Abstract
OCRL, whose mutations are responsible for Lowe syndrome and Dent disease, and INPP5B are two similar proteins comprising a central inositol 5-phosphatase domain followed by an ASH and a RhoGAP-like domain. Their divergent NH2-terminal portions remain uncharacterized. We show that the NH2-terminal region of OCRL, but not of INPP5B, binds clathrin heavy chain. OCRL, which in contrast to INPP5B visits late stage endocytic clathrin-coated pits, was earlier shown to contain another binding site for clathrin in its COOH-terminal region. NMR structure determination further reveals that despite their primary sequence dissimilarity, the NH2-terminal portions of both OCRL and INPP5B contain a PH domain. The novel clathrin-binding site in OCRL maps to an unusual clathrin-box motif located in a loop of the PH domain, whose mutations reduce recruitment efficiency of OCRL to coated pits. These findings suggest an evolutionary pressure for a specialized function of OCRL in bridging phosphoinositide metabolism to clathrin-dependent membrane trafficking.
Keywords: AP-2; APPL; endocytosis; PI(4,5)P2 ; Rab5
Introduction
Phosphatidylinositol, a phospholipid concentrated at cytosolic leaflet of cellular membranes, has a central function in cell regulation (Hokin, 1985). Reversible phosphorylation of its cytoplasmically exposed phosphorylated inositol ring generates seven phosphoinositides that provide a membrane-binding platform for a variety of protein modules. As a result, phosphorylation–dephosphorylation of its ring controls a wide variety of cell processes. Furthermore, as each of the seven phosphoinositides has its own distinctive subcellular localization, these phospholipids have an important function in determining specificities of membrane interaction and in defining and maintaining organelle identity. For example, PI(4,5)P2 and PI(3,4,5)P3 are primarily enriched in the plasma membrane and control many important reactions that occur at this membrane, including generation of intracellular second messengers, transport function between the extracellular and intracellular space, exocytosis, endocytosis, and actin nucleation (Odorizzi et al, 2000; De Matteis and Godi, 2004; Di Paolo and De Camilli, 2006). The selective concentration of specific phosphoinositide species on certain membranes is controlled by the spatial segregation of kinases and phosphatases that act on the inositol ring. Despite their physiological importance, the properties of many of these enzymes remain poorly characterized. Mutations of some of them are responsible for human diseases (Di Paolo and De Camilli, 2006; Vicinanza et al, 2008; McCrea and De Camilli, 2009). One such enzyme is OCRL.
OCRL (also called OCRL1), one of the 10 inositol 5-phosphatases encoded by mice and human genomes, was originally identified as the product of the gene responsible for the oculocerebrorenal syndrome of Lowe (Attree et al, 1992). Lowe syndrome is an X-linked disorder involving congenital cataracts, mental retardation, and renal Fanconi syndrome, a renal proximal tubulopathy (Lowe et al, 1952). As shown recently, mutations in the OCRL gene can also cause Dent disease (Hoopes et al, 2005), another X-linked condition characterized by a kidney dysfunction very similar to that of Lowe syndrome. Surprisingly, Dent syndrome patients have no obvious eye or neurological defects (Thakker, 2000).
OCRL is a multi-domain protein comprising a central inositol 5-phosphatase domain that favours PI(4,5)P2 and PI(3,4,5)P3 as substrates (Zhang et al, 1995; Schmid et al, 2004). This domain is flanked at its C-terminal side by an ASH (Ponting, 2006) and a catalytically inactive RhoGAP-like domain (Erdmann et al, 2007) and at its NH2-terminal side by an ∼220 amino-acid region that remains uncharacterized. The structure of the inositol 5-phosphatase domain can be predicted from the reported structure of a homologous 5-phosphatase module in Schizosaccharomyces pombe (Tsujishita et al, 2001). The crystal structure of the ASH/RhoGAP-like domains was recently solved, revealing a classical RhoGAP fold and the first structure of an ASH domain (Erdmann et al, 2007). OCRL is closely related to another inositol 5-phosphatase, INPP5B (also called OCRL2), which has similar substrate preferences (Matzaris et al, 1994; Jefferson and Majerus, 1995). The two proteins have very substantial primary sequence similarity in the central and C-terminal regions, but no sequence homology in the NH2-terminal region, which also in the case of INPP5B remains uncharacterized.
OCRL and INPP5B share several interacting partners that may help recruit them to their sites of action and/or regulate their activities. The ASH-RhoGAP-like domains of both proteins bind Rac and Cdc42, the endosomal protein Rab5 and the endocytic adaptor and Rab5 effector APPL1 (Shin et al, 2005; Hyvola et al, 2006; Erdmann et al, 2007). They also bind other small GTPases (Fukuda et al, 2008). An interesting difference between them is the presence in OCRL, but not in INPP5B, of two binding sites for clathrin coat components. A loop of the RhoGAP-like domain of OCRL, which is absent in the RhoGAP-like domain of INPP5B, contains a classical clathrin-box motif (LIDID) that bind the clathrin heavy chain (Ungewickell et al, 2004; Choudhury et al, 2005). An FxDxF motif within the NH2-terminal region of OCRL, also missing in INPP5B, binds the endocytic clathrin adaptor AP-2 (Ungewickell et al, 2004).
In agreement with the endocytic functions of at least some of its partners, growing consensus indicates that OCRL, like INPP5B (Shin et al, 2005) has a critical function in the endocytic pathway (Ungewickell et al, 2004; Choudhury et al, 2005; Hyvola et al, 2006; Erdmann et al, 2007). OCRL, originally described as a Golgi complex protein (Olivos-Glander et al, 1995; Faucherre et al, 2003), has now also been detected at the plasma membrane (Erdmann et al, 2007), on early endosomes (Ungewickell et al, 2004; Choudhury et al, 2005; Hyvola et al, 2006; Erdmann et al, 2007) and at late stage endocytic clathrin-coated pits (Erdmann et al, 2007), all sites where its enzymatic activity may help to couple endocytosis to PI(4,5)P2 and PI(3,4,5)P3 dephosphorylation, as first shown for another inositol 5-phosphatase, synaptojanin (Cremona et al, 1999; Wenk and De Camilli, 2004; Perera et al, 2006). Other intracellular locations of OCRL may help prevent ectopic accumulations of these phosphoinositides. This may explain the significant accumulation of total cellular PI(4,5)P2 levels observed in OCRL-deficient fibroblasts (Zhang et al, 1998; Wenk et al, 2003). Defects in the endocytic pathway may account for the kidney manifestations of Lowe syndrome and Dent disease, two conditions that involve impaired proximal tubule reabsorption defects (Lowe, 2005). Likewise, defects in the endocytic pathway in neurons may account for mental retardation in Lowe syndrome, given the essential role of membrane traffic in neuronal function and neuronal signalling. INPP5B shares some of its intracellular locations with OCRL but, consistent with the lack of clathrin and INPP5B binding sites, is not found at endocytic clathrin-coated pits (Erdmann et al, 2007).
