Abstract
A fundamental question in ecology is how many species occur within a given area. Despite the complexity and diversity of different ecosystems, there exists a surprisingly simple, approximate answer: the number of species is proportional to the size of the area raised to some exponent. The exponent often turns out to be roughly 1/4. This power law can be derived from assumptions about the relative abundances of species or from notions of self-similarity. Here we analyze the largest existing data set of location-mapped species: over one million, individually identified trees from five tropical forests on three continents. Although the power law is a reasonable, zeroth-order approximation of our data, we find consistent deviations from it on all spatial scales. Furthermore, tropical forests are not self-similar at areas ≤50 hectares. We develop an extended model of the species-area relationship, which enables us to predict large-scale species diversity from small-scale data samples more accurately than any other available method.
A primary motivation for modern ecological research is the effort to save as many species as possible from the sixth great mass extinction that currently threatens them (1, 2). How does habitat loss and destruction of tropical forests relate to species extinction? How many tree species must remain in an exploited forest if primate species are to survive in it? What is the best possible design of a natural reserve that maximizes the number or genetic diversity of surviving species? All of these questions underscore the necessity to understand the relationship between species diversity and sampled area (3–8)—a longstanding and controversial subject in ecology (9–11).
The earliest model of the species–area relationship (SAR) was introduced by Arrhenius in 1921 and posits a power law: the number of species, S, found in a census area, A, is given by
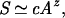 |
1 |
where c and z are constants (12). Empirical observations suggest that z is about 1/4 for many ecosystems (13). The power law is a cornerstone for theories of biogeography (14–16). In 1975 May (17) derived the power law by assuming that species' abundances follow a lognormal distribution. The canonical lognormal distribution implies that S ≃ cN1/4, where N is the total number of individuals and c is a constant. Assuming that N is proportional to the area A, we immediately obtain Eq. 1 with z = 1/4.
More recently, Harte et al. (18) have shown that the power law is equivalent to self-similarity. If the fraction of species in an area A that are also found in one-half of that area is independent of A, then the spatial distribution of species is self-similar. Let Ai = A0/2i denote the area of a rectangular patch obtained after i bisections of the total sampled area A0. Denote by Si the average number of species found in a patch Ai. If the ratio ai = Si/Si−1 does not depend on i then the assemblage is self-similar (18). Self-similarity is equivalent to Si = cAiz with z = −log2a. Unlike the canonical lognormal, self-similarity does not provide an a priori estimate of the exponent z.
Tropical Forest Data
To test the basic principles of SARs—with an aim toward generalizing the power law—we have analyzed five 50-hectare (ha) plots of tropical forests across the globe. Although tropical forests cover only 7% of the Earth's land surface, they contain more than half of the world's species (6). Tropical forests are well known as the most genetically diverse, terrestrial communities on Earth (19). Moreover, animal diversity in tropical forests depends crucially on the diversity of plants (20).
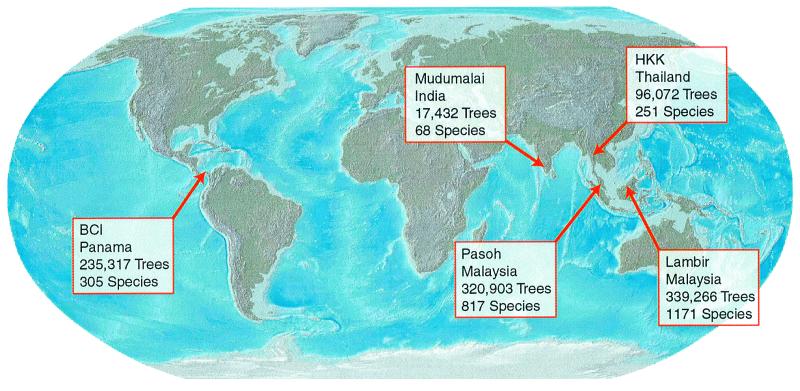
Each of the 50-ha plots that we analyze is part of a long-term research program coordinated by the Smithsonian's Center for Tropical Forest Science. The plots are located in the following forests: Huai Kha Khaeng (HKK) Wildlife Sanctuary, Thailand (first census); Lambir Hills National Park, Sarawak, Malaysia (third census); Pasoh Forest Reserve, Peninsular Malaysia (third census); Barro Colorado Island (BCI), Panama (first census); and Mudumalai Wildlife Sanctuary, India (second census). For each of our plots, every free-standing, woody stem over 1 cm in diameter has been identified to species. We include all such stems in our analyses. (Our qualitative results are unchanged if we include, instead, stems over 5 cm in diameter.) The number of such stems, and the number of species among them, varies greatly from plot to plot (Fig. 1).
