Abstract
Background
Pancreatic cancer is an almost uniformly fatal disease, and early detection is a critical determinant of improved survival. A variety of noninvasive precursor lesions of pancreatic adenocarcinoma have been identified, which provide a unique opportunity for intervention prior to onset of invasive cancer. Biomarker discovery in precursor lesions has been hampered by the ready availability of fresh specimens, and limited yields of proteins suitable for large scale screening.
Methods
We utilized Liquid Tissue®, a novel technique for protein extraction from archival formalin-fixed material, and mass spectrometry to conduct a global proteomic analysis of an intraductal papillary mucinous neoplasm (IPMN). Tissue microarrays comprised of 38 IPMNs were used for validation of candidate proteins.
Results
The proteomic analysis of the IPMN Liquid Tissue lysate resulted in identification of 1,534 peptides corresponding to 523 unique proteins. A subset of 25 proteins was identified that had previously been reported as upregulated in pancreatic cancer. Immunohistochemical analysis for two of these, deleted in malignant brain tumors 1 (DMBT1) and tissue transglutaminase 2 (TGM2), confirmed their overexpression in IPMNs.
Conclusion
Global proteomics analysis using the Liquid Tissue workflow is a feasible approach for unbiased biomarker discovery in limited archival material, particularly applicable to precursor lesions of cancer.
Key Words: Pancreatic cancer, Intraductal papillary mucinous neoplasms, Proteomics, Mass spectrometry
Introduction
Pancreatic cancer is the fourth most common cause of cancer-related mortality in the United States, accounting for 32,000 deaths annually [1]. The overwhelming majority of patients present with advanced metastatic disease, rendering their tumors inoperable. Diagnosis of pancreatic cancer at an early stage provides the best option for successfully treating and improving the dire prognosis of this malignancy. It is now well established that pancreatic adenocarcinomas do not arise de novo, but rather progress through a series of histologically distinct non-invasive precursor lesions [2, 3]. The three most common subtypes of pancreatic cancer precursors include pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasm (IPMN), and mucinous cystic neoplasm (MCN). Surgical resection of noninvasive cystic precursors (IPMN and MCNs) is typically curative, while the presence of any invasive component confers an adverse prognosis on outcome [4, 5]. The identification of biomarkers of pancreatic cancer precursors has the potential to yield diagnostic candidates that will facilitate the early detection of these lesions in at-risk individuals [6].
Global proteomic analysis has emerged as a promising strategy to elucidate potential biomarkers in various cancer subtypes [7,8,9,10]. Unfortunately, these analyses typically mandate the use of snap-frozen or fresh cancer tissues, or the use of cancer cell lines, precluding the direct analysis of smaller, precursor lesions. The ability to perform global proteomic analyses from microscopic precursor lesions in archival tissues would greatly expand the repertoire of specimens for cancer biomarker discovery. Standard formalin-fixed, paraffin-embedded (FFPE) samples are abundant in surgical pathology archives and permit the accurate histological designation of precursor lesions in these cases, without the presence of contaminating cancer cells. Several approaches have recently been described for mass spectrometric analysis of peptides from FFPE specimens, including cancer [11,12,13], as well as non-cancerous tissues [14, 15]. The Liquid Tissue® platform is a technology that permits facile global proteomic analysis of archival material by mass spectrometry [16] and has been utilized for identifying candidate biomarkers of prostate cancer from microdissected, formalin-fixed radical prostatectomy specimens [17], and most recently, from archival sections of head and neck squamous cell carcinomas (HNSCCs) [18]. Notably, these reports confirm that formalin fixation does not significantly alter the ability to generate tryptic peptides for subsequent global proteomic analysis by mass spectrometry.
The objectives of this pilot study were twofold. The first goal was to establish that global proteomic approaches are not limited to cancer cells, but can also be extended to the archival precursor lesions, using the Liquid Tissue platform. The second goal was toward elucidation of specific proteins whose dysregulation is associated with an early stage in the multistep progression of pancreatic cancer, by performing this analysis on a noninvasive IPMN. Two significant candidate marker proteins for IPMN were identified by mass spectrometry, deleted in malignant brain tumors 1 (DMBT1) and tissue transglutaminase 2 (TGM2). The expression levels of these proteins were validated in a larger series of IPMNs by immunohistochemistry on tissue microarrays (TMAs). These results confirm that global proteomic analysis of archival precursor lesions is feasible, and can facilitate the identification of candidate biomarkers applicable in subsequent translational research.