An overlapping function of OCRL and INPP5B is supported by the finding that while KO mice for either protein have no major obvious defects (with the exception of fertility problems in INPP5B KO mice (Hellsten et al, 2001)), double KO mice die embryonically (Janne et al, 1998). Clearly, in humans INPP5B cannot fully compensate for lack of OCRL either because of its different pattern of expression or because of differences from OCRL in properties that are critical for the function of selected organs. Thus, further elucidating potential differences between the properties of OCRL and INPP5B is an important priority towards a greater understanding of pathogenetic mechanisms in Lowe syndrome and Dent disease. Their divergent NH2-terminal regions are likely to account for specific functions. Hence, we performed a comparative analysis of the structural and functional properties of these two protein regions. We report that, despite lack of primary sequence similarity, the NH2-terminal region of both proteins contains a PH domain. We also provide new evidence for a tight functional link between OCRL and clathrin, strongly supporting the hypothesis that abnormalities in clathrin-dependent protein trafficking may be implicated in at least some of the phenotypic manifestations of Lowe syndrome and Dent disease.
Results
The NH2-terminal region of OCRL, but not of INPP5B, binds clathrin
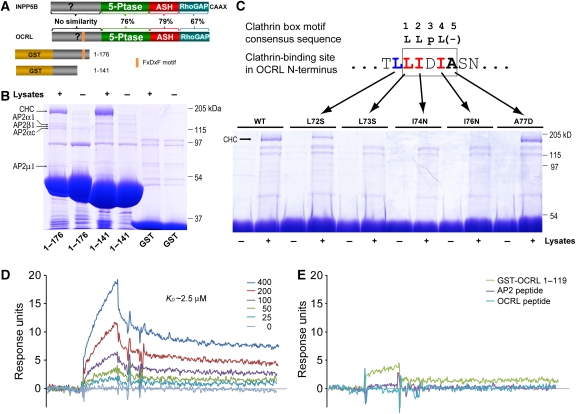
As a step towards the functional characterization of the NH2-terminal region of OCRL, we searched for novel interacting partners of this region. NH2-terminal fragments of OCRL were generated as GST-fusion proteins and used as bait in pull-downs from rat brain lysates (Figure 1A). The OCRL fragment comprising residues 1–176, which contains the known AP-2 binding site (151FEDNF155) (Lowe, 2005) pulled-down the AP-2 complex as expected. In addition, this construct, as well as the 1–141 construct (OCRL1−141), which does not contain the AP-2 binding site, specifically and massively purified a 180-kDa molecule (Figure 1B). As revealed by mass spectrometry, and then verified by western blot, this band is clathrin heavy chain. In similar experiments using GST fusions of the NH2-terminal portion of INPP5B as bait (amino acids 1–148, 1–156, 1–181), neither clathrin nor AP-2 were found in the affinity purified material (data not shown).
Figure 1.
The NH2-terminal region of OCRL binds to clathrin. (A) Schematic diagram showing domain structure of INPP5B and OCRL and the GST-fusion constructs used for pull-downs. Sequence similarity between INPP5B and OCRL is listed for each pair of homologous domains. (B) GST and GST-OCRL NH2-terminal regions were incubated with rat brain lysate, and bound proteins were analysed by SDS–PAGE, followed by Coomassie blue staining. Protein bands were identified by mass spectrometry and confirmed by western blotting. CHC, clathrin heavy chain; AP-2 followed by a Greek symbol, subunits of the clathrin adaptor AP-2. (C) GST pull-downs as in (B) using GST-OCRL1−141 wild-type (WT) or with single point mutations in the sequence 72LLIDIA77. Binding to clathrin (CHC) is abolished by mutations of hydrophobic residues of the motif 73LIDI76, and partially impaired by mutation of the leucine upstream of the motif. (D) SPR analysis of the binding of various concentrations (0–400 nM) of the NH2-terminal β-propeller domain of clathrin heavy chain to immobilized GST-OCRL1−119. The calculated KD value is indicated. (E) Binding of clathrin's propeller domain (400 nM) to GST-OCRL1−119 after preincubation with the module itself (GST-OCRL1−119, 4 μM), with the clathrin-box peptide contained in this module (ETLLIDIASNS) (8 μM) or with a peptide from the clathrin adaptor AP-2 comprising a classical clathrin-box motif (GDLLNLDLGP) (8 μM).
Incubation of GST-OCRL1−141 with recombinant His-tagged clathrin heavy chain fragments mapped the OCRL-binding site to the NH2-terminal β-propeller domain of clathrin (Supplementary Figure S1). As most clathrin-binding partners engage this domain through a clathrin-box motif (Dell'Angelica, 2001), we inspected the primary sequence of the NH2-terminal region of OCRL for any such motif. The sequence 73LIDIA77 matches the first four positions of the classical clathrin-box motif sequence consensus ‘LLDLD' (Figure 1C). Replacement of three hydrophobic residues of the consensus (L73S, I74N, and I76N) with hydrophilic ones abolished clathrin binding, confirming this site as the relevant site (Figure 1C). Notably, mutation of the fifth amino acid of the sequence 73LIDIA77, which does not fit the consensus, did not have an obvious effect on clathrin binding. Mutation of the alanine to a proline did not seem to affect binding and mutation to an aspartate, as in the consensus, did not enhance binding (Figure 1C). In contrast, the L72S mutation partially impaired clathrin binding (Figure 1C), suggesting a partial contribution of this leucine 72 to the interaction.
Results obtained by GST-pull-down assays (Figure 1B and C) were further validated by the surface plasma resonance (SPR) method. This technique revealed an affinity between the NH2-terminal region of OCRL and clathrin's NH2-terminal β-propeller domain of about KD∼2.5 μM (Figure 1D) and confirmed specificity of binding as the I74N mutant had no detectable affinity (data not shown). Furthermore, binding was blocked by preincubation of clathrin's NH2-terminal β-propeller domain with a peptide comprising the clathrin-binding site in the NH2-terminal region of OCRL and with a clathrin box containing peptide derived from the clathrin adaptor protein AP-2 (Figure 1E). The latter result suggests that the NH2-terminal clathrin box of OCRL binds to clathrin in a similar fashion to classical clathrin boxes, even though it lacks the negatively charged residue at the fifth position.
OCRL contains a previously characterized clathrin heavy chain binding site, a conventional clathrin box (LIDLE), in its C-terminal region, within a loop of the RhoGAP-like domain (Erdmann et al, 2007). Our identification of a second clathrin heavy chain binding site in the NH2-terminal region of the protein raises the possibility that OCRL may simultaneously engage two distinct clathrin triskelia. This would be reflected in the property to facilitate clathrin cage assembly in vitro, as shown for clathrin adaptor complexes (Zaremba and Keen, 1983; Owen et al, 2000). To test this possibility, full-length OCRL, or a truncated OCRL lacking the NH2-terminal region, were produced in sf9 cells. Either protein was then mixed with clathrin, which had been purified from bovine brain, and the reaction mixtures were applied to EM grids and visualized by negative staining. Numerous clathrin baskets with a diameter in the 70 nm range were observed in the sample of full-length OCRL mixed with clathrin. No such structure was observed on grids prepared under the same conditions but using a deletion mutant of OCRL lacking the NH2-terminal region (Supplementary Figure S2). Such results further validate the novel clathrin-binding site identified in the NH2-terminal region of OCRL and support a critical link between OCRL and clathrin function.
The NH2-terminal clathrin box of OCRL is contained within a PH domain
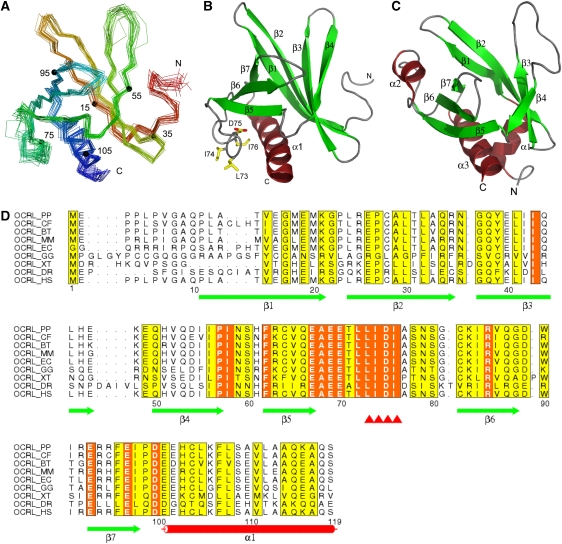
To gain further insight into the properties of the NH2-terminal region of OCRL, we sought to acquire structural information. Extensive crystallization trials using various fragment of this region of human OCRL did not yield crystals, leading us to consider 3D-NMR spectroscopy. Initial NMR spectra analysis revealed a minimal folded structure within the first NH2-terminal 119 residues (OCRL1−119), which comprises the clathrin-box motif. Constructs containing longer C-terminal extensions did not change the overall pattern of the spectra, but yielded additional peaks characteristic of nonstructured peptides (data not shown). Thus, fragment OCRL1−119 was used for further analysis. As a first step towards structure determination, the 1H, 13C, and 15N NMR chemical shifts of this fragment were assigned to near completion. Secondary sets of chemical shifts were observed for residues in several regions (an example was shown in Supplementary Figure S3), which explained the resistance of the samples to form crystals and made the analysis more challenging. Careful choice of spectral signals from the predominant population allowed us to solve the solution structure of OCRL1−119. Tertiary structures were calculated using the CYANA software package (Guntert et al, 1997) and refined by using xplor-nih (Schwieters et al, 2003) based on a total of 1627 NOE-derived distance restraints, 53 hydrogen bond restraints, and 69 backbone torsion angle restraints. The final ensemble of tertiary structures is presented in Figure 2A and B and structural statistics are listed in Supplementary Table 1.
Figure 2.
NMR structure of the NH2-terminal PH domain of OCRL. (A) Superimposed Cα backbone traces of the 20 lowest-energy calculated structures. Traces are rainbow coloured with the NH2-terminus in red and the COOH-terminus in blue. Numbers and corresponding small filled circles indicate residues multiple of 20. (B) Ribbon diagram of a representative structure from (A) shown in the same orientation. Residues that are involved in clathrin binding are shown in sticks and labelled. (C) Ribbon diagram of the structure of the PH domain of PLCδ (PDB ID 1mai). (D) Sequence alignment of the NH2-terminal PH domains of OCRL orthologs. Secondary elements are drawn under the alignment. The clathrin-biding motif is marked by red triangles. This motif, as well as the preceding leucine that partially contributes to binding (see Figure 1C) is completely conserved among all OCRL orthologs. Entrez database accession numbers are as follow: OCRL_PP, GI: 55729733; OCRL_CF, GI: 74008389; OCRL_BT, GI: 156121029; OCRL_MM, GI: 45768389; OCRL_EC, GI: 149745642; OCRL_GG, GI: 118089331; OCRL_XT, GI: 115529017; OCRL_DR, GI: 118150544; OCRL_HS, GI: 13325072. PP, Pongo pygmaeus; CF, Canis familiaris; BT, Bos Taurus; MM, Mus musculus; EC, Equus caballus; GG, Gallus gallus; XT, Xenopus tropicalis; DR, Danio rerio; HS, Homo sapiens.
The core of OCRL1−119 is composed of seven β-strands and one α-helix (Figure 2A and B). The seven β-strands are arranged in an antiparallel configuration defining two sheets (β1 to β4 and β5 to β7, respectively), which are almost perpendicular to each other. The only α-helix, which is located at the very C-terminus of this domain, seals the gap between the two β-sheets. Despite the lack of detectable primary sequence similarity to any other proteins, a search for homologous structures using the Dali algorithm (Holm and Sander, 1995) showed that the fold of this module is similar to that of PH domains. A classical PH domain, the PH domain of PLCδ, is illustrated in Figure 2C. This domain as well comprises seven β-strands that form a two-layered sheet and one α-helix between the two β-sheets. The RMSD between the PH domain of OCRL and that of PLCδ is about 2.2 Å over 65 overlapping residues.
Interestingly, the clathrin-binding sequence 73LIDI76 (clathrin box) is surface exposed within the loop between β5 and β6, which also contains an evolutionarily conserved acidic amino-acid stretch upstream of the clathrin box (Figure 2B and D). This loop is flexible, as judged by the lack of long-range NOESY distance restraints, hence the increased heterogeneity seen for the region in the final structural ensemble (Figure 2A). Its flexible nature makes it an optimal peptide to engage the β-propeller domain of clathrin heavy chain.
The NH2-terminal region of INPP5B also contains a PH domain
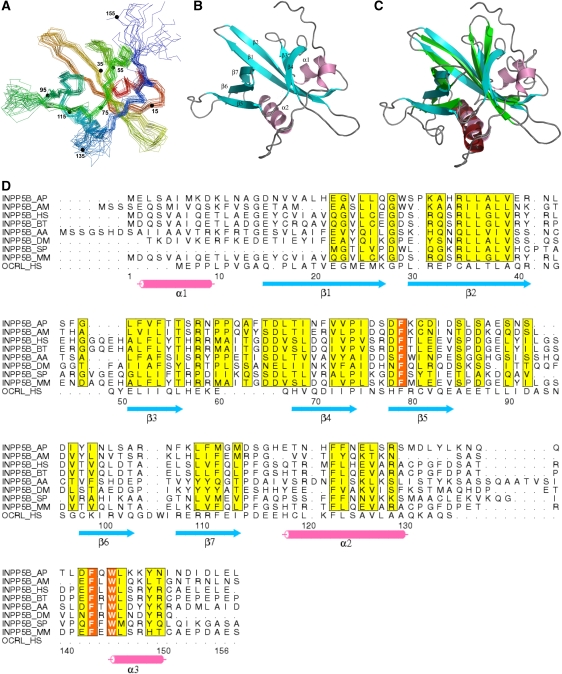
We next investigated the structure of the NH2-terminal region of INPP5B. Pilot experiments by NMR showed that the fragment comprising amino acids 1 to 156 (INPP5B1−156) was folded and that additional C-terminal deletions of this fragment resulted in aggregation within hours. Hence, the structure of this fragment was determined using the same NMR methodology described above for the NH2-terminal region of OCRL (Supplementary Table 1). Surprisingly, despite its primary sequence divergence from the corresponding region of OCRL, INPP5B1−156 also has a typical PH domain fold with seven β-strands and a C-terminal long α-helix (Figure 3A and B). Its structure matches well with the structure of the OCRL PH domain with an RMSD of 1.8 Å over 78 residues (Figure 3C). The PH domain of INPP5B, however, has an additional α-helix at its NH2-terminus and an extension at its C-terminus whose deletion resulted in instability of the entire PH domain. In fact, side chains of the two bulky hydrophobic residues (Trp148 and Leu149) within this extension are buried in a hydrophobic pocket on the outer surface of the two-layered β-sheets. The PH domain of INPP5B also differs from the PH domain of OCRL in the loop between β5 and β6 that does not contain a clathrin-box motif (Figure 3D), consistent with lack of clathrin binding.
Figure 3.
NMR structure of the NH2-terminal PH domain of INPP5B. (A) Cα backbone traces of the 20 lowest-energy structures of the NH2-terminal PH domain of INPP5B in superimposition. Traces are rainbow coloured with the NH2-terminus in red and the COOH-terminus in blue. Numbers and corresponding small filled circles indicate residues multiple of 20. (B) Ribbon diagram of a representative structure from (A). (C) Structure comparison of the PH domains from OCRL (in green and red) and from INPP5B (in blue and pink). (D) Sequence alignment of the NH2-terminal PH domains of INPP5B orthologs in several species. The human OCRL PH domain sequence is alignment on the bottom line based on 3D structure. Secondary elements are drawn under the alignment. Entrez database accession numbers are as follow: INPP5B_AP, GI: 193671757; INPP5B_AM, GI: 110761623; INPP5B_HS, GI: 55666459; INPP5B_ BT, GI: 194665894; INPP5B_AA, GI: 157134484; INPP5B_ DM, 24639220; INPP5B_ SP, GI: 115947205; INPP5B_MM, GI: 30231213; OCRL_HS, GI: 13325072. AP, Acyrthosiphon pisum; AM, Apis mellifera; HS, Homo sapiens, BT, Bos Taurus; AA, Aedes aegypti; DM, Drosophila melanogaster; SP, Strongylocentrotus purpuratus; MM, Mus musculus.
Neither PH domain binds phospholipids
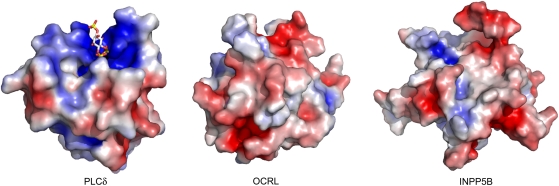
Several, but not all, PH domains bind phosphoinositides (Lemmon, 2004). Both PIP strips based binding assays and liposome pull-down assays (data not shown) were used to assess a potential binding of the PH domains of OCRL and INPP5B to lipids and phosphoinsotides. Neither assay detected specific lipid binding (Supplementary Figure S4 and data not shown). Lack of phosphoinositide binding is in agreement with the surface electrostatic potentials of these two domains. Unlike the PH domain of PLCδ, a well-characterized PI(4,5)P2 binding module that has a large positively charged pocket optimally suited to accommodate the head group of PIP2, the PH domains of both OCRL and INPP5B lack such pocket (Figure 4). We cannot rule out weak binding to lipids beyond the sensitivity of the methods we used in our experiments.
Figure 4.
The PH domains of OCRL and INPP5B lack the basic PI(4,5)P2 binding pocket present in the PH domain of PLCδ. Positive and negative electrostatic surface potentials are indicated in blue and red, respectively. The PH domain of PLCδ with bound IP3 (PDB ID: 1mai) is from Ferguson et al (1995).
The PH domain of OCRL helps recruiting OCRL to endocytic clathrin-coated pits
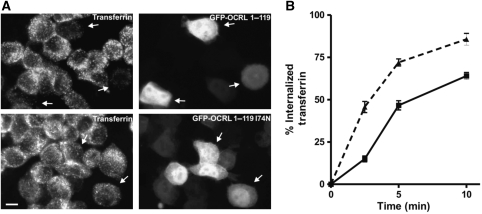
To provide a physiological context for our findings, we examined the importance of the PH domain of OCRL for its interaction with clathrin in intact cells. First, we investigated whether the binding of the NH2-terminal domain of OCRL to clathrin can occur in living cells. Typically, when expressed at high levels, protein fragments containing clathrin boxes compete with endogenous clathrin adaptors at sites of endocytosis and impair clathrin-mediated endocytosis. When GFP-tagged NH2-terminal domain of OCRL (a.a 1–119) was overexpressed in Hela cells, uptake of transferrin-Alexa594, an assay that reports clathrin-mediated endocytosis, was inhibited relative to control cells overexpressing GFP-I74N OCRL (a.a 1–119) (Figure 5A and B). This result indicates that the PH domain of OCRL does bind clathrin within the context of the cell cytosol.
Figure 5.
Overexpression of EGFP-OCRL1−119 impairs endocytosis of transferrin. (A) Hela cells were transfected with expression vectors for the indicated GFP-fusion proteins. At 48 h after transfection, cells were incubated with transferrin-Alexa594 for 10 min. After washing and acid stripping, cells were fixed and inspected by fluorescence microscopy. Arrows indicate transfected cells demonstrating reduced transferrin uptake after EGFP-OCRL1−119 transfection and unchanged transferrin uptake after EGFP-OCRL1−119 I74N transfection. Scale bar is 10 μm. (B) Quantification of transferrin uptake. Hela cells were transfected with expression vectors for EGFP-OCRL1−119 (filled squares) or EGFP-OCRL1−119 I74N (filled triangles). After 48 h, uptake of biotinylated transferrin was measured at the indicated time points using an ELISA assay. Values are the mean±s.e. (n=3).
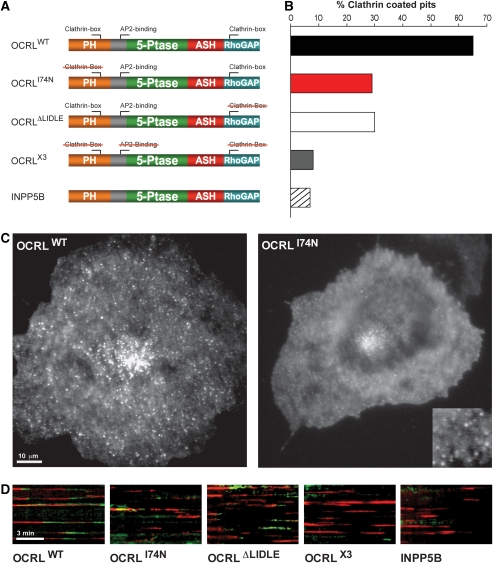
Next, the contribution of the clathrin box within the PH domain in OCRL in the recruitment of OCRL to endocytic clathrin-coated pits was examined by total internal reflection fluorescence (TIRF) microscopy of live cells. Cos-7 cells were cotransfected with RFP clathrin light chain (LCa-mRFP) and with GFP fusions of WT OCRL, of OCRL harbouring the point mutation I74N (OCRL174N) in the NH2-terminal clathrin box and of OCRL with a deletion of the COOH-terminal clathrin box, that is the sequence 702LIDLE706 within the RhoGAP-like domain (OCRLΔLIDLE). As a control, a GFP fusion of OCRL (referred to as OCRLX3) with mutations in each of the three motifs responsible for binding to clathrin coat components, that is the mutations I74N, ΔLIDLE, and F151S (a mutation in the AP-2 binding FxDxF motif) was also used (Figure 6A). Only cells with low levels of expression were chosen to avoid dominant negative effects. In the TIRF field, wild-type OCRL had a punctate localization (Figure 6C) that overlapped to a large extent with clathrin spots (not shown and Erdmann et al, 2007; see also Figure 6D). In the case of GFP-OCRLI74N, which does not bind clathrin through its NH2-terminal region, a diffuse cytosolic localization was very prominent (Figure 6C). Spots were still visible within this diffuse fluorescence (Figure 6C, inset), but many of these spots were highly mobile and probably reflected OCRL bound to endosomes through Rab5 (Hyvola et al, 2006; Erdmann et al, 2007; McCrea et al, 2008). Quantification of the number of endocytic clathrin-coated pits (LCa-mRFP fluorescence) that were clearly visited by GFP-OCRL fluorescence before their disappearance showed that the majority of the pits were visited by wild-type OCRL (Figure 6B, see also kymographs of Figure 6D), but the percentage of these pits was strongly decreased by the single I74N (Figure 6B and D) mutation. A similar decrease was produced by the ΔLIDLE mutation. This result indicates that both clathrin boxes of OCRL are important in clathrin-coated pit targeting. The percentage of pits positive for OCRL3X was only near background similar to what could be observed with GFP-INPP5B, which lack binding sites for the clathrin coat (Figure 6B and D).
Figure 6.
Recruitment of OCRL to clathrin-coated endocytic pits as assessed by TIRF microscopy. (A) Schematic representation of the domain architecture of OCRL/INPP5B and of the mutant constructs used in TIRF microscopy experiments. Red strikethrough indicates disruption of the corresponding clathrin coat-binding site by mutagenesis (see Materials and methods). (B) Percentage of clathrin-coated pits (LCa-mRFP) that clearly become positive for GFP construct indicated during their final stages of maturation. (C) Representative TIRF microscopy images of wild-type and mutant (I74N) OCRL. The I74N mutation dramatically disrupts OCRL localization resulting in a loss of peripheral dots (clathrin-coated pits) and a higher cytosolic pool. Many of the dots still visible (inset) are highly mobile, most likely representing Rab5-bound OCRL on early endosomes. (D) Kymographs of cells coexpressing the indicated GFP-OCRL/INPP5B proteins (green) and LCa-mRFP (red). Individual clathrin-coated pits appear as horizontal red traces. A green end of the trace indicates the recruitment of the GFP-fusion protein before pit disappearance.
Discussion
This study completes the characterization of the modular structure of OCRL and INPP5B and corroborates evidence for a role of OCRL in membrane traffic, and, in particular, clathrin-dependent endocytic membrane traffic. It provides new evidence for the hypothesis that the pathological manifestations of Lowe syndrome and Dent disease result, at least in part, from abnormal function of the endocytic pathway due to abnormal metabolism of phosphoinositides along this pathway.
Our structural data show that OCRL and INPP5B are more similar to each other than previously thought based on primary sequence similarity. Although the NH2-terminal regions of these two otherwise similar proteins have no primary sequence homology, they contain a folded module, a PH domain, which is remarkably similar in the two proteins. Presence of a clathrin box in a loop between two β-strands is a unique feature of the PH domain of OCRL. Typically, clathrin boxes are located within long unfolded linker regions (ter Haar et al, 2000). In the case of OCRL PH domain, the clathrin-box motif is unusual as it is located within a short loop projecting from a folded module. Interestingly, the closest example of another clathrin box located in the loop of a folded domain is the other clathrin box of OCRL, the one located in its C-terminal RhoGAP-like domain (Erdmann et al, 2007), suggesting the importance of close proximity between OCRL modules and clathrin.
Neither of the two OCRL clathrin boxes are present in INPP5B, which also lacks the binding site for the clathrin adaptor AP-2. Sequence comparisons of proteins in the database reveal that invertebrates contain only one protein with the OCRL/INPP5B structure, whose NH2-terminal region is more similar to INPP5B, and that such invertebrate proteins lack clathrin-binding motifs. Conversely, OCRL orthologs, which appear in vertebrates and are defined by an NH2-terminal region substantially different from the NH2-terminal region of INPP5B, typically contain clathrin and AP-2 binding sites. This indicates a specialized function of OCRL in bridging phosphoinositide metabolism to clathrin-dependent membrane trafficking. However, the presence of both in OCRL and INPP5B of an NH2-terminal PH domain points to additional shared function(s) of the PH domains of OCRL/INPP5B independent of clathrin binding. Our results suggest that such a function is not binding to phosphoinositides or to other acidic phospholipids. This PH domain may mediate yet to be discovered protein–protein interactions or be involved in intramolecular interactions that regulate INPP5B/OCRL function.
The structural data shown here conclusively resolve a controversy about the translation starting codon of OCRL (http://research.nhgri.nih.gov/lowe/ocrl1_mut_db.shtml). Some studies used a short version of OCRL that starts with a methionine 17 amino acids downstream of the OCRL methionine that we used here as the starting amino acid. It is now clear that these 17 residues contribute to the first β-strand of the PH domain (Figure 2B and C). In fact, attempts to generate an NH2-terminal fragment of OCRL lacking the first 17 amino acids yielded no soluble recombinant protein.
The clathrin-binding sequence in the PH domain of OCRL only partially fits the classical definition of the clathrin-box motif, whose consensus is LΦpΦ(−), where Φ denotes a bulky hydrophobic residue, p is a polar residue, and (−) is a negatively charged residue (i.e. aspartate or glutamate) (Dell'Angelica, 2001). However, in human OCRL PH domain, the fifth position is not an acidic residue but an alanine residue. Furthermore, some OCRL orthologs have a proline at this position (Figure 2D), and we have found that the human OCRL PH domain, in which this alanine is mutated to proline, can still bind to clathrin (data not shown). Thus, the fifth position does not have a dominant function in clathrin binding, whereas the leucine (L72) upstream of the motif may partially contribute to binding. Similar noncanonical clathrin boxes have been identified earlier (Dell'Angelica, 2001; Teo et al, 2001; Lafer, 2002). On the basis of this definition, previously overlooked clathrin boxes can be identified. For example, the C-terminal portion of the 170 kDa isoform of synaptojanin 1 binds clathrin but was reported not to contain clathrin-box motif (Haffner et al, 2000). Inspection of the same protein region reveals the sequence 1354LIQL1357 that matches the LΦpΦ.
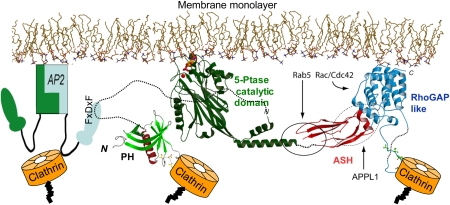
The new structural data allow updating our model of full-length OCRL at the membrane interface (Figure 7). As suggested earlier (Erdmann et al, 2007), the C-terminal ASH and RhoGAP domains, which have multiple interactions with endocytic proteins, help positioning and orienting the catalytic module at the membrane interface. Such arrangement allows catalytic residues to interact with the phosphorylated inositol ring of phosphoinositides (Erdmann et al, 2007). The NH2-terminal PH domain is tethered to the NH2-terminal end of the catalytic module through a predicted unfolded, and thus flexible, linker region that contains the AP-2 binding FxDxF motif. The newly described clathrin box allows a new binding interface with clathrin, in addition to those mediated by the clathrin box in the Rho-GAP domain and, indirectly, by the binding to AP-2.
Figure 7.
Model of the full-length OCRL at the membrane interface. The inositol 5-phosphatase domain together with the ASH and RhoGAP-like domains are docked at the membrane interface as proposed earlier (Erdmann et al, 2007). The NH2-terminal PH domain of OCRL is connected to the 5-phosphatase domain by a long flexible linker region. An FxDxF motif within this region mediates binding to the endocytic clathrin adaptor AP-2. The β-propeller NH2-terminal region of clathrin binds directly to OCRL through two clathrin boxes in the NH2-terminal and C-terminal region of OCRL, respectively, and indirectly through AP-2. Binding site for Rab5 and APPL1 in OCRL are also indicated.
Although the multiplicity of clathrin-binding motifs could suggest a clathrin assembly role for OCRL, a possibility that is consistent with our in vitro studies, the late arrival of OCRL at endocytic clathrin-coated pits demonstrated by TIRF microscopy studies (Erdmann et al, 2007; and this study) indicates that OCRL is recruited to an already grown clathrin lattice. In assembled endocytic clathrin coats, a fraction of clathrin NH2-terminal domains (β-propeller-like domains) are bound to clathrin boxes of the clathrin adaptors (Dell'Angelica, 2001). Yet, the number of clathrin heavy chain molecules in a clathrin coat exceeds the number of adaptors (Fotin et al, 2004; Rapoport et al, 2008), thus leaving free binding sites for additional clathrin boxes, such as those of OCRL. Similarly to what we have shown for synaptojanin 1, another protein containing both an inositol 5-phosphatase domain and binding sites for clathrin coat components (McPherson et al, 1996; Cremona et al, 1999; Haffner et al, 2000; Perera et al, 2006), a function of OCRL may be to couple clathrin-mediated endocytosis to PI(4,5)P2 and PI(3,4,5)P3 dephosphorylation. This action would facilitate clathrin uncoating after vesicle fission and regulate intracellular phosphoinositide-mediated signalling by endocytic membranes. Additional clathrin-related function along the endocytic pathway may also occur, as clathrin coats also participate in membrane sorting on endosomes (Stoorvogel et al, 1996; Brodsky et al, 2001) and in the trans-Golgi network (Robinson and Bonifacino, 2001).
In conclusion, following duplication of the single protein with the modular structure of INPP5B/OCRL during evolution, the NH2-terminal region of OCRL has diverged. This divergence correlated with the appearance within OCRL of multiple binding sites for the clathrin coat, which are all missing in INPP5B. Further elucidation of how this difference between OCRL and INPP5B impacts cell physiology will help shed new light on disease mechanisms in Lowe syndrome and Dent disease.
Materials and methods
Cloning and mutagenesis
Full-length cDNA of OCRL and LCa-mRFP were cloned as described (Erdmann et al, 2007). Full-length and partial cDNAs of OCRL were subcloned in either pEGFPC1 for TIRF microscopy experiments or a self-reconstructed baculovirus transfer vector for protein production. All mutations were introduced into plasmids by using the QuickChange site-directed mutagenesis kit (Stratagene). cDNA constructs encoding NH2-terminal fragments of OCRL were subcloned into the pGEX-6P-1 vector (GE Healthcare). GFP-INPP5B was a kind gift of Martin Lowe (University of Manchester). Clathrin heavy chain full-length cDNA was purchased from ATCC, and clathrin heavy chain fragments (amino acids 1–363, 1–544, 545–1074, 1076–1630) were subcloned into the pET21a vector (Qiagen).
Protein purification
Full-length OCRL (residues 1–901) and OCRL C-terminal fragment (residues 219–901) were expressed in sf9 cells as GST-fusion proteins. Fusion proteins were purified on glutathione Sepharose beads and GST was removed by PreScission (GE Healthcare). Recombinant NH2-terminal fragments of both OCRL and INPP5B and clathrin fragments were expressed in Escherichia coli BL21 as GST or HIS-tagged fusion, respectively. Fusion proteins were purified on glutathione Sepharose beads according to standard protocols. Clathrin was purified from bovine brain according to Keen et al (1979).
For NMR sample preparation, the NH2-terminal fragments of OCRL (residues 1–119) and INPP5B (residues 1–156) was expressed as a GST fusion. Isotopically labelled protein was produced using the appropriate Spectra-9 bacterial growth medium (SpetraStableIsotopes). The GST tag was removed by PreScission protease from the fusion proteins bound to glutathione beads and the eluted protein was further purified using gel filtration chromatography. NMR samples of 1.0 mM proteins were prepared in a 20 mM potassium phosphate (pH 6.4), 1 mM DL-1,4-dithiothreitol-d10 (SpetraStableIsotopes), 0.05% azide, 5% 2H2O, 10 μM Dss solution containing 1 μM of the protease inhibitors PMSF, leupeptin, and pepstatin.
Pull-downs
Adult rat brain extracts were prepared by homogenization in lysis buffer [PBS, 0.5% Triton (v/v), protease inhibitor mixture (Roche)] followed by ultracentrifugation (100 000 g, 60 min, 4°C) to remove insoluble material. Bacterial lysates containing recombinant clathrin heavy chain fragments were prepared by sonication in PBS buffer followed by 40 000 g centrifugation for 20 min. Cleared lysates were then incubated with GST-fusion proteins on beads for 2 h at 4 °C. After extensive wash, bound proteins were analysed by SDS–PAGE.
SPR analysis
GST–OCRL1−119 or GST–OCRL1−119 I74N-coupled CM5 sensor chip was prepared using an amine coupling kit (BIAcore, Piscataway, NJ), as described earlier (Jin et al, 2006). SPR was measured using Biacore 2000 (GE Healthcare). Clathrin's heavy chain NH2-terminal β-propeller domain was injected over the chip in 20 mM Hepes, pH 7.3, 150 mM NaCl, 0.05% Tween 20 (running buffer) at a flow rate of 10 μl/min at room temperature. Clathrin box containing peptides were dissolved in DMSO at a concentration of 10 mM and then diluted at a ratio of 1:100 in the running buffer. The diluted peptides were mixed with clathrin's β-propeller protein at a molar ratio of protein:peptide =1:20 (400 nM protein with 8 μM peptide) and incubated at room temperature for 3 h before injection. The dissociation constant (KD) was determined by fitting the first-order Langmuir-binding model to the data.
Transferrin uptake assay
For the light microscopic analysis of transferrin-Alexa594 uptake, Hela cells were cultured on coverslips and transfected with expression vectors for EGFP–OCRL1−119 or EGFP–OCRL1−119 I74N using Polyfect (Qiagen) according to the manufacturer's protocol. At 48 h after transfection, cells were serum starved for 1 h and incubated with transferrin-Alexa594 at 25 μg/ml for 10 min. Cells were then acid stripped with 10 mM HCl, 150 mM NaCl and, after neutralization with PBS, fixed with paraformaldehyde. For quantification of transferrin uptake, cells were transfected as described above. At 48 h after transfection, uptake of biotinylated transferrin was quantified as described using an ELISA assay (Engqvist-Goldstein et al, 2004).
TIRF microscopy and image analysis
Cos7 cells (ATCC, Rockvile, MD) were cultured at 37 °C in 10% CO2 in DMEM supplemented with 10% FBS and 100 mg/ml penicillin/streptomycin (Invitrogen, Calsbad, CA). Fluorescently tagged proteins were coexpressed in Cos7 cells by transfecting 2–3 mg of DNA with the Amaxa Nucleofector Kit (Amaxa, Cologne, Germany). Transfected cells were plated in glass-bottom 35 mm dishes (Mattek, Ashland, MA) and imaged ∼15–24 h later.
Before imaging, culture medium was replaced with an imaging buffer containing 136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, and 10 mM Hepes at pH 7.4. TIRF microscopy was performed at 37 °C with an Olympus IX81 (Olympus, Melville, NY) inverted microscope fitted with a 60 × 1.49 N.A. TIRFM oil immersion objective and controlled by Andor iQ software (Andor Technologies, Belfast, N. Ireland). The 488 and 568 nm laser lines from argon and argon/krypton lasers (Melles Griot, Carlsbad, CA) were coupled to the TIRF condenser through a single optical fibre. The evanescent field depth was ∼100 nm. Cells were imaged without binning with 0.3–0.5 s exposures and detected with a back illuminated iXon EMCCD camera (512 × 512, 16-bit, Andor Technologies).
ImageJ (National Institutes of Health, Bethesda, MD) and Andor iQ software (Andor Technologies) were used to analyse raw images. Colocalization between LCa and OCRL proteins was determined by randomly selecting 100 clathrin-coated pits spots in the RFP channel, followed by manually scoring for colocalization of OCRL in the GFP channel (from three to five different cells). Kymographs were generated with Andor iQ software.
Lipids binding assays
Phospholipid Dot-blot–PIP strips were purchased from Echelon Biosciences. Dot-blot experiments were carried out according to the manufacturer's protocol. Strips were incubated for 60 min in TBST (0.15 M NaCl, 10 mM Tris–HCl, 0.05% Tween 20, pH 8.0) with 5% nonfat milk at room temperature, then transferred to the bacterially purified tagged protein solution at 0.5 μg/ml in TBST overnight at 4°C. Each strip was then washed three times in TBST buffer before incubation with anti-GST antibody and a secondary HRP conjugate antibody solution. Antibody binding was detected using ECL.
In vitro clathrin cage formation
A measure of 20 μl of purified clathrin (5 mg/ml in a buffer containing 20 mM HEPES, pH 7.4, 0.5 M NaCl, 1 mM MgCl2) were mixed with 20 μl recombinant OCRL-FL or OCRL-CT proteins (1 mg/ml in Buffer A: 20 mM HEPRES, pH 7.4, 0.2 mM NaCl, 4 mM β-ME) and subsequently add buffer A to 200 μl. The reaction was incubated at room temperature for 20 min. Samples were analysed by uranyl acetate negative stain electron microscopy.
NMR assignments and structure determination
All NMR spectra were collected at 25 °C on a Varian INOVA 600 MHz spectrometer and used pulse sequences available in the Varian BioPack User Library. Sequential and aliphatic side chain assignments were determined by analysis of 3D-HNCO, HNCA, HNCACB, HN(CO)CA, HCACO, CBCA(CO)NH, CC(CO)NH, HCC(CO)NH, and HCCH-TOCSY NMR experiments. All spectra were processed with NMRPipe (Delaglio et al, 1995) and subjected to visual analysis in Sparky (Kneller and Goddard, 1997).
Restraints on the backbone dihedral angles were derived from an analysis of backbone chemical shifts with the TALOS program (Cornilescu et al, 1999). Hydrogen bonds were identified during the later stages of structure determination based on the consistent proximity of hydrogen bonding partners in the calculated ensembles. NOE correlations between nearby protons were identified in 3D 15N-NOESYHSQC, 13C-NOESYHSQC (aromatic), and 13C-NOESYHSQC (aliphatic) NMR spectra.
Structure calculations were performed using the CYANA software package (Guntert et al, 1997) based on NOEs that were interpreted and calibrated by the program CANDID (Herrmann et al, 2002). The final series of structure calculations used the CANDID-derived NOE restraints and the earlier described hydrogen bond and backbone torsion angle restraints. Structures, we further refined with the XPLOR-NIH program (Schwieters et al, 2003) with the restraints converted from the output NOE restraints from CYANA, the dihedral angle restraints calculated by TALOS, and the hydrogen bonds restraints. During the iterative refinement cycles, violated NOE restraints were loosened, deleted, or reassigned based on the experimental NOE spectra. The final 20 structures with the lowest target function values from 200 independently calculated structures were retained as the final ensemble.
Miscellaneous methods
Structure representations were drawn by using MolScript (Kraulis, 1991) or PyMol (DeLano Scientific LLC). Multiple sequence alignments were generated by ClustralW 2 (Labarga et al, 2007) and coloured by ALSCRIPT (Barton, 1993). LC MS/MS was done at the Keck Research Facility at Yale University.
Data deposition
The chemical shift assignments of both OCRL and INPP5B are deposited in the BioMagRedBank with the BMRB accession number 12671 and 12673. Coordinates for the ensemble of the NH2-terminal domain of OCRL and INPP5B structures have been deposited in the Protein Data Bank with the PDB ID 2kie and 2kig, respectively.
Addendum
While this paper was in review, a manuscript was published online (Choudhury et al, 2009) reporting the identification of the same novel clathrin-binding motif in the NH2-terminal region of OCRL that we report here.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Material
Review Process File
Acknowledgments
We thank Laura Swan and Peter McPherson for critical discussion and suggestions, Derek Toomre for advice, and Louise Lucast, Summer Paradise, and Frank Wilson for technical help. This work was supported in part by the G Harold and Leila Y Mathers Charitable Foundation and NIH grants to PDC (NS36251, CA46128, DK45735, and DA018343), grants from the Lowe Syndrome Association to PDC and YM, a Howard Hughes Medical Institute fellowship from the Life Sciences Research Foundation to YM, an NIH MSTP TG 5T32GM07205 for DMB, and an NIH grant (CA 108992) to MH. We thank Yale Center for High Performance Computation in Biology and Biomedicine, which is supported by NIH grant 1 S10 RR19895.
Footnotes
The authors declare that they have no conflict of interest.
References
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL (1992) The Lowe's oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358: 239–242 [DOI] [PubMed] [Google Scholar]
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, Greene LE, Lowe M (2005) Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell 16: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M (2009) Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem 284: 9965–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Cremona O, Di Paolo G, Wenk MR, Luthi A, Kim WT, Takei K, Daniell L, Nemoto Y, Shears SB, Flavell RA, McCormick DA, De Camilli P (1999) Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99: 179–188 [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Godi A (2004) PI-loting membrane traffic. Nat Cell Biol 6: 487–492 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC (2001) Clathrin-binding proteins: got a motif? Join the network!. Trends Cell Biol 11: 315–318 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositids in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG (2004) RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell 15: 1666–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P (2007) A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell 13: 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O, Gacon G (2003) Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet 12: 2449–2456 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison SC, Kirchhausen T (2004) Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature 432: 649–653 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T (2008) Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics 7: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298 [DOI] [PubMed] [Google Scholar]
- Haffner C, Di Paolo G, Rosenthal JA, de Camilli P (2000) Direct interaction of the 170 kDa isoform of synaptojanin 1 with clathrin and with the clathrin adaptor AP-2. Curr Biol 10: 471–474 [DOI] [PubMed] [Google Scholar]
- Hellsten E, Evans JP, Bernard DJ, Janne PA, Nussbaum RL (2001) Disrupted sperm function and fertilin beta processing in mice deficient in the inositol polyphosphate 5-phosphatase Inpp5b. Dev Biol 240: 641–653 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319: 209–227 [DOI] [PubMed] [Google Scholar]
- Hokin LE (1985) Receptors and phosphoinositide-generated second messengers. Annu Rev Biochem 54: 205–235 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Hoopes RR Jr, Shrimpton AE, Knohl SJ, Hueber P, Hoppe B, Matyus J, Simckes A, Tasic V, Toenshoff B, Suchy SF, Nussbaum RL, Scheinman SJ (2005) Dent disease with mutations in OCRL1. Am J Hum Genet 76: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M (2006) Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J 25: 3750–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Suchy SF, Bernard D, MacDonald M, Crawley J, Grinberg A, Wynshaw-Boris A, Westphal H, Nussbaum RL (1998) Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest 101: 2042–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson AB, Majerus PW (1995) Properties of type II inositol polyphosphate 5-phosphatase. J Biol Chem 270: 9370–9377 [DOI] [PubMed] [Google Scholar]
- Jin M, Song G, Carman CV, Kim YS, Astrof NS, Shimaoka M, Wittrup DK, Springer TA (2006) Directed evolution to probe protein allostery and integrin I domains of 200,000-fold higher affinity. Proc Natl Acad Sci USA 103: 5758–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen JH, Willingham MC, Pastan IH (1979) Clathrin-coated vesicles: isolation, dissociation and factor-dependent reassociation of clathrin baskets. Cell 16: 303–312 [DOI] [PubMed] [Google Scholar]
- Kneller DG, Goddard TD (1997) SPARKY, 3.105 edit. University of California: San Francisco [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst 24: 946–950 [Google Scholar]
- Labarga A, Valentin F, Anderson M, Lopez R (2007) Web services at the European bioinformatics institute. Nucleic Acids Res 35: W6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer EM (2002) Clathrin-protein interactions. Traffic 3: 513–520 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2004) Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32: 707–711 [DOI] [PubMed] [Google Scholar]
- Lowe CU, Terrey M, Mac LE (1952) Organic-aciduria, decreased renal ammonia production, hydrophthalmos, and mental retardation; a clinical entity. AMA Am J Dis Child 83: 164–184 [DOI] [PubMed] [Google Scholar]
- Lowe M (2005) Structure and function of the Lowe syndrome protein OCRL1. Traffic 6: 711–719 [DOI] [PubMed] [Google Scholar]
- Matzaris M, Jackson SP, Laxminarayan KM, Speed CJ, Mitchell CA (1994) Identification and characterization of the phosphatidylinositol-(4,5)-bisphosphate 5-phosphatase in human platelets. J Biol Chem 269: 3397–3402 [PubMed] [Google Scholar]
- McCrea HJ, De Camilli P (2009) Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology 24: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea HJ, Paradise S, Tomasini L, Addis M, Melis MA, De Matteis MA, De Camilli P (2008) All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding. Biochem Biophys Res Commun 369: 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang X, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, De Camilli P (1996) A presynaptic inositol-5-phosphatase. Nature 379: 353–357 [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (2000) Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci 25: 229–235 [DOI] [PubMed] [Google Scholar]
- Olivos-Glander IM, Janne PA, Nussbaum RL (1995) The oculocerebrorenal syndrome gene product is a 105-kD protein localized to the Golgi complex. Am J Hum Genet 57: 817–823 [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR (2000) The structure and function of the beta 2-adaptin appendage domain. EMBO J 19: 4216–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Zoncu R, Lucast L, De Camilli P, Toomre D (2006) Two synaptojanin 1 isoforms are recruited to clathrin-coated pits at different stages. Proc Natl Acad Sci USA 103: 19332–19337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP (2006) A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Rapoport I, Boll W, Yu A, Bocking T, Kirchhausen T (2008) A motif in the clathrin heavy chain required for the Hsc70/auxilin uncoating reaction. Mol Biol Cell 19: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS (2001) Adaptor-related proteins. Curr Opin Cell Biol 13: 444–453 [DOI] [PubMed] [Google Scholar]
- Schmid AC, Wise HM, Mitchell CA, Nussbaum R, Woscholski R (2004) Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett 576: 9–13 [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Shin HW, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, Frankel WN, Solimena M, De Camilli P, Zerial M (2005) An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol 170: 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W, Oorschot V, Geuze HJ (1996) A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol 132: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo M, Tan L, Lim L, Manser E (2001) The tyrosine kinase ACK1 associates with clathrin-coated vesicles through a binding motif shared by arrestin and other adaptors. J Biol Chem 276: 18392–18398 [DOI] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T (2000) Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA 97: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker RV (2000) Molecular pathology of renal chloride channels in Dent's disease and Bartter's syndrome. Exp Nephrol 8: 351–360 [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH (2001) Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105: 379–389 [DOI] [PubMed] [Google Scholar]
- Ungewickell A, Ward ME, Ungewickell E, Majerus PW (2004) The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc Natl Acad Sci USA 101: 13501–13506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, D'Angelo G, Di Campli A, De Matteis MA (2008) Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci 65: 2833–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR, De Camilli P (2004) Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc Natl Acad Sci USA 101: 8262–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P (2003) Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat Biotechnol 21: 813–817 [DOI] [PubMed] [Google Scholar]
- Zaremba S, Keen JH (1983) Assembly polypeptides from coated vesicles mediate reassembly of unique clathrin coats. J Cell Biol 97: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hartz PA, Philip E, Racusen LC, Majerus PW (1998) Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J Biol Chem 273: 1574–1582 [DOI] [PubMed] [Google Scholar]
- Zhang X, Jefferson AB, Auethavekiat V, Majerus PW (1995) The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci USA 92: 4853–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Table 1
Supplementary Material
Review Process File