Figure 1.
The locations of five tropical forest plots across the globe. Each census encompasses 50 ha of forest within which every woody stem greater than 1 cm in diameter has been identified to species, measured for girth, and spatially mapped to <1-m accuracy. The name, country, number of trees, and number of species is indicated for each plot. The forests vary widely in species diversity and environment. Pasoh and Lambir (Malaysia) are evergreen, dipterocarp rainforests; BCI (Panama) is a lowland, moist forest, with a 4-month dry season; HKK (Thailand) and Mudumalai (India) are the only forests that are regularly subject to fires. For a complete list of references, consult the Center for Tropical Forest Science web site at http://www.stri.org.
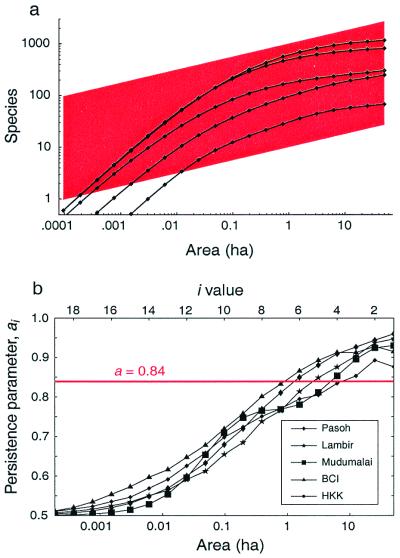
Fig. 2a shows the species-area relationship for the five tropical forests compared with the best-fit power law. The average slope for the forests is z≃0.25. As suggested by May, the power law tends to overestimate the slope at large areas and underestimate the slope at small areas (17). But the extent to which the power law fails is often poorly recognized in the ecological literature (3, 18, 21). Previous research has uncovered the power law's failure for small areas, but has downplayed its deviations for areas larger than 2 ha (22). Fig. 2b shows the dependence of the parameter ai = Si/Si−1 on area. The ratio ai describes the average fraction of species that persist upon the ith bisection; therefore we call ai the spatial persistence parameter. Self-similarity would require that this parameter be independent of area (18). As the figure shows, however, ai is not constant for any range of areas between 1 m2 and 50 ha. Hence, tropical forests are conclusively not self-similar at these scales. The empirical form of the spatial persistence curve, and its departure from self-similarity, may result in part from aggregation of conspecifics—a possibility that we explore in detail elsewhere (29).
Figure 2.
Graphs of the SAR and the spatial persistence parameter for each of five tropical forests. (a) Log-log graph of the observed species-area data. Each plot encompasses a total area A0 = 50 ha. We measure the mean species diversity, Si, found in disjoint patches obtained by repeated bisections of A0. The log-log species-area data are concave down for all five plots. The SAR is approximated loosely by the power law, z = 0.25, whose slope is indicated by the trapezoid (red). (b) The persistence parameter, ai = Si/Si−1, provides a sensitive tool for analyzing SARs and testing self-similarity. Self-similarity would predict constant a ≃ 2−0.25 ≃ 0.84, shown in red. All five persistence curves are seen to depart from the power-law model over the entire range of areas.
A Differential Equation Approach
Instead of self-similarity we find a consistent functional relationship between ai and the area, A, in all five forests (Fig. 2b). This observation is striking in light of the forests' disparate geographic locations, climates, and overall species diversities. We now introduce the spatial persistence function, a(A), as a continuous extension of ai. In the Appendix, we use the persistence values of our data to derive a canonical, two-parameter model of a(A). Once this function has been derived, we obtain the SAR by solving the differential equation
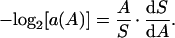 |
2 |
Using the diversity measured in a small area as the initial condition in Eq. 2, we may predict the diversity of a much larger area, if we know a(A).