Materials and Methods
Tissue Processing and Sample Preparation
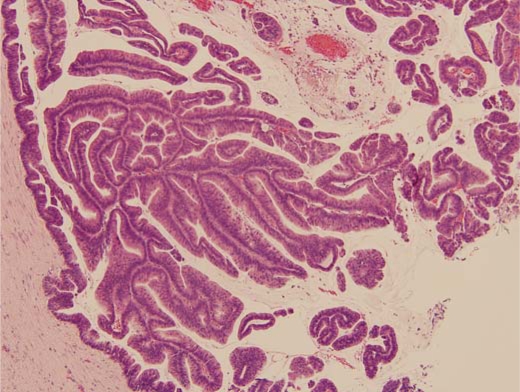
An FFPE block of a noninvasive IPMN with carcinoma in situ was selected for obtaining epithelial cells. A representative hematoxylin and eosin stained section was utilized to confirm the pathological diagnosis of IPMN by established criteria [19] (fig. 1). For tissue collection and protein preparation, a single 10-μm-thick tissue section was cut from the tissue block, placed on a standard microscope slide, and heated for 1 h at 60°C. Paraffin was removed with SubX (Surgipath Medical Industries, Richmond, Ill., USA) followed by tissue rehydration through a series of graded ethanol solutions and distilled water. Approximately 30,000 cells from the relevant region were procured by manual (needle) microdissection for processing, as described previously [16,17,18]. The collected tissue was processed with the Liquid Tissue MS Protein Prep Kit according to manufacturer's recommendations (Expression Pathology, Gaithersburg, Md., USA). Briefly, the tissue was suspended in 20 μl of Liquid Tissue buffer, incubated at 95°C for 90 min, then cooled on ice for 2 min, at which time 1 μg of sequencing-grade porcine trypsin (Promega, Madison, Wisc., USA) was added followed by incubation at 37°C for 16 h. Dithiothreitol was added to a final concentration of 10 mM and the sample was heated for 5 min at 95°C to reduce cysteine residues. The protein digest was stored at −20°C until MS analysis. Prior to MS analysis, the tryptic digest was desalted using a C-18 ZipTip micro-column (Millipore, Billerica, Mass., USA), lyophilized to dryness, and re-suspended in 5 μl of 0.1% trifluoroacetic acid.
Fig. 1.
Photomicrograph of HE-stained, noninvasive IPMN used for microdissection and Liquid Tissue lysate preparation.
Liquid Chromatography–Tandem Mass Spectrometry and Bioinformatic Analysis
Liquid chromatography (LC) was performed using a Dionex Ultimate 3000 nanoflow LC system (Dionex Corporation, Sunnyvale, Calif., USA) coupled on-line to a linear ion trap (LIT) mass spectrometer (MS) (LTQ; ThermoFisher Scientific Inc., San Jose, Calif., USA). Separation of the sample was performed using a 75-μm inner diameter ×360 outer diameter ×10 cm-long fused silica capillary column (Polymicro Technologies, Phoenix, Ariz., USA) packed in house with 5 μm, 300 Å pore size Jupiter C-18 stationary phase (Phenomenex, Torrance, Calif., USA). After injecting 250 ng of the global protein digest, the column was washed with 98% mobile phase A (0.1% formic acid in water) for 30 min and peptides were eluted by development of a linear gradient of 2% mobile phase B (0.1% formic acid in acetonitrile) to 42% mobile phase B in 140 min, then to 98% B in an additional 20 min, all at a constant flow rate of 250 nl/min. The LIT-MS was operated in a data-dependent MS/MS mode in which each full MS scan (precursor ion selection scan range of m/z 350–1,800) was followed by seven MS/MS scans where the seven most abundant peptide molecular ions dynamically determined from the MS scan were selected for tandem MS using a relative collision-induced dissociation (CID) energy of 35%. Dynamic exclusion was utilized to minimize redundant selection of peptides for CID. Tandem mass spectra were searched against the UniProt Homo sapiens proteome database (http://www.expasy.org) using SEQUEST (ThermoFisher Scientific, Inc.). Peptides were considered legitimately identified if they achieved specific charge state and proteolytic cleavage-dependent cross-correlation (Xcorr) scores of 1.9 for [M+H]1+, 2.2 for [M+2H]2+, and 3.1 for [M+3H]3+, and a minimum delta correlation score (ΔCn) of 0.08. Further manual annotation of the identified proteins was performed using publicly available internet-accessible tools, including the NCBI CGAP ‘Batch Gene Finder’ (http://cgap.nci.nih.gov/Genes/BatchGeneFinder). Gene ontology (GO) annotation was performed using a full-featured version of the Expression Analysis Systematic Explorer (EASE, version 2.0 for Windows operating systems) available at http://david.abcc.ncifcrf.gov [20, 21].
Immunohistochemistry
TMAs comprising tissue cores from 38 IPMNs were used for immunohistochemistry as previously described [22, 23]. The archival IPMN samples for TMA construction were obtained from surgically resected specimens at The Johns Hopkins Hospital; all samples were de-linked from direct patient identifiers. Briefly, 5-μm sections of the IPMN TMAs were deparaffinized by routine techniques. Antigen retrieval was performed by incubating with DAKO low pH antigen retrieval reagent (1:10 dilution) for 20 min at 95°C, followed by 20 min of cooling to ambient temperature. Primary and secondary incubation steps for anti-deleted in malignant brain tumor 1 (anti-DMBT1h12, 1/100 dilution, mouse monoclonal) and anti-tissue transglutaminase 2 (anti-TGM2, 1/100 dilution of rabbit polyclonal; Neomarkers; LabVision Corp., Fremont, Calif., USA) were performed as described previously [24, 25]. Scoring of TMAs utilized a four tier classification (0, 1, 2 and 3) where less than 5% of labeled epithelial cells was scored as ‘0’, 5–10% of epithelium labeling was scored as ‘1’, 10–30% of epithelium labeling was scored as ‘2’, and >30% of cells labeling was scored as ‘3’.
Results
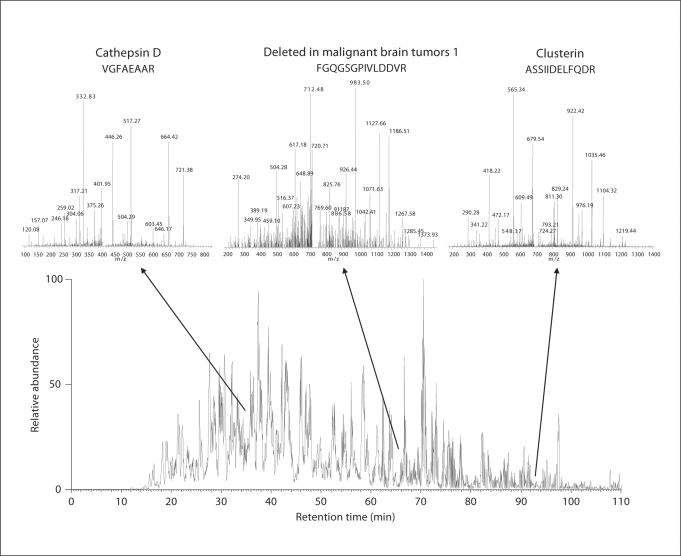
Proteomic analysis of the IPMN Liquid Tissue lysate using nanoLC-MS/MS yielded a total of 1,534 peptides, corresponding to 869 total proteins and 523 unique proteins, which are presented in their entirety in online suppl. 1 (www.karger.com/doi/10.1159/000161012). Further, 95 proteins (∼18%) were identified by two or more unique tryptic peptides. In online supplement 2, we have provided the SEQUEST parameters (Xcorr, ΔCn, Sp, RSp, and the coverage of y- and b-ions), which were determined in order to reduce the numbers of false positives, and increase the proportion of true positives. The calculated false-positive rate was 3.26%. Representative tandem mass spectra, including peptides corresponding to three of the proteins identified as expressed in IPMN (cathepsin D, DMBT1, and clusterin), are shown in figure 2. Additional annotation for each unique protein is provided in online supplement 1, including NCBI Entrez ID, unigene cluster for the corresponding transcript, cytogenetic location, and gene ontology (GO) classification by molecular function, process, and component. Summarized graphically in online supplement figure 1 is the GO molecular function, demonstrating that the top categories of functional proteins identified include those involved in structural and catalytic activity, and nucleic acid, DNA and protein binding.
Table 1.
Subset of pancreatic cancer-associated proteins identified by Liquid Tissue technology in IPMN
| Protein | Gene symbol | Unigene ID | Number of peptides | References |
|---|---|---|---|---|
| Annexin A1 | ANXA1 | Hs.494173 | 2 | [25, 26, 27, 31, 33] |
| Annexin A2 | ANXA2 | Hs.511605 | 1 | [32, 33] |
| Caldesmon | CALD1 | Hs.490203 | 4 | [33] |
| Cathepsin D | CTSD | Hs.654447 | 2 | [32] |
| Cytokeratin-7 | KRT7 | Hs.670221 | 4 | [25, 26, 28] |
| Cytokeratin-8 | KRT8 | Hs.533782 | 8 | [27] |
| Cytokeratin-19 | KRT19 | Hs.654568 | 4 | [25, 27, 28] |
| Cofilin | CFL1 | Hs.170622 | 2 | [32] |
| Collagen type I, alpha 1 | COL1A1 | Hs.172928 | 32 | [26, 30] |
| Collagen type I, alpha 2 | COL1A2 | Hs.489142 | 19 | [26, 28] |
| Collagen type IV, alpha 3 | COL4A3 | Hs.570065 | 3 | [32] |
| Deleted in malignant brain tumors 1 | DMBT1 | Hs.279611 | 1 | [24] |
| Fibrillin 1 | FBN1 | Hs.591133 | 4 | [32] |
| Fibrinogen gamma chain | FGG | Hs.546255 | 1 | [32] |
| Gelsolin | GSN | Hs.522373 | 5 | [32] |
| Lumican | LUM | Hs.406475 | 5 | [32] |
| Peroxiredoxin 1 | PRDX1 | Hs.180909 | 1 | [27] |
| Phosphoglycerate kinase 1 | PGK1 | Hs.654578 | 5 | [27] |
| Plectin 1 | PLEC1 | Hs.434248 | 3 | [25, 32] |
| Rho GDP Dissociation Inhibitor alpha | ARHGDIA | Hs.159161 | 2 | [31] |
| Transglutaminase 2 | TGM2 | Hs.517033 | 4 | [25, 26, 27, 29, 32] |
| Transgelin 2 | TAGLN2 | Hs.517168 | 1 | [32] |
| Tropomysin 4 | TPM4 | Hs.631618 | 1 | [27] |
| Tubulin alpha-1 chain | TUBA1A | Hs.654422 | 4 | [27] |
| Vimentin | VIM | Hs.642813 | 21 | [29] |
Table 2.
Immunohistochemical analysis of DMBT1 and TGM2 in IPMN TMAs
| Antigen labeling | Percent positive |
|---|---|
| DMBT1 | |
| 0 | 7/38 (18%) |
| 1+ | 16/38 (42%) |
| 2+ | 6/38 (16%) |
| 3+ | 9/38 (24%) |
| TGM2 | |
| 0 | 13/33 (40%) |
| 1+ | 8/33 (24%) |
| 2+ | 7/33 (21%) |
| 3+ | 5/33 (15%) |
Five TMA cores dropped out and were therefore not evaluable in the TGM2 series. DMBT1 labeling is restricted essentially to the IPMN epithelium, while TGM2 labeling is observed in both the neoplastic epithelium and the juxta-tumoral stroma.
Fig. 2.
Representative tandem mass spectra of peptides identified in the IPMN Liquid Tissue lysate preparation. Inset spectra illustrate three identified peptides corresponding to cathepsin D, DMBT1, and clusterin.
It is likely that the vast majority of proteins whose peptides were identified by LC-MS/MS in the IPMN Liquid Tissue lysate are constitutively expressed ‘native’ proteins in the pancreas (including those expressed in the contaminating stromal component). Upon manual curation, however, the list of expressed proteins in the noninvasive IPMN included several whose mRNA transcripts are overexpressed in invasive pancreatic cancer by global mRNA expression profiling technologies (SAGE, cDNA and oligonucleotide microarrays) [24,25,26,27,28,29,30,31]. Comparison with quantitative proteomic studies of invasive pancreatic cancer [32, 33] also revealed additional examples of upregulated proteins that were detected in the IPMN. This ‘enriched’ subset of 25 previously identified cancer-associated proteins in IPMN (table 1) represents a source of markers with aberrant expression at the earliest stages of pancreatic neoplasia. For further validation in a larger series of IPMN TMAs we selected two proteins, DMBT1 and TGM2, which have – to the best of our knowledge – so far never been reported to be overexpressed in IPMNs. However, we and others have previously described their upregulation in invasive pancreatic cancers [24, 25,34,35,36].
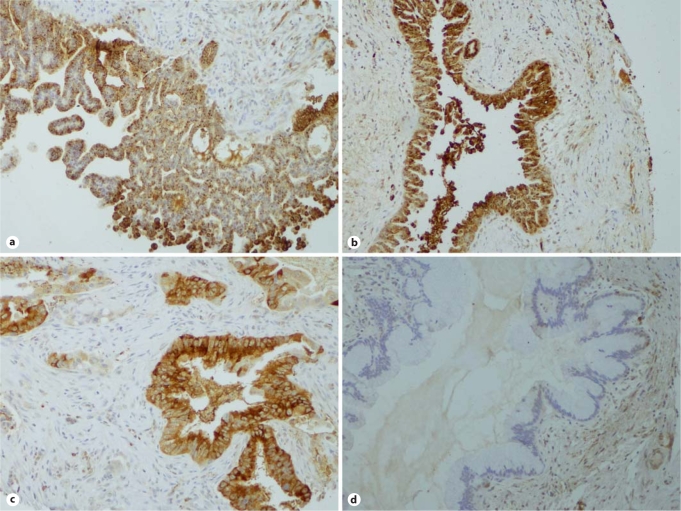
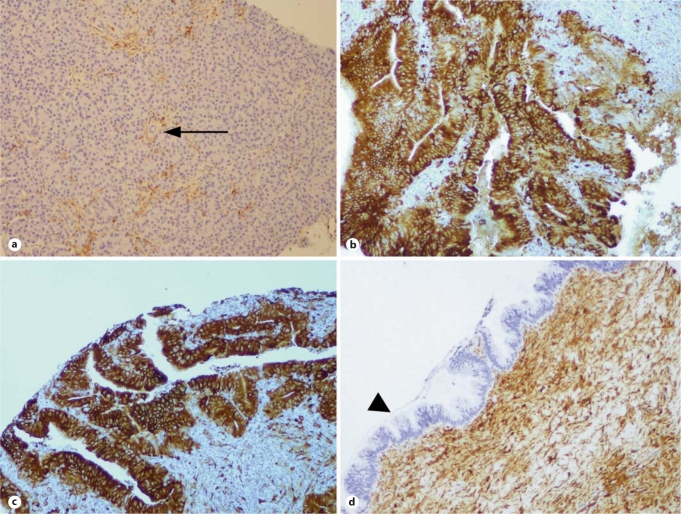
Immunohistochemical analysis confirmed the overexpression of both DMBT1 and TGM2 in IPMN tissues (fig. 3, 4). Specifically, 2–3+ staining was observed in 15 of 38 (39%) IPMNs labeled with anti-DMBT1 antibody, and in 12 of 33 (36%) cases labeled with anti-TGM2 (five IPMN cores stained with anti-TGM2 were not able to be evaluated) (table 2). While DMBT1 was primarily expressed in the IPMN epithelium (fig. 3), TGM2 expression was also seen in the juxta-tumoral stroma, including in occasional cases where the epithelium itself did not express TGM2 (fig. 4). In contrast, minimal or no DMBT1 or TGM2 expression was observed in the non-neoplastic pancreas, including sections of normal intralobular ducts when these were available for evaluation. Although the numbers of cases are limited, we did not observe any significant correlation between aberrant protein expression and histologic grade of the IPMN epithelium (adenoma, moderate dysplasia, or carcinoma in situ) [19, 37].
Fig. 3.
DMBT1 labeling in IPMN tissue microarrays. a, b Two examples of noninvasive IPMNs with intense DMBT1 expression in the neoplastic epithelium. c A representative example of a DMBT1-expressing invasive adenocarcinoma arising in an IPMN. Notice the absence of labeling in the stroma. d Example of IPMN with no expression of DMBT1 protein.
Fig. 4.
TGM2 labeling in IPMN TMAs. a Absence of TGM2 labeling in non-neoplastic pancreatic acini and in small intra-lobular ducts (arrow); labeling is observed only in endothelial cells. b, c Two examples of noninvasive IPMNs with intense TGM2 expression in the neoplastic epithelium. Unlike DMBT1, TGM2 expression is often observed in the juxta-tumoral stroma (d), even in the absence of demonstrable labeling in the IPMN epithelium (arrowhead).
Discussion
Few public health measures have had as much success in ameliorating cancer mortality as early detection at an operable, and hence potentially curable, stage [38, 39]. The National Cancer Institute recognizes this and has therefore invested considerable efforts in early detection strategies [40, 41]. In order to implement effective early diagnosis of cancer, two prerequisite tenets need to be met. First, there must be a recognizable precursor stage before invasive cancer develops which provides a window of opportunity for preventative intervention. This criterion has now been met for most common cancers [42,43,44,45,46], including pancreatic cancer, where tangible noninvasive precursor lesions are recognized, and for which standardized diagnostic criteria have been established [2, 37]. The second requirement is the identification of sensitive and specific surrogate biomarkers that can provide ancillary information to routine clinical assays for diagnosing the presence of high-risk precursor lesions [41, 47]. The latter remains a work in evolution. For example, in the case of pancreatic cancer, the most commonly utilized serological marker – CA19-9 – has suboptimal performance in the diagnosis of early pancreatic neoplasia, and is probably best utilized in monitoring disease recurrence in patients who have already received therapy [48, 49].
Large-scale proteomic approaches have provided a major impetus to the search for new biomarkers in cancers and in their precursor lesions [8, 50]. One of the major limitations of proteomic applications in clinical samples has been the requirement for high-quality snap-frozen or live tissue specimens [51], essentially excluding the repositories of fixed, archival paraffin-embedded tissues available in most institutions. Further, the current approach to biomarker discovery in precursor lesions involves the retrospective analysis of tumor antigens which are first identified in invasive cancers (‘reverse proteomics’) [52]. An unbiased ‘forward’ proteomics analysis of precursor lesions might directly elucidate biomarkers that are aberrantly expressed at the earliest stages of multistep cancer progression. The Liquid Tissue platform has previously been applied to the discovery of differentially expressed proteins in formalin-fixed paraffin embedded tissues [16,17,18]. In this study, we have applied the Liquid Tissue platform to elucidate the proteome of a noninvasive precursor lesion of pancreatic cancer (IPMN). We have confirmed the upregulation of two of the expressed proteins (DMBT1 and TGM2) in IPMNs versus normal pancreatic tissues by immunohistochemistry, underscoring the validity of our approach for tumor marker discovery in precursor lesions. Although our proof-of-principle analysis in IPMNs was not quantitative in nature, the Liquid Tissue platform has been shown to be amenable to quantitative proteomic analyses (e.g. stable isotope labeling) for biomarker discovery in prostate cancer [17]. Nevertheless, by simple manual curation, we were able to focus on those proteins most likely to be aberrantly expressed in IPMNs compared to normal pancreas based on their known association with the invasive pancreatic cancer proteome (table 1). One of the validated proteins, DMBT1, belongs to the superfamily of scavenger receptor cysteine-rich proteins and is a secreted glycoprotein [53]. Loss of DMBT1 expression was first recognized in brain tumors, but overexpression of this protein has now been documented in pancreatic cancers [24, 34]. Hence, our findings presented here, demonstrating for the first time overexpression of DMBT1 in IPMNs, are in line with these previous reports and suggest that upregulation of DMBT1 is a comparably early event in pancreatic carcinogenesis. TGM2 plays a critical role in catalyzing the calcium-dependent transamidation of polypeptides, but is unique in that it also has protein isomerase and kinase activities [54]. Several lines of evidence support a role for TGM2 in cancer biology [55], with distinct and potentially contrasting effects of expression in neoplastic cells [56] and in the juxta-tumoral stroma [57]. We and others have established TGM2 overexpression as a phenotype of invasive pancreatic cancer [25, 36], and this study extends this finding to IPMNs as well.
In conclusion: In the present study, we show that our recently described technology for protein extraction from archival tissue sample material is amenable to mass spectrometric analysis of tryptic peptides for analyzing the proteome of noninvasive pancreatic cancer precursor lesions. We hope that application of the Liquid Tissue platform to proteomic analysis of archival formalin-fixed tissues will prove to be a valuable additional tool for biomarker discovery in the future.
Acknowledgements
The Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation, and the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer P50CA62924. H.A. received generous support from the D’Amato Family. G.F. was supported by a fellowship grant within the postdoctoral program of the German Academic Exchange Service (DAAD). J.M. received support from the Future Award in Health Sciences 2005 of the HGF.
Marlene M. Darfler and David B. Krizman are full-time employees of Expression Pathology Incorporated, Gaithersburg, Md. Other than that, the authors declare no conflict of interest.
References
- 1.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 3.Singh M, Maitra A. Precursor lesions of pancreatic cancer: molecular pathology and clinical implications. Pancreatology. 2007;7:9–19. doi: 10.1159/000101873. [DOI] [PubMed] [Google Scholar]
- 4.Sohn TA, Yeo CJ, Cameron JL, Hruban RH, Fukushima N, Campbell KA, Lillemoe KD. Intraductal papillary mucinous neoplasms of the pancreas: an updated experience. Ann Surg. 2004;239:788–797. doi: 10.1097/01.sla.0000128306.90650.aa. discussion 797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilentz RE, Albores-Saavedra J, Zahurak M, Talamini MA, Yeo CJ, Cameron JL, Hruban RH. Pathologic examination accurately predicts prognosis in mucinous cystic neoplasms of the pancreas. Am J Surg Pathol. 1999;23:1320–1327. doi: 10.1097/00000478-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Goggins M. Molecular markers of early pancreatic cancer. J Clin Oncol. 2005;23:4524–4531. doi: 10.1200/JCO.2005.19.711. [DOI] [PubMed] [Google Scholar]
- 7.Srinivas PR, Srivastava S, Hanash S, Wright GL., Jr Proteomics in early detection of cancer. Clin Chem. 2001;47:1901–1911. [PubMed] [Google Scholar]
- 8.Verma M, Wright GL, Jr, Hanash SM, Gopal-Srivastava R, Srivastava S. Proteomic approaches within the NCI early detection research network for the discovery and identification of cancer biomarkers. Ann NY Acad Sci. 2001;945:103–115. doi: 10.1111/j.1749-6632.2001.tb03870.x. [DOI] [PubMed] [Google Scholar]
- 9.Lohr M, Faissner R. Proteomics in pancreatic disease. Pancreatology. 2004;4:67–75. doi: 10.1159/000077212. [DOI] [PubMed] [Google Scholar]
- 10.Papachristou GI, Malehorn DE, Lamb J, Slivka A, Bigbee WL, Whitcomb DC. Serum proteomic patterns as a predictor of severity in acute pancreatitis. Pancreatology. 2007;7:317–324. doi: 10.1159/000105497. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, Yang L, Wang W, Shi SR, Liu C, Liu Y, Fang X, Taylor CR, Lee CS, Balgley BM. Antigen retrieval for proteomic characterization of formalin-fixed and paraffin-embedded tissues. J Proteome Res. 2008;7:1098–1108. doi: 10.1021/pr7006768. [DOI] [PubMed] [Google Scholar]
- 12.Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J Proteome Res. 2008;7:969–978. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 13.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 14.Bagnato C, Thumar J, Mayya V, Hwang SI, Zebroski H, Claffey KP, Haudenschild C, Eng JK, Lundgren DH, Han DK. Proteomics analysis of human coronary atherosclerotic plaque: a feasibility study of direct tissue proteomics by liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:1088–1102. doi: 10.1074/mcp.M600259-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, Fernald GH, Gerlitz B, Robinson WH, Baranzini SE, Grinnell BW, Raine CS, Sobel RA, Han DK, Steinman L. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 2008;451:1076–1081. doi: 10.1038/nature06559. [DOI] [PubMed] [Google Scholar]
- 16.Prieto DA, Hood BL, Darfler MM, Guiel TG, Lucas DA, Conrads TP, Veenstra TD, Krizman DB. Liquid Tissue: proteomic profiling of formalin-fixed tissues. Biotechniques. 2005;(suppl):32–35. doi: 10.2144/05386su06. [DOI] [PubMed] [Google Scholar]
- 17.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14:1002–1014. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa T, Klöppel G, Volkan Adsay N, Albores-Saavedra J, Fukushima N, Horii A, Hruban RH, Kato Y, Klimstra DS, Longnecker DS, Lüttges J, Offerhaus GJ, Shimizu M, Sunamura M, Suriawinata A, Takaori K, Yonezawa S. Classification of types of intraductal papillary-mucinous neoplasm of the pancreas: a consensus study. Virchows Arch. 2005;447:794–799. doi: 10.1007/s00428-005-0039-7. [DOI] [PubMed] [Google Scholar]
- 20.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 22.Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue CA, van Heek NT, Cameron JL, Yeo CJ, Hruban RH, Goggins M. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004;164:903–914. doi: 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hustinx SR, Hruban RH, Leoni LM, Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Brown PN, Argani P, Asfaq R, Fukushima N, Goggins M, Kern SE, Maitra A. Homozygous deletion of the mtap gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol Ther. 2005;4:83–86. doi: 10.4161/cbt.4.1.1380. [DOI] [PubMed] [Google Scholar]
- 24.Hustinx SR, Cao D, Maitra A, Sato N, Martin ST, Sudhir D, Iacobuzio-Donahue C, Cameron JL, Yeo CJ, Kern SE, Goggins M, Mollenhauer J, Pandey A, Hruban RH. Differentially expressed genes in pancreatic ductal adenocarcinomas identified through serial analysis of gene expression. Cancer Biol Ther. 2004;3:1254–1261. doi: 10.4161/cbt.3.12.1238. [DOI] [PubMed] [Google Scholar]
- 25.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res. 2003;63:8614–8622. [PubMed] [Google Scholar]
- 26.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, Jaffee E, Ryu B, Jones J, Eshleman JR, Yeo CJ, Cameron JL, Kern SE, Hruban RH, Brown PO, Goggins M. Exploration of global gene expression patterns in pancreatic adenocarcinoma using CDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 29.Missiaglia E, Blaveri E, Terris B, Wang YH, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, Lemoine NR. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer. 2004;112:100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 30.Ryu B, Jones J, Blades NJ, Parmigiani G, Hollingsworth MA, Hruban RH, Kern SE. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- 31.Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 32.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, Aebersold R, Brentnall TA. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, Costello E, Neoptolemos J, Lemoine NR. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki K, Sato K, Akiyama Y, Yanagihara K, Oka M, Yamaguchi K. Peptidomics-based approach reveals the secretion of the 29-residue COOH-terminal fragment of the putative tumor suppressor protein DMBT1 from pancreatic adenocarcinoma cell lines. Cancer Res. 2002;62:4894–4898. [PubMed] [Google Scholar]
- 35.Gr⊘nborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 36.Verma A, Wang H, Manavathi B, Fok JY, Mann AP, Kumar R, Mehta K. Increased expression of tissue transglutaminase in pancreatic ductal adenocarcinoma and its implications in drug resistance and metastasis. Cancer Res. 2006;66:10525–10533. doi: 10.1158/0008-5472.CAN-06-2387. [DOI] [PubMed] [Google Scholar]
- 37.Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, Biankin SA, Compton C, Fukushima N, Furukawa T, Goggins M, Kato Y, Klöppel G, Longnecker DS, Lüttges J, Maitra A, Offerhaus GJ, Shimizu M, Yonezawa S. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 38.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ. Cancer Intervention and Surveillance Modeling Network (CISNET) Collaborators: Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 39.Smith RA, Cokkinides V, Eyre HJ. American Cancer Society guidelines for the early detection of cancer, 2006. CA Cancer J Clin. 2006;56:11–25. doi: 10.3322/canjclin.56.1.11. quiz 49–50. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava S, Kramer BS. Early detection cancer research network. Lab Invest. 2000;80:1147–1148. doi: 10.1038/labinvest.3780122. [DOI] [PubMed] [Google Scholar]
- 41.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 42.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 43.Shaheen N, Ransohoff DF. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: Scientific review. JAMA. 2002;287:1972–1981. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 44.Brewer MA, Johnson K, Follen M, Gershenson D, Bast R., Jr Prevention of ovarian cancer: intraepithelial neoplasia. Clin Cancer Res. 2003;9:20–30. [PubMed] [Google Scholar]
- 45.Wistuba II. Genetics of preneoplasia: lessons from lung cancer. Curr Mol Med. 2007;7:3–14. doi: 10.2174/156652407779940468. [DOI] [PubMed] [Google Scholar]
- 46.Bostwick DG. Prostatic intraepithelial neoplasia. Curr Urol Rep. 2000;1:65–70. doi: 10.1007/s11934-000-0037-x. [DOI] [PubMed] [Google Scholar]
- 47.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 48.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC., Jr ASCO, ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 49.Rosty C, Goggins M. Early detection of pancreatic carcinoma. Hematol Oncol Clin North Am. 2002;16:37–52. doi: 10.1016/s0889-8588(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 50.Negm RS, Verma M, Srivastava S. The promise of biomarkers in cancer screening and detection. Trends Mol Med. 2002;8:288–293. doi: 10.1016/s1471-4914(02)02353-5. [DOI] [PubMed] [Google Scholar]
- 51.Wulfkuhle JD, Sgroi DC, Krutzsch H, McLean K, McGarvey K, Knowlton M, Chen S, Shu H, Sahin A, Kurek R, Wallwiener D, Merino MJ, Petricoin EF, 3rd, Zhao Y, Steeg PS. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–6749. [PubMed] [Google Scholar]
- 52.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 53.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, von Deimling A, Poustka A. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3–26.1 is deleted in malignant brain tumours. Nat Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 54.Facchiano F, Facchiano A, Facchiano AM. The role of transglutaminase-2 and its substrates in human diseases. Front Biosci. 2006;11:1758–1773. doi: 10.2741/1921. [DOI] [PubMed] [Google Scholar]
- 55.Mangala LS, Mehta K. Tissue transglutaminase (TG2) in cancer biology. Prog Exp Tumor Res. 2005;38:125–138. doi: 10.1159/000084237. [DOI] [PubMed] [Google Scholar]
- 56.Verma A, Mehta K. Tissue transglutaminase-mediated chemoresistance in cancer cells. Drug Resist Updat. 2007;10:144–151. doi: 10.1016/j.drup.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Xu L, Hynes RO. GPR56 and TG2: possible roles in suppression of tumor growth by the microenvironment. Cell Cycle. 2007;6:160–165. doi: 10.4161/cc.6.2.3760. [DOI] [PubMed] [Google Scholar]