Abstract
To determine whether oxidative stress after cerebral ischemia–reperfusion affects genetic stability in the brain, we studied mutagenesis after forebrain ischemia–reperfusion in Big Blue transgenic mice (male C57BL/6 strain) containing a reporterlacI gene, which allows detection of mutation frequency. The frequency of mutation in this reporter lacI gene increased from 1.5 to 7.7 (per 100,000) in cortical DNA after 30 min of forebrain ischemia and 8 hr of reperfusion and remained elevated at 24 hr reperfusion. Eight DNA lesions that are characteristic of DNA damage mediated by free radicals were detected. Four mutagenic lesions (2,6-diamino-4-hydroxy-5-formamidopyrimidine, 8-hydroxyadenine, 5-hydroxycytosine, and 8-hydroxyguanine) examined by gas chromatography/mass spectrometry and one corresponding 8-hydroxy-2′-deoxyguanosine by a method of HPLC with electrochemical detection increased in cortical DNA two- to fourfold (p < 0.05) during 10–20 min of reperfusion. The damage to γ-actin and DNA polymerase-β genes was detected within 20 min of reperfusion based on the presence of formamidopyrimidine DNA N-glycosylase-sensitive sites. These genes became resistant to the glycosylase within 4–6 hr of reperfusion, suggesting a reduction in DNA damage and presence of DNA repair in nuclear genes. These results suggest that nuclear genes could be targets of free radicals.
Keywords: apoptosis, brain, mutation, dementia, oxidative stress, stroke
Ischemic neuronal injuries may be initiated by a number of mediators. Among these are reactive oxygen species (ROS), calcium ions, and extracellular glutamate (Beauchamp and Fridovich, 1970; Chance et al., 1979; Halliwell and Gutteridge, 1984; Dugan and Choi, 1994). A majority of ROS (superoxide ions, hydroxyl radicals, and others) are products of aerobic and arachidonic acid metabolism, lipid peroxidation, or activated phagocytes. Mutagenesis initiated by ROS, especially the hydroxyl radical, has been reported in bacterial and mammalian cell culture systems (Kasai et al., 1986; Kuchino et al., 1987; Moraes et al., 1990; Wood et al., 1990; Akman et al., 1991;McBride et al., 1991; Shibutani et al., 1991; Tkeshelashvili et al., 1991; Wink et al., 1991; Nguyen et al., 1992; Marragos et al., 1993;Feig et al., 1994; Schmutte et al., 1994). Interactions between DNA and the hydroxyl radical produce DNA strand breaks and base modifications, which are frequently assessed by measurement of the nucleoside 8-hydroxy-2′-deoxyguanosine (oh8-dG) (Fraga et al., 1990) corresponding to the base lesion 8-hydroxyguanine (8-OH-Gua). In addition to 8-OH-Gua, at least seven other base lesions have been reported in human cells after ionizing radiation (Dizdaroglu, 1992) or exposure to hydrogen peroxide (Jaruga and Dizdaroglu, 1996): 8-hydroxyadenine (8-OH-Ade), 4,6-diamino-5-formamidopyrimidine (FapyAde), 2-hydroxyadenine (2-OH-Ade), 5-hydroxycytosine (5-OH-Cyt), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua), 5-hydroxy-5-methylhydantoin (5-OH-5-MeHyd), and 5-hydroxyhydantoin (5-OH-Hyd). Both the extent of deletion (Corral-Debrinski et al., 1992;Cortopassi et al., 1992; Zhang et al., 1992) and the number of oh8-dG lesions in mtDNA of human brains increase with age (Mecocci et al., 1993). Together, these findings raise the possibility that an increase in oh8-dG content may be linked to mtDNA deletion in humans (Hayakawa et al., 1992). Positive correlations between germline mutation and familial forms of premature neurodegeneration have been reported (Goate et al., 1991; Schellenberg et al., 1992; Levy-Lahad et al., 1995a,1995b; Sherrington et al., 1995). The etiology of sporadic cases of premature neurodegeneration at the molecular level is not understood totally. An association between premature neurodegenerative disorders and traumatic head injuries raises the possibility that ROS generated during oxidative stress may damage nuclear DNA, initiate somatic mutation (Martin, 1991), and accelerate aging of the brain in a manner similar to that found in familial cases.
Transient cerebral ischemia–reperfusion affects the oxygen supply and perturbs energy metabolism (oxidative stress). Stroke is associated with an increase in brain ROS content (Liu et al., 1989; Kinouchi et al., 1991; Sakamoto et al., 1991; Althaus et al., 1993; Barone et al., 1993; Malinski et al., 1993; Sen and Phillis, 1993; Dugan et al., 1995;Suzuki et al., 1995). Several investigators have clearly demonstrated ROS-mediated toxicity that affects virtually all cellular components in the in vivo model of cerebral ischemia–reperfusion (Imlay and Linn, 1988; Moraes et al., 1989; Floyd and Carney, 1992; Grammas et al., 1993; Andrus et al., 1994; Salminen et al., 1995), with one notable exception: DNA damage by cerebral ischemia–reperfusionin vivo has not yet been explored. The present study was undertaken to determine whether nuclear DNA can be damaged after forebrain ischemia–reperfusion and whether the repair process adequately maintains the genomic stability.
MATERIALS AND METHODS
Materials. All chemicals of molecular biology grade were obtained from Sigma (St. Louis, MO) unless specified otherwise. Radioactive nucleotides [35S-dATP (1500 Ci/mmol),32P-dATP (3000 Ci/mmol)] and GenScreen Plus membranes for DNA transfer were obtained from DuPont NEN (Wilmington, DE).Escherichia coli formamidopyrimidine DNAN-glycosylase (Fpg protein), modified via insertion of its coding sequence into the Nde/XhoI sites of the pET22b (Novagen, San Diego, CA), contained a six amino acid C-terminal addition including six terminal histidines. No difference was detected in the activity between the original and the modified protein (L. E. Rabow and Y. W. Kow, unpublished observation). The protein was purified to >95% purity using a Ni2+ affinity column and procedures similar to those described by Boiteux et al. (1990).Taq DNA polymerase (5 U/μl) was obtained from Perkin-Elmer Cetus (Norwalk, CT). Human placenta RNasin, EcoRI restriction endonucleases, T7 RNA polymerase, and agarose were obtained from Promega (Madison, WI). Silica gel–resin columns and proteinase K were obtained from Qiagen (Chatsworth, CA). Acrylamide and bis-acrylamide were obtained from Boehringer Mannheim Corporation (Indianapolis, IN). Sequenase II for DNA sequencing was from Amersham Life Science/United States Biochemicals (Cleveland, OH). The ApopTaq kit for in situ apoptosis detection using terminal transferase dUTP nick-end labeling (TUNEL) assay was obtained from Oncor (Gaithersburg, MD). Ketaset (ketamine HCl) and xylazine were obtained from Fort Dodge Laboratories (Fort Dodge, IA) and Lloyd Laboratories (Shenandoah, IA), respectively. Transpack for packaging shuttle vector, RNase cocktail (RNace-it), SCS-8 strain of E. coli, primers for lacI DNA amplification, and 5-bromo-4-chloro-3-indol-β-galactopyranoside (X-gal) were obtained from Stratagene (La Jolla, CA). RNAzol B was obtained from Biotecx Laboratories (Houston, TX). Random hexamer, SuperScript II reverse transcriptase (RT-RNase H−), and DNA size markers were from Life Technologies (Gaithersburg, MD). DNA primers were custom-synthesized from Ransom Hill Bioscience (Ramona, CA). Single-stranded 32P-cDNA probe for the Southern blot hybridization was synthesized from the cloned partial cDNA using the downstream primer (see below) and Prime-A-Probe kit (Ambion, Austin, TX).
A mouse model of forebrain ischemia–reperfusion. Male Big Blue transgenic C57BL/6 mice by Stratagene (6–10 weeks of age) and nontransgenic litter mates of matching strain, sex, and age were obtained from Taconic Farms (Germantown, NY). Mice were anesthetized with ketamine (100 mg/kg, i.p.) plus xylazine (10 mg/kg, i.p.). A midline ventral incision was made in the neck. Both common carotid arteries (CCAs) were isolated, freed of nerve fibers, and occluded using nontraumatic aneurysm clips. Complete interruption of blood flow was confirmed under an operating microscope. After 30 min of ischemia, the aneurysm clips were removed from both CCAs. Restoration of blood flow (reperfusion) was observed directly under the microscope. Sham-operated controls were subjected to the same surgical procedures except that CCAs were not occluded. All procedures have been approved by the institutional animal welfare committee in accordance with the Public Health Service Guide for the Care and Use of Laboratory Animals. The body temperature was monitored and maintained at 37°C ± 0.5°C during surgery and during the immediate postoperative period until the animals recovered fully from anesthesia. The mortality rate in this mouse model was 12%, and all of the deaths occurred in animals with reperfusion of 16–48 hr. The mortality rate was similar to that reported in mice by Barone et al. (1993). At the designated reperfusion time points, animals were killed for collection of brain samples.
Brain sample collection. At the end of reperfusion, the animals were decapitated under anesthesia, and the brain was removed quickly. The forebrain was immediately separated from the cerebellum and brain stem. The cerebral cortex was sampled by removing the subcortical structures. Each sample was frozen quickly in liquid nitrogen and then transferred to a freezer (−70°C) for storage.
Mutation analysis. Mutations generally occur in 1/105 cells. To facilitate the detection of this low-frequency effect, we adapted a mutation system using Big Blue transgenic mice carrying the E. coli lacI gene in shuttle vectors integrated in chromosome 4. The lacI gene codes a repressor protein. This protein forms a tetramer and binds thelac operator to prevent the transcription of theα-lacZ gene (N terminus of galactosidase). After being packaged in Transpack, transduced to the host SCS-8 strain of E. coli, and plated in agar plates with X-gal, the expression of nonmutated lacI genes in bacteriophage suppresses the expression of a functional galactosidase, and X-gal in the agar medium is not used as a substrate. The plaque is colorless. When mutated, the suppressor protein is incapable of inhibition; therefore, the host bacteria produce a functional galactosidase: the host with mutantlacI vector produces blue plaques. The number of blue plaques compared with the total number of plaques is the mutant frequency. Mutations that are detected in the lacI gene in the cell nucleus are somatic mutants.
Isolation of the reporter gene lacI. Each cerebral cortex was homogenized in 3 ml of ice-cold STE buffer (500 mm NaCl, 10 mm Tris HCl, 1 mm EDTA, pH 8.0, RNase-it 20 μl/ml) using a Dounce homogenizer. High molecular weight DNA from each animal was purified by adding 3 ml of proteinase K (2 mg/ml proteinase K, 2% SDS, 0.1 m EDTA, pH 7.5) and incubated at 50°C for 3 hr. The homogenates were extracted in equilibrated phenol/chloroform (1:1, pH 8.0) and then in a mixture of phenol/chloroform/isoamyl alcohol (50:48:2, v/v), as described by Liu et al. (1993). The DNA was precipitated by a gradual addition of 100% ethanol (12 ml). The DNA threads were separated from ethanol by using a fire-polished Pasteur pipette. The DNA was dislodged into 200 μl of 70% ethanol in a microfuge vial and centrifuged (1000 × g, 15 min, room temperature), and the ethanol was removed. The DNA pellet was air-dried briefly and dissolved in 500 μl TE buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.0). DNA concentrations were measured using a spectrophotometer (model DU-64) with the Nucleic Acid Soft-pac module (Beckman Instruments, Fullerton, CA) for calculations of the optical density ratio at 260 and 280 (OD260/280) for each sample. The final DNA concentration was adjusted to 500 μg/ml. DNA from three or more mice per reperfusion time point were mixed as a pool. The λ shuttle vector was recovered using 4 μg of pooled DNA per packaging extract (Transpack) and was followed by transduction into the SCS-8 strain of E. coli. The transduced E. coli was plated in NZY agar and incubated at 37°C for 16 hr to allow plaque formation. The density of plaques per plate averaged 50 plaque-forming units (pfu) per cm2. We obtained a packaging efficiency of 2–4 × 106 pfu/mg genomic DNA. More than one packaging extract was used to recover at least 130,000 colorless plaques per expression time. The mutants of lacI were isolated based on the formation of blue plaques in NZY agar plates supplemented with X-gal at 1.5 mg/ml. Each blue plaque was retested to confirm its ability to produce only blue plaques. The mutants were stored for further verification of the molecular bases of mutation. Mutant lacI genes (1.08 kb) were isolated as described by Kohler et al. (1991). The mutant frequency was calculated by the sum of blue plaques divided by the sum of total plaques for each time point from all packaging experiments.
Single-stranded conformation polymorphism (SSCP) of mutant RNA transcripts (SSCP-RNA analysis). The lacI DNA was amplified using seven sets of PCR primers in seven short DNA fragments of 200–300 bp each (Danenberg et al., 1992). The RNA transcript of each short DNA fragment was obtained for analysis using T7 RNA polymerase and was resolved in 8% nondenaturing polyacrylamide gel by electrophoresis (1500 volt-hr) at 4°C. The primer set for the RNA transcript, which is presented in this report, is 5′-T7 promoter plus tgtaaagcggcggtgcac-3′ and 5′-agaacttaatgggcccgc-3′. The mutation in the fragment was confirmed by direct DNA sequencing using Sequenase II (Liu, 1993).
DNA lesions after forebrain ischemia–reperfusion. DNA from control and ischemic animals was isolated using the same batch of buffers. Cerebral cortex of each brain was homogenized in 9.5 ml of G2 buffer (800 mm guanidine hydrochloride, 30 mm EDTA, 30 mm Tris-HCl, 5% Tween 20, 0.5% Triton X-100, pH 8.0, nuclease-free RNase 200 μg/ml) on ice according to the Qiagen genomic DNA purification procedure (Qiagen). Briefly, proteinase K was added immediately to individual homogenates in the same buffer (final concentration 1 mg/ml) and incubated for 4 hr at 40°C. DNA of each homogenate was loaded onto a modified silica gel–resin column previously equilibrated in QBT buffer (750 mm NaCl, 50 mm MOPS, 15% ethanol, 0.15% Triton X-100, pH 7.0). After two washes using QC buffer (1 m NaCl, 50 mm MOPS, 15% ethanol, pH 7.0), the DNA fraction was eluted using QF buffer (1.25 m NaCl, 50 mm Tris-HCl, 15% ethanol, pH 8.5). The purified DNA fraction was precipitated by the addition of 0.7× vol of isopropanol. The DNA threads were recovered (see above), washed, and air-dried as before. The DNA pellet was resuspended in TE buffer. DNA was never exposed to phenol or chloroform. The amount of mtDNA in the total DNA was estimated to be ≤1%. The OD260/280 was 1.74 ± 0.05 (mean ± SEM, n = 46). Aliquots of DNA (40 μg) from each animal were labeled with a code and shipped in a double-blinded fashion as dried pellets to Oklahoma City, OK, or in TE buffer to Gaithersburg, MD, for determination of DNA damage using HPLC with electrochemical detection (HPLC–EC) or gas chromatograph/mass spectrometry with the selected ion monitoring (GC/MS-SIM), respectively. The conversion of 1 nmol of a modified base per milligram of DNA corresponds to ∼32 modified bases per 105 bases. The first set of 10 animals was employed in a pilot study to optimize the chromatographic systems for DNA samples from animals. In addition, nine animals were used to determine the effect of centrifugation during sample preparation. Once the conditions were selected, 27 animals were used in the subsequent studies. More than three animals (n ≥ 3) were used in each time point or treatment. Unless specified otherwise, data were analyzed using one-way ANOVA followed by Tukey test (DataMost, Salt Lake City, UT) and plotted using the GraphPad Prism Intuitive Software for Science program (GraphPad Software, San Diego, CA).
Gene probe cloning using RNA isolation and reverse transcription (RT). To obtain mouse DNA probe for the detection of damage to nuclear genes (see below), we isolated genes from mouse brain using RT-PCR as described in Liu et al. (1993). Briefly, total RNA was isolated by RNAzol B, and total RNA (5 μg) from cortex was reverse-transcribed to cDNA as described (Liu, 1993). The reaction was carried out at 35°C for 1 hr using 200 U of SuperScript II reverse transcriptase in a volume of 100 μl containing 1 μmol primer dT15, 20 U RNasin, 75 nmol dNTP, 200 nmol dithiothreitol (DTT), and the manufacturer’s buffer. The resulting cDNA was extracted with phenol/chloroform, precipitated using 80% ethanol, and then resuspended in 50 μl H2O. As described by Liu et al. (1993), 3 μl of the cDNA was amplified using PCR to obtain the target genes in 30 cycles of denaturation (94°C, 45 sec), annealing (55°C, 30 sec), and extension (70°C, 1 min). Two nuclear genes were isolated: the γ-actin and DNA polymerase-β genes. The latter is a housekeeping gene located on chromosome 8 (McBride et al., 1990). The forward and backward primers for the actin cDNA were from Colotta et al. (1992). A partial cDNA (125 bp) of mouse DNA polymerase-β was obtained using the upstream primer 5′-gaattcatgagcaaacgcaaggc-3′ and the downstream primer 5′-gaatctgtacgcattgtacttgt-3′ from the mouse DNA polymerase-β sequence (Yamaguchi et al., 1987). The identity of the amplified product was confirmed using diagnostic sequence analyses (Liu, 1993).
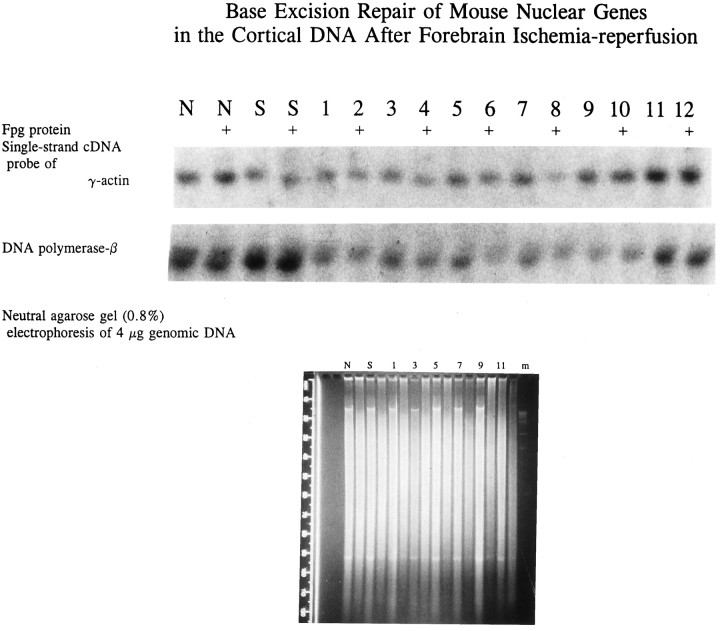
DNA damage in nuclear genes. Cortical DNA from four animals in each time point was pooled, and 12 μg was incubated in a buffer (20 mm Tris HCl, pH 7.5, 50 mm KCl) with or without Fpg protein (turnover number = 35 pmol · min−1 · μg−1 protein average of an oh8-dG-containing, double-strand oligonucleotide at a dose of 0.04 μg Fpg protein per microgram of DNA) at 37°C for 60 min. The enzyme was inactivated at 90°C for 10 min. After they had cooled at room temperature for 30 min, DNA samples in all vials were incubated with EcoRI restriction endonuclease (2 U/μg DNA) in buffer supplied by the manufacturer for 2 hr at 37°C. Digested DNA was purified using TE-equilibrated phenol/chloroform/isoamyl alcohol, pH 8.0, and then in chloroform/isoamyl alcohol. DNA was precipitated in 70% ethanol at −20°C for 16 hr. The DNA pellet was dissolved in TE buffer. DNA (4 μg/lane) was loaded onto each well and resolved in 0.8% neutral agarose gel (field-inversion electrophoresis at 50 V for 800 volt-hr at room temperature) in TBE buffer (45 mm Tris, 45 mm boric acid, and 0.5 mm EDTA, pH 8.0) using a power inverter [PPI-200 (five steps, with 0.02 sec initial reverse time, 0.02 sec reverse increment, 0.05 sec initial forward time, 0.05 sec forward increment, 0.02 sec reverse increment increment, and 0.05 sec forward increment increment), MJ Research, Watertown, MA] to enhance separation of DNA fragments. At least two gels were made from each digestion to obtain duplicate blots. Resolved DNA in each gel was stained with ethidium bromide (0.5 μg/ml) for 30 min and then electrotransferred to a membrane (GenScreen Plus) and hybridized at 65°C for 16 hr with a gel-purified, single-stranded32P-cDNA probe (2 × 106 cpm, specific activities = 5 × 109 cpm/μg). The blot was washed for 30 min at room temperature (twice at 2× SSC/0.1% SDS) and at 60°C (twice at 1× SSC/0.1% SDS). The autoradiogram was developed at −70°C for 10 d. The autoradiographic signals, as shown in Results, were scanned and stored as digitized images in a computer using a desktop scanner (Color/Grey Scanner; Microtek, San Jose, CA). The amount (pixel) of each gene was measured four times using an image analysis program (DESQView/X; Quarterdeck Office Systems, Santa Monica, CA). The amount of zero class and the frequency of Fpg protein-sensitive sites (FPGSS) in the actin genes were calculated as described previously (Bohr et al., 1985; Mellon et al., 1987). To assure reproducibility of detection, DNA damage in each nuclear gene was measured twice in each of the duplicate blots using the same probe. Moreover, one of the duplicate blots was stripped at 100°C for 10 min to remove radioactivity in the blot before additional hybridizations with the probe of a second gene.
DNA fragmentation after forebrain ischemia–reperfusion. The incorporation of digoxigenin-dUTP (dig-dUTP) into the 3′-OH terminal of DNA fragments using TUNEL stain was examined in coronal sections (5 μm) from formalin-fixed, paraffin-embedded mouse brains (three animals each for sham-operated and normal control samples, and 4–10 animals each for 30 min of ischemia with 1, 2, 3, 4, and 8 d of reperfusion). Coronal sections of the forebrain and cerebellum (5 μm) from paraffin-embedded brains were treated with xylene and then with chloroform and xylene. The incorporated dig-dUMP was detected using fluorescein conjugates of antibodies against digoxigenin followed by 0.1 μg/ml propidium iodide (PI) staining in the presence of heat-treated RNase A (0.5 μg/ml) as described by Oncor.
RESULTS
Increased mutations in nuclear genes of the brain
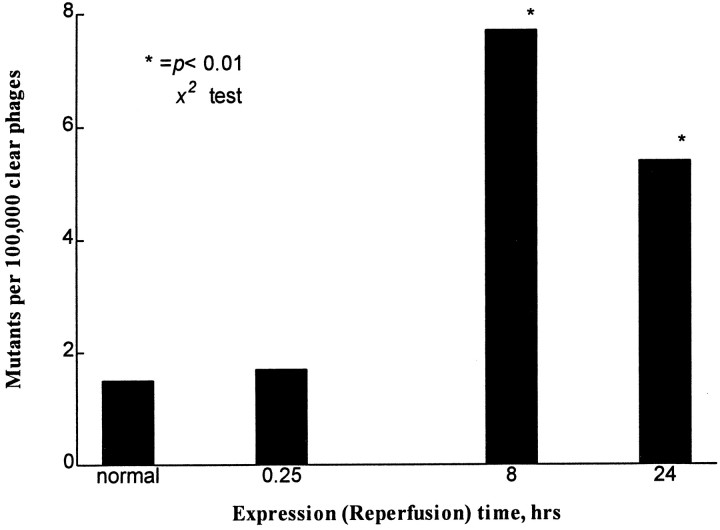
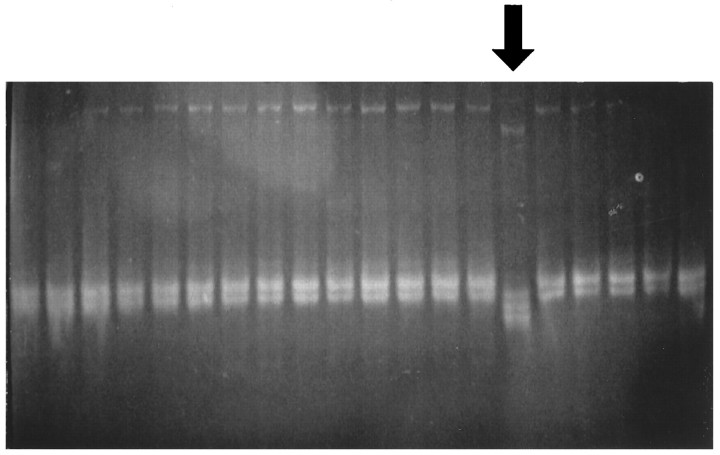
Using the reporter lacI gene system in Big Blue transgenic mice, we obtained a spontaneous frequency of 1.5 mutants per 105 plaques based on screening cortical DNA of three animals with sham operation (Fig. 1). This frequency was similar to reports by others (Kohler et al., 1991). There was a significant increase in the mutant frequency (7.7 × 10−5) after 30 min of ischemia with 8 hr of reperfusion time (Fig. 1). The mutant frequency remained elevated at 6 × 10−5 at 24 hr reperfusion; however, there was no significant increase in the frequency from animals with 30 min of ischemia and 15 min of reperfusion as compared with that of the normal animals. The mutation in the mutant lacI gene was identified using SSCP-RNA analysis (Fig. 2) and direct DNA sequencing (Table 1). In a summary from the SSCP-RNA analysis (Table 1), we observed that 6 of 19 ischemia-induced mutants occurred on the 5′-terminus [nucleotide (nt) −49 to 251], and the other nine occurred around the sequence toward the 3′-end of thelacI gene (nt 723–963). Direct DNA sequencing revealed that four lacI mutants from three normal nonischemic mice contained one transversion and two transitions (Table 1). The sequence changes in 19 ischemia-induced mutants contained 23 different mutations in the lacI gene: results suggested multiple hits within thelacI gene. There were 14 mutants that each contained a single-base mutation (i.e., excluding clones 6, 9, 14, 22, and 40). Twelve of these mutations (86%) originated in G or C sequence. Three mutants contained either multiple deletions or a combination of deletion and insertion (clone 6 or 22, Table 1). These 23 mutations involved seven transitions, five transversions, nine deletions, and two insertions. Sequencing data have confirmed that these clones are mutants. The locations of mutations in the lacI gene mutants thus identified using direct DNA sequencing assay agree with SSCP-RNA analysis.
Fig. 1.
Mutant frequency of a nuclear gene (lacI). The number of phages assayed was 205,430 in three animals with sham operation, and the numbers assayed after 30 min of ischemia were 292,280 (0.25 hr of reperfusion, three animals), 338,875 (8 hr of reperfusion, seven animals), and 130,220 (24 hr of reperfusion, four animals). Two of four mutants from the sham operation have the same mutation at the same location (Table 1) and are regarded as having the same clonal origin. (* denotes statistical significance at p < 0.01 by χ2 test; DataMost).
Fig. 2.
SSCP-RNA analysis. A mutant RNA transcript of theE. coli lacI mutant was identified by its altered mobility in 8% nondenaturing PAGE (arrow).
Table 1.
Mutation analysis using single-strand conformation polymorphism in RNA and direct DNA sequencing in the reporter genelacI
| SSCP-RNA | Direct DNA sequencing | ||
|---|---|---|---|
| Spontaneous lacI mutants | |||
| Clone no. 50 | nt 548–763 | 569-C to t | (CAG t AAA) |
| 52/54 | nt 50–163 | 86-G to t | (ACC t TTT) |
| 53 | nt 50–163 | 54-T to c | (ATG c CGC) |
| Forebrain ischemia-induced lacI mutants | |||
| 30 min ischemia + 8 hr reperfusion | |||
| Clone no. 10 | nt 723–963 | d813-T | (GGC - GCG) |
| 12 | nt 723–963 | d781-G | (GCT - GGC)* |
| 14 | nt 50–163 | 90-CCC to aag | (TTT aag GCG)* |
| 22 | nt 723–963 | d761-G | (GTT - CCA) |
| d764-AA | (GCC -- CGA) | ||
| d800-A | (ATT - CCG) | ||
| 31 | nt 723–963 | 819-T to a | (GCG a TGG) |
| 35 | nt −49 to 163 | 49-C to a | (ATA a GAT) |
| 39 | nt 723–963 | 843-G to a | (TGG a ATA)* |
| 40 | nt 181–403 | d273-cgattaaatctcgcg-287 | (CGC-n15-CCG) |
| 41 | nt 723–963 | 792-G to a | (TGC a CGC) |
| 42 | nt 723–963 | d927-G | (CCA - CGT) |
| 48 | nt −49 to 163 | 49-C to a | (ATA a GAT) |
| RC-8 | nt −49 to 163 | 92-C to t | (TCC t GCG)* |
| 30 min ischemia + 24 hr reperfusion | |||
| Clone no. 4 | nt 50–251 | 95-G to a | (CGC a TGG) |
| 5 | nt 723–963 | 853-T to g | (CGA g ACC) |
| 6 | nt 50–163 | d50-G | (TAC - ATG) |
| i54-TgC-55 | (TGT g CGC) | ||
| nt 347–601 | 419-C to t | (GAC t AGG) | |
| 7 | nt 548–763 | i675-GgG-676 | (ACG g GGA)* |
| 8 | nt 548–763 | 630-G to a | (GCT a GCA) |
| 9 | nt 548–763 | d624-ctgg-627 | (TGG ---- CTG) |
| 33 | nt 723–963 | 880-G to a | (CCC a CCG) |
Repeatable and identical base alterations were accepted as the mutation of that clone. Upper cases = parental sequences; lower cases = mutant sequences; d = deletion; i = insertions; nt = nucleotide number of lacI gene; * = mutation in the triplet CCC (or GGG) sequences; these mutations were similar to those using nitric oxide (Wink et al., 1991; Nguyen et al., 1992; Marragos et al., 1993;Schmutte et al., 1994).
Oxidative damage in cellular DNA
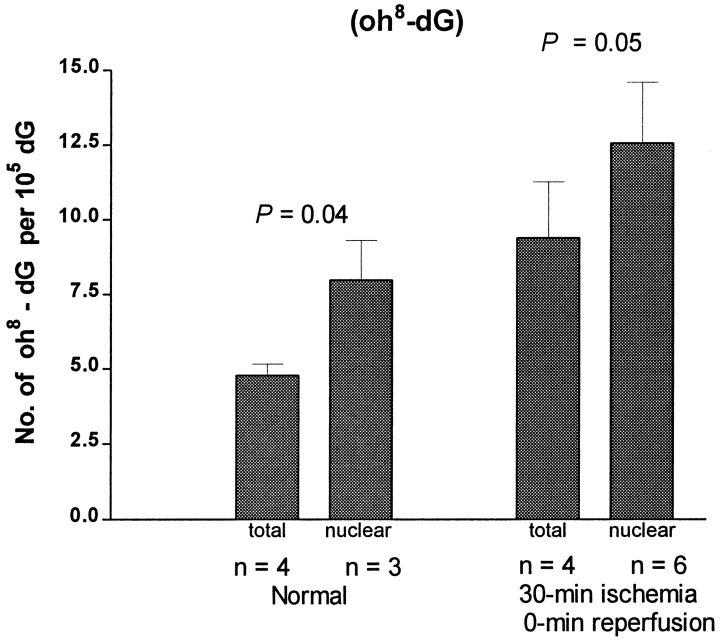
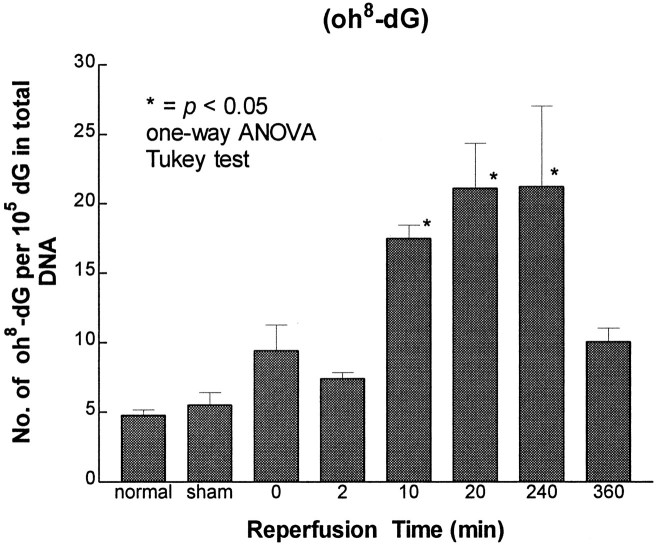
An increase in the frequency of mutation in the lacI gene of the Big Blue transgenic mice suggests that nuclear genes of the brain were damaged in this forebrain ischemia model. The damage could have resulted from oxidative stress. To demonstrate that forebrain ischemia–reperfusion induced oxidative stress, we measured oxidative lesions in cellular DNA. At least 20 DNA lesions that are believed to be results of the interaction between oxygen free radicals and DNA have been identified in organs, including the brain. The HPLC–EC method detects modified nucleoside oh8-dG (Mecocci et al., 1993), whereas the GC/MS-SIM method detects several modified bases including oh8-dG in its free-base form 8-OH-Gua (Dizdaroglu, 1994). Among these lesions, oh8-dG determined by the HPLC–EC method is considered to be an indicator of DNA damage (Fraga et al., 1990; Floyd and Carney, 1992). In preliminary studies, an attempt was made to remove mitochondria in the homogenate by centrifugation at 3000 × g (4°C for 30 min) before proteinase K digