EMBO J 28, 1878–1889 (2009); published online 08 July 2009
DNA repair in the context of chromatin represents a challenge both in terms of accessibility to the lesion and maintenance of genome stability. Histones, the main protein component of chromatin, can be subjected to a variety of post-translational modifications (PTMs) that impact on genome function by either directly affecting nucleosome stability or providing a docking site for distinct regulatory proteins. In this issue of EMBO Journal, Tjeertes et al investigate histone PTMs with a specific involvement in the DNA damage response (DDR).
After DNA damage, a prominent change at the chromatin level is the increase in H2A.X phosphorylation (YH2A.X) known to elicit recruitment and activation of many effector proteins (Figure 1) (van Attikum and Gasser, 2009). Interestingly, early work showed that after UV damage in human fibroblasts, a wave of hyperacetylation followed by subsequent deacetylation occurred on histones at a global level (Ramanathan and Smerdon, 1986). Such connection with histone acetylation has been further substantiated in the context of DSB with the Tip60 HAT complex leading to the notion that histone acetylation might assist the repair process itself and/or participate in the chromatin restoration step (van Attikum and Gasser, 2009). However, a survey to examine the specific contribution of a wide range of histone PTMs to the DDR was still missing especially in mammals.
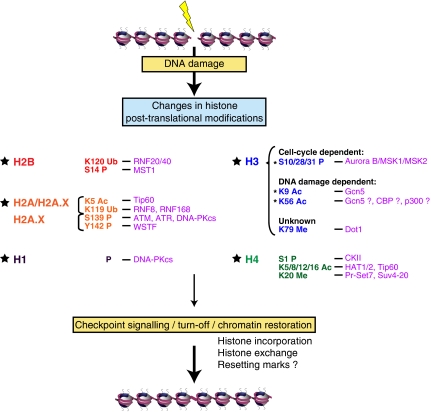
Figure 1.
A histone code for the DNA damage response in mammalian cells. After DNA damage, histones undergo many different post-translational modifications (PTMs) that can impact on the DNA repair process itself or during the chromatin restoration step. Tjeertes et al report new findings concerning changes in histone PTMs on DNA damage (marked with *). In contrast, methylation of H3K79 and H4K20 while unchanged on DSB formation rather become accessible thereby facilitating the recruitment of checkpoint signalling proteins such as 53BP1. In addition to the changes in PTMs imposed by specific enzymatic complexes (in purple), chromatin-remodelling complexes also participate in the DNA damage response by making chromatin more accessible through histone eviction or exchange (not shown). Ac, acetylation; Me, methylation; P, phosphorylation; Ub, ubiquitination. K, lysine; S, serine; Y, tyrosine.
In this issue of EMBO Journal, Tjeertes et al, using human cell lines that they subjected to various DNA damaging agents, systematically analysed changes in histone PTMs with commercially available PTM-specific histone antibodies. They report a global reduction of H3K9ac and H3K56ac on DNA damage and discuss the potential roles of these modifications during the DNA damage response. In contrast with many PTMs that are imposed on the free N-terminal part of histones, H3K56 acetylation, like H3K79 methylation, is located in the globular domain of histone H3. This region is close to the DNA entry–exit point of the nucleosome core particle, leading to the hypothesis that acetylation could potentially affect nucleosome stability (Groth et al, 2007). Although until recently, the presence of H3K56ac in mammals had remained elusive, using both mass spectrometry and antibodies in western blot analysis, three independent groups have now detected H3K56ac in human cell lines (Das et al, 2009; Xie et al, 2009; Yuan et al, 2009).
In their survey, although 6 antibodies out of 32 proved non-specific, Tjeertes et al choose to focus on a subgroup of eight antibodies (YH2A.X, H3R2me2, H3S10p, H3S28p, H3S31p, H3K9ac, H3K14ac, H3K56ac), which showed a common response to replicative stress or to DSB-induced DNA damage. Importantly, they took into account that DNA damage induces alterations in cell-cycle progression and/or changes in transcription, and checked in each case whether changes in histone PTMs observed in their assays were a consequence of cell-cycle arrest or truly induced DNA damage effects. This analysis showed that decrease of H3S10p, H3S28p and H3s31p modifications after DNA damage simply reflected the cell-cycle arrest and depletion in mitotic cells in which these modifications are normally highly represented (Figure 1) (Tjeertes et al, 2009). However, H3K9ac and H3K56ac levels, under their experimental conditions and in the cell line studied, did not change significantly during cell cycle nor on transcription inhibition, yet they were reduced after damage. In yeast, H3K56ac is upregulated in S-phase (Groth et al, 2007), whereas in human cells this is a matter of debate (Yuan et al, 2009). Thus, the extent to which H3K56ac properties in Saccharomyces cerevisiae can apply to mammals will have to be examined closer. It will surely be important to take into account the physiological state of the cells, as senescence state or aging may possibly affect the observations. Future studies should shed light on these issues.
In S. cerevisiae, acetylation of H3K56 depends on the complex containing Rtt109 HAT (KAT11) together with a histone chaperone, Vps75 (a Nap-1-related protein) or Asf1. Interestingly, Rtt109 can also catalyse H3K9 acetylation in S. cerevisiae, in addition to Gcn5 (KAT2A) (Corpet and Almouzni, 2009). Here, Tjeertes et al show that human Gcn5 can act as an acetyltransferase for both H3K9 and H3K56 in vitro on recombinant histone H3. They further provide arguments in vivo by RNAi depletion experiments on human cells in culture, even though possible indirect effects may explain results in RNA interference. Given the potential structural homology between S. cerevisiae Rtt109 and human p300 (Tang et al, 2008), the human HAT p300 has to be also considered as a candidate. Interestingly, Das et al show that the simultaneous knockdown of two human HATs, p300 and CBP, leads to a major decrease in H3K56ac levels. However, DNA damage induced by p300 knockdown in Tjeertes work prevented further analysis. How these multiple HATs act on H3K56ac in mammalian cells will be an interesting issue to explore.
The present work enabled to specifically assign changes in histone PTMs to the presence of DNA damage per se at least for a set of modifications (Figure 1). Although extending these studies to other modifications will be an obvious next step, determining the functional importance of these changes in the DDR pathway represents a major challenge for future research. The parallel changes in H3K9 and K56 acetylation suggest a possible linked regulation and function. This is reminiscent of the diacetylation of H4 at lysines 5 and 12, typical of newly synthesized histones (Loyola and Almouzni, 2007), which has been reported at DSB sites. Whether drop in the levels of H3K9 and K56 aceylation levels reflect events related to nucleosome assembly or rather relate to signalling pathways downstream of DSB, remains to be determined. An example of decrease in a modification on DNA damage is the phosphorylation of tyrosine 142 on H2A.X (Y142 P) (Xiao et al, 2009). This modification, imposed by WSTF, was proposed to regulate the maintenance/disappearance rate of YH2A.X (Xiao et al, 2009). A comparison with H3K9ac and K56ac awaits further characterization of the kinetics of their variations on DNA damage. The global reduction in acetylation levels at H3K9 and K56 could also reflect a short-term effect due to the loss of histones from the DSB site followed by their subsequent degradation. As new histones can be incorporated at DSB sites after repair in human cells (Groth et al, 2007), one could also imagine that H3K9ac and H3K56ac levels would increase on chromatin at later time points after completion of DNA damage repair.
Nevertheless, a careful comparison of cell lines, their physiological condition (passage number), antibodies, extract preparation and damage treatment will be necessary to fully comprehend how H3K56ac, and any other modification to consider, behave during the DNA damage response in mammals and reconcile contradictory findings reported on H3K56ac levels upon DNA damage in the recent literature (Das et al, 2009; Tjeertes et al, 2009; Yuan et al, 2009). Overall, at this point in time, a take home message is certainly to be extremely careful with commercially available antibodies, and to distinguish changes that relate to cell cycle or transcription with a specific code for the DNA damage response or any other signal one wants to study. This work will undoubtedly set the stage to investigate in a specific way the importance of key modifications related to DNA damage. Further experiments using advanced mass spectrometry should help confirm this first screening approach and, therefore, deepen our understanding of histone PTMs in relation to DDR, which we have only begun to unravel.
Footnotes
The authors declare that they have no conflict of interest.
References
- Corpet A, Almouzni G (2009) Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol 19: 29–41 [DOI] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK (2009) CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128: 721–733 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G (2007) Marking histone H3 variants: how, when and why? Trends Biochem Sci 32: 425–433 [DOI] [PubMed] [Google Scholar]
- Ramanathan B, Smerdon MJ (1986) Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis 7: 1087–1094 [DOI] [PubMed] [Google Scholar]
- Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, Marmorstein R (2008) Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol 15: 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP (2009) Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J 28: 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD (2009) WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M (2009) Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Pu M, Zhang Z, Lou Z (2009) Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle 8: 1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]