In the Appendix, we derive Eq. 2 and find a general solution of the form S = cAzexp[P(A)], where P(A) is an infinite polynomial in A. We can truncate after the first n terms to obtain an approximate solution. Truncating after the first term leads to the expression
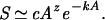 |
3 |
Here c, z, and k are constants determined by a(A). This approximate solution is less accurate than the complete solution to Eq. 2, and it is only valid for a limited range of areas. Nevertheless, the approximation has the obvious advantage of simplicity. If we let n → ∞, then we recover the full solution to Eq. 2; if we let n → 0, then we reduce to the power law. Hence the power law is a zeroth-order, special case of our general model for the SAR.
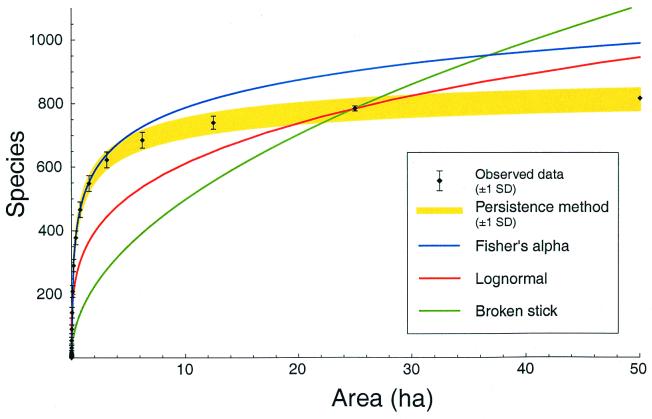
Eq. 2 accurately predicts diversity given only a small amount of data. Because the persistence curves are similar across the five plots (Fig. 2b), we may use the canonical form of a(A) fit at one plot to predict diversity for another plot. For example, using BCI's persistence curve to determine a(A) and using the diversity in a single ha of Pasoh as the initial condition, we can predict the 50-ha diversity of Pasoh within 3% on average (Fig. 3). Conversely, Pasoh's persistence function predicts BCI's total diversity with 4% average error, and Lambir's diversity with 9% error, from a single ha of data.
Figure 3.
The actual SAR at Pasoh (black) compared with the SAR predicted by our model and three classical models. Assuming that individuals scale linearly with area, MacArthur's “broken stick” distribution of relative abundances, the canonical lognormal distribution, and Fisher's log series each yields a one-parameter model of the SAR (green, red, and blue, respectively). The first two of these models were parameterized by using 25 ha of Pasoh data; 1 ha was used to parameterize the log series. The log series provides a fairly accurate model, but it overestimates 50-ha diversity by 21%. The canonical log normal accounts for steeper slopes at small areas and gentler slopes at large areas, and hence it is more accurate than the broken stick (17). Our persistence method (yellow) requires two parameters, fit by using any other forest, and an initial condition obtained from 1 ha of Pasoh data. The persistence method extrapolates 50-ha diversity with 3% average error. The figure indicates a 1-SD confidence interval around the extrapolation.
Fig. 3 illustrates the extrapolative ability of Eq. 2 as compared with the classical models of the SAR. The precision of our method—namely, the ability to predict 50-ha diversity within 5% at Pasoh and BCI and 10% at Lambir—is an improvement over other previous methods. It is 1- to 7-fold more precise than Fisher's alpha (23), and 5- to 10-fold more precise than the power law. On the one hand, the increased precision of our method is not surprising; we have used two parameters to describe a(A), as opposed to the classical models, which generally require one parameter. On the other hand, given the interplot similarity of persistence curves, in practice we need only measure one parameter—the diversity of a single ha—to extrapolate diversity via Eq. 2.
Implications and Conclusions
We have analyzed the largest existing data set of location-mapped trees in tropical forests. We find that the SAR shows consistent deviations from the power law on all spatial scales that were studied, ranging from 1 m2 to 50 ha. Hence, self-similarity does not hold over this range of areas. (There is the possibility that tropical forests are self-similar over scales greater than 50 ha, but in the absence of further data this remains speculative.) These results might have some bearing on the longstanding controversy surrounding SARs. Previous research has focused on why the SAR has different slopes in different ecosystems (11), but in our extensive data the SAR does not possess a constant slope whatsoever.
Instead of self-similarity we propose a model of the SAR, based on the spatial persistence function, which generalizes the power law. This framework allows us to predict 50-ha diversity from small-scale samples with greater accuracy than ever before. Candidate logging protocols often are assayed at the 50- to 100-ha scale, and they are evaluated on the proportion of diversity that regenerates, as estimated from a small census (24). Hence, an accurate method to extrapolate 50-ha diversity from a small census will greatly benefit in the formulation of protocols for sustainable forestry and for biodiversity surveys (25). Furthermore, our methods may be extended to estimate landscape-scale diversity (see Appendix). These advances may induce ecologists to focus on the persistence curve itself as a unifying concept. The search for a biological mechanism that explains the observed persistence patterns offers an important challenge to ecology. In the meantime, our theory provides a valuable tool for conservation planning and a practical method for estimating diversity in the field.
Acknowledgments
We sincerely thank Alice Chen, Jay Rosenheim, Stuart Davies, Egbert Leigh, Bill Bossert, Burt Singer, Steve Pacala, Jerome Chave, and Simon Levin for many helpful discussions. We are especially grateful to Helene Muller-Landau, Bob Holt, and Robert May for their conceptual and editorial input. J.B.P. is supported by a National Science Foundation graduate fellowship.
Abbreviations
- SAR
species–area curve
- ha
hectare
- BCI
Barro Colorado Island
- HKK
Huai Kha Khaeng
Appendix
In this appendix we provide the details behind the derivation of Eq. 2, its solution, and its application to extrapolating diversity.
Derivation of Eq. 2.
Given the definition of the spatial persistence parameter, ai = Si/Si−1, we start by deriving its relationship to the slope of the SAR. All logarithms are henceforth taken base two:
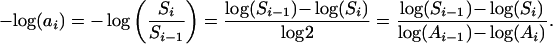 |
4 |
The last equality follows because Ai−1/Ai= 2. The final quantity in Eq. 4 measures the slope of a small chord on the log-log SAR to the right of log(Ai). We conclude that
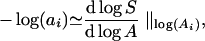 |
5 |
where i is now a continuous variable. To be more precise, the chord to the right of log(Ai) is an estimate of the derivative at log(A(i+(i−1))/2). Note that Eq. 5 clearly illustrates the equivalence between self-similarity (constant a) and the power law (constant dlogS/dlogA).
Eq. 2 follows easily from Eq. 5, using the fact that dlogS/dlogA = A/S ⋅ dS/dA. Note that Eq. 2 is a strict generalization of self-similarity: if a(A) is constant, then Eq. 2 reduces to the power law. If a(A) = exp(−1/logA), then Eq. 2 reduces to S(A)∝logA. Hence Eq. 2 also generalizes the logarithmic law suggested by Gleason (26).
Diversity Extrapolation.
Eq. 2 together with the interplot similarity of the persistence curves provide a method for extrapolating diversity over many spatial scales. For example, to extrapolate diversity from a subsample of Pasoh, we use BCI data to fit a(A), and then we solve Eq. 2 according to the small, initial condition measured at Pasoh. In effect, this process translates BCI's log-log SAR so that it coincides with Pasoh's initial condition; nevertheless, the universality between the forests is described more simply in terms of the persistence parameter, ai.
We choose to model the persistence values
ai with the simple, two-parameter
family of curves ¼Φ[(α −
i)/β] + ¾. (Other choices, such
as a cubic model, are also possible and produce accurate predictions at
these scales and beyond. See below.) Here Φ(x) is the
“error function” given by the cumulative distribution of the
Gaussian: Φ(x) =
(2/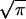 )⋅∫0xe−t2dt.
The parameter α moves the inflection point of the persistence curve
horizontally, and β determines the slope at the inflection point.
Hence β measures the maximal “acceleration” of diversity with
area, and α measures the spatial scale at which acceleration is
maximized. The best-fit at BCI is given by α = 8.56, β =
8.08. For Pasoh, α = 7.73, β = 7.41; for Mudumalai,
α = 7.06 and β = 7.76.
)⋅∫0xe−t2dt.
The parameter α moves the inflection point of the persistence curve
horizontally, and β determines the slope at the inflection point.
Hence β measures the maximal “acceleration” of diversity with
area, and α measures the spatial scale at which acceleration is
maximized. The best-fit at BCI is given by α = 8.56, β =
8.08. For Pasoh, α = 7.73, β = 7.41; for Mudumalai,
α = 7.06 and β = 7.76.
We may express all solutions to Eq. 2 in the form S = cAzexp[P(A)], where P(A) is a polynomial in A of arbitrary degree n, with no constant term. Once we specify α and β, we expand −log(a(A)) in a Taylor series of order n around the point A = 25 ha. The resulting separable equation can always be solved in closed form, yielding P. For example, using Pasoh's α and β to determine a(A), the approximate solution of order n =1 is given by S(A) = S(1ha)⋅AzekA, where z = 0.125 and k = −5.66⋅10−4. For Mudumalai, z = 0.161 and k = −5.41⋅10−4. This approximation is only valid for A ≤ 50 ha, but its accuracy compares well with the complete numerical solution: it predicts 50-ha diversity with average error 4% at Pasoh, 9% at BCI, and 16% at Lambir.
In practice, a numerical solution of Eq. 2 yields the most accurate SAR. We used the Fehlberg-Runge-Kutta method to generate the prediction shown in Fig. 3. The initial condition S(1ha) for Fig. 3 was determined by the diversity of a single, random, 1-ha subplot of Pasoh. The confidence interval was constructed from 1,000, independently sampled, 1-ha initial conditions.
We have divided our five plots into two categories: those that suffer regular disturbance and those that do not. The two tropical forests subject to regular fires, HKK and Mudumalai, generally should not be modeled via the persistence curve from a more stable, moist tropical forest. The values of a1 to a6 are generally smaller at HKK and Mudumalai than the other three forests. This reflects the fact that HKK and Mudumalai are subject to more disturbances, causing greater patchiness. The persistence curve at Mudumalai can predict HKK and conversely within 17%, given 1 ha of data. Compared with BCI, Pasoh, and Lambir, the accuracy of Eq. 2 has been decreased by the disturbances at Mudumalai and HKK. Nevertheless, 17% error is still preferable to the 28% error-rate or worse given by Fisher's alpha or a power law on these disturbed forests. In practice, when estimating diversity in a new forest, the ecologist should first determine the frequency of disturbances (e.g., fires, hurricanes, or roaming elephants) and choose a model forest, where α and β are known, accordingly.
Extrapolation Beyond 50 ha.
For 50-ha predictions, such as would be useful to assess logging protocols, Φ provides a simple, two-parameter model of the persistence curve. For larger areas, however, a cubic model (which works as well as Φ at 50 ha) is often more effective. For example, we can use a cubic persistence curve calibrated at Pasoh to extrapolate the diversity of the entire BCI, which occupies 1,500 ha, from a 1-ha sample. Using Eq. 2, the predicted diversity for all of BCI is 436 ± 32 species (1 SD). This estimate compares favorably with Croat's floral count of 450 tree and shrub species on the island (27). For even larger areas, the persistence curve should be parameterized by using multiple, small censuses spread across the landscape (as in ref. 28), although such techniques require further development.
References
- 1.May R. Philos Trans R Soc London B. 1999;354:1951–1959. doi: 10.1098/rstb.1999.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid W, Miller K. Keeping Options Alive: The Scientific Basis for Conservation Biology. Washington, DC: World Resources Institute; 1989. [Google Scholar]
- 3.Reid W. In: Tropical Deforestation and Species Extinction. Whitmore T, Sayer J, editors. London: Chapman & Hall; 1992. pp. 55–73. [Google Scholar]
- 4.Simberloff D. In: Dynamics of Extinction. Elliot D, editor. New York: Wiley; 1986. pp. 165–180. [Google Scholar]
- 5.Raven P. In: Biodiversity. Wilson E O, Peter F, editors. Washington, DC: Natl. Acad. Press; 1988. pp. 119–122. [Google Scholar]
- 6.Wilson E O. In: Biodiversity. Wilson E O, Peter F, editors. Washington, DC: Natl. Acad. Press; 1988. pp. 3–18. [Google Scholar]
- 7.May R, Lawton J, Stork N. In: Extinction Rates. Lawton J, May R, editors. Oxford: Oxford Univ. Press; 1995. pp. 1–24. [Google Scholar]
- 8.Pimm S, Raven P. Nature (London) 2000;403:843–845. doi: 10.1038/35002708. [DOI] [PubMed] [Google Scholar]
- 9.Connor E, McCoy E. Am Nat. 1979;113:791–833. [Google Scholar]
- 10.McGuinness K. Biol Rev. 1984;59:423–440. [Google Scholar]
- 11.Durrett R, Levin S. J Theor Biol. 1996;179:119–127. [Google Scholar]
- 12.Arrhenius O. J Ecol. 1921;9:95–99. [Google Scholar]
- 13.Rosenzweig M. Species Diversity in Space and Time. Cambridge: Cambridge Univ. Press; 1995. [Google Scholar]
- 14.Preston F. Ecology. 1962;43:185–215. [Google Scholar]
- 15.MacArthur R, Wilson E O. Island Biogeography. Princeton: Princeton Univ. Press; 1967. [Google Scholar]
- 16.Hubbell S. The Unified Theory of Biogeography and Biodiversity. Princeton: Princeton Univ. Press; 2000. , in press. [Google Scholar]
- 17.May R. In: Ecology and Evolution of Communities. Cody M, Diamond J, editors. Cambridge: Belknap; 1975. pp. 81–120. [Google Scholar]
- 18.Harte J, Kinzig A, Green J. Science. 1999;284:334–336. doi: 10.1126/science.284.5412.334. [DOI] [PubMed] [Google Scholar]
- 19.Hubbell S, Foster R. In: Tropical Rain Forests: Ecology and Management. Sutton S, Whitmore T, editors. London: Blackwell; 1983. pp. 24–40. [Google Scholar]
- 20.Huston M. Biological Diversity. Cambridge: Cambridge Univ. Press; 1994. [Google Scholar]
- 21.Lamb D, Parrotta J, Keenan R, Tucker N. In: Tropical Forest Remnants. Laurance W, Bierregaard R, editors. Chicago: Univ. of Chicago Press; 1997. pp. 366–385. [Google Scholar]
- 22.Condit R, Hubbell S, LaFrankie J, Sukumar R, Manokaran N, Foster R, Ashton P. J Ecol. 1996;84:549–562. [Google Scholar]
- 23.Condit R, Foster R, Hubbell S, Sukumar R, Leigh E, Manokaran N, Loo de Lao Z, LaFrankie J, Ashton P. In: Measuring and Monitoring Forest Biodiversity: The International Network of Biodiversity Plots. Dallmeier F, editor. Washington, DC: Smithsonian Institution; 1996. pp. 247–268. [Google Scholar]
- 24.Stork N, Samways M, Bryant D. In: Global Biodiversity Assessment. Heywood V, editor. Univ. Press, Cambridge: Cambridge; 1995. pp. 461–472. [Google Scholar]
- 25.Balmford A, Gaston K. Nature (London) 1999;398:204–205. [Google Scholar]
- 26.Gleason H. Ecology. 1922;3:158–162. [Google Scholar]
- 27.Croat T. Flora of Barro Colorado Island. Stanford, CA: Stanford Univ. Press; 1978. [Google Scholar]
- 28.Harte J, McCarthy S, Taylor K, Kinzig A, Fischer M. Oikos. 1999;86:45–54. [Google Scholar]
- 29.Plotkin, J. B., Potts, M., Leslie, N., Manokaran, N. & Ashton, P. (2000) J. Theor. Biol., in press. [DOI] [PubMed]