35/p12, C9-p35/p12-R56A, ProC9, or ProC9-TM±ProC9*-R56A; 200 nM each or as described), and Cc/dATP, along with DEVD-AMC (15 μM) in a final volume of 25 μl. The reaction was initiated with the addition of a saturating amount of ProC3 (500 nM), and the activation of ProC3 was monitored by measuring DEVDase activity as previously described (Cain et al, 2000). It should be noted that similar coupled assays have previously been utilized to measure apoptosome activity, using lysates immunodepleted of caspase-9 as the source of Apaf-1 and caspase-3 (Stennicke et al, 1999; Bratton et al, 2001; Pop et al, 2006).
In some cases where indicated, apoptosome-reconstitution assays were carried out using recombinant caspase-9 proteins and naïve lysates prepared from caspase-9−/− MEFs (Figure 5A). Briefly, caspase-9-deficient MEFs were harvested by trypsinization, washed with PBS, resuspended in PIPES buffer containing protease inhibitors (0.5 M PIPES (KOH), pH 6.5; 0.5 M EDTA, pH 8.0; 0.1% CHAPS; 10 mg/ml leupeptin; 10 mg/ml pepstatin; 10 mg/ml aprotinin; 200 mM phenylmethylsulfonyl fluoride), and lysed using a 1-ml syringe and 26 G needle (30 plunges). The lysed cells were then centrifuged at 1000 g for 15 min at 4°C, and the resulting supernatant was further centrifuged at 20 000 g for 20 min. The final supernatant (lysate) was then diluted to 20 mg/ml with caspase-assay buffer, and caspase-activation assays were carried out at 37°C for 30 min using 20 μl of the cell lysate, Cc/dATP, and increasing concentrations of wild-type ProC9 or ProC9-TM. The activation of endogenous ProC3 by the apoptosome was assayed by measuring the release of AMC from the caspase-3 substrate DEVD-AMC.
Displacement assay
Apoptosome complexes were reconstituted as before using Cc/dATP (10 μM), Apaf-1 (300 nM or 43 nM of heptameric apoptosome complexes), and caspase-9 (C9-p35/p12 or ProC9-TM; 200 nM). Catalytically inactive (C287A) ProC9 (ProC9*) was then added at increasing concentrations (25–1600 nM) and incubated for an additional 5 min. Finally, a saturating amount of ProC3 (500 nM) was added to the samples and DEVDase activity was determined as before.
Caspase-9-inhibition assay with XIAP
Apaf-1 (300 nM or 43 nM of heptameric apoptosome complexes), caspase-9 (ProC9-TM, C9-p35/p12 or wild-type ProC9; 200 nM) and GST–BIR3-RING (200 nM) were incubated along with ProC3 (500 nM) and the fluorescent caspase-3 substrate DEVD-AMC. Reactions were then initiated by adding Cc/dATP (10 μM), and the release of AMC was monitored using a Victor3 plate reader.
Lentiviral re-expression of caspase-9 in caspase-9-deficient MEFs
The cDNA encoding wild-type ProC9 and non-cleavable ProC9-TM were cloned (BamHI/NotI sites) into the FG9 lentiviral vector (generously provided by Dr Casey Wright, The University of Texas at Austin). The viral plasmids were then co-transfected into HEK293 cells, along with three helper plasmids—pMDLg/pRRE (gag and pol), pRSV-Rev (rev), and pMD2.G (vsv-g)—to generate caspase-9-expressing lentiviruses. Caspase-9-deficient MEFs (kindly provided by Dr Keisuke Kuida, Vertex Pharmaceuticals; Kuida et al, 1998), were then transformed using the NIH 3T3 protocol and infected with the vector control, ProC9, or ProC9-TM-expressing lentiviruses. Finally, stable clones expressing varied levels of either wild-type ProC9 or non-cleavable ProC9-TM, were obtained through selection with hygromycin and were subsequently UV irradiated on a transilluminator for 8 min. At the indicated time points, the cells were collected, stained with annexin V-FITC/propidium iodide, and assayed for cell death by flow cytometry (Milleron and Bratton, 2006).
Supplementary Material
Review Process File
Acknowledgments
We thank Drs Walter Fast and Kevin Dalby for helpful discussions and review of the paper. The authors are also grateful to Drs Casey Wright and Keisuke Kuida for kindly providing the FG9 lentiviral plasmid and caspase-9-deficient MEFs, respectively. This work was supported in part by grants from The American Cancer Society RSG-05-029-01-CCG, PhRMA Foundation, and the NCI/NIH CA129521 (all to SBB).
Footnotes
The authors declare that they have no conflict of interest.
References
- Acehan D, Jiang X, Morgon DG, Heuser JE, Wang X, Akey CW (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR (2007) Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310 [DOI] [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR (2003) Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5: 647–654 [DOI] [PubMed] [Google Scholar]
- Benedict MA, Hu Y, Inohara N, Nunez G (2000) Expression and functional analysis of Apaf-1 isoforms. Extra WD-40 repeat is required for cytochrome c binding and regulated activation of procaspase-9. J Biol Chem 275: 8461–8468 [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS (2003) A unified model for apical caspase activation. Mol Cell 11: 529–541 [DOI] [PubMed] [Google Scholar]
- Bratton SB, Lewis J, Butterworth M, Duckett CS, Cohen GM (2002) XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ 9: 881–892 [DOI] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula S, Sun X-M, Butterworth M, Alnemri ES, Cohen GM (2001) Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20: 998–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain K, Bratton SB, Langlais C, Walker G, Brown DG, Sun XM, Cohen GM (2000) Apaf-1 oligomerizes into biologically active ∼700 kDa and inactive ∼1.4 MDa apoptosome complexes. J Biol Chem 275: 6067–6070 [DOI] [PubMed] [Google Scholar]
- Cain K, Brown DG, Langlais C, Cohen GM (1999) Caspase activation involves the formation of the aposome, a large (∼700 kDa) caspase-activating complex. J Biol Chem 274: 22686–22692 [DOI] [PubMed] [Google Scholar]
- Cain K, Langlais C, Sun XM, Brown DG, Cohen GM (2001) Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J Biol Chem 276: 41985–41990 [DOI] [PubMed] [Google Scholar]
- Chandra D, Bratton SB, Person MD, Tian Y, Martin AG, Ayres M, Fearnhead HO, Gandhi V, Tang DG (2006) Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome C and inhibiting apoptosome. Cell 125: 1333–1346 [DOI] [PubMed] [Google Scholar]
- Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y (2005) Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol 3: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR (2008) How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol 18: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault JB, Eckelman BP, Shin H, Pop C, Salvesen GS (2007) Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9. Biochem J 405: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomyo Y, Sasaki J, Branch C, Roth JA, Mukhopadhyay T (2004) 5-aza-2′-deoxycytidine upregulates caspase-9 expression cooperating with p53-induced apoptosis in human lung cancer cells. Oncogene 23: 6779–6787 [DOI] [PubMed] [Google Scholar]
- Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ (2004) Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J 23: 2134–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang X (2000) Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J Biol Chem 275: 31199–311203 [DOI] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X (2005) Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA 102: 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HE, Jiang X, Du F, Wang X (2008) PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell 30: 239–247 [DOI] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA (1998) Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94: 325–337 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X (1997) Cytochrome c and dATP-dependent formation of Apaf-1·caspase-9 complex initiates an apoptotic protease cascade. Cell 91: 479–489 [DOI] [PubMed] [Google Scholar]
- Milleron RS, Bratton SB (2006) Heat shock induces apoptosis independently of any known initiator caspase-activating complex. J Biol Chem 281: 16991–17000 [DOI] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS (2006) The apoptosome activates caspase-9 by dimerization. Mol Cell 22: 269–275 [DOI] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y (1999) Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399: 549–557 [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y (1999) Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev 13: 3179–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES (1999) Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J Biol Chem 274: 17941–17945 [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Kornbluth S (2006) The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell 10: 549–561 [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Parrish AB, Wright KM, Margolis SS, Marks JR, Deshmukh M, Kornbluth S (2006) Enhanced sensitivity to cytochrome c-induced apoptosis mediated by PHAPI in breast cancer cells. Cancer Res 66: 2210–2218 [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Shi Y (2002) Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA 99: 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS (2005) The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci 1: 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES (2001) A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410: 112–116 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS (1999) Caspase-9 can be activated without proteolytic processing. J Biol Chem 274: 8359–8362 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS (2000) Caspase assays. Methods Enzymol 322: 91–100 [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ (2008) Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9: 231–241 [DOI] [PubMed] [Google Scholar]
- Yanamandra N, Kondraganti S, Srinivasula SM, Gujrati M, Olivero WC, Dinh DH, Rao JS (2004) Activation of caspase-9 with irradiation inhibits invasion and angiogenesis in SNB19 human glioma cells. Oncogene 23: 2339–2346 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW (2005) A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure 13: 1725–1735 [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X (1999) An Apaf-1·cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 274: 11549–11556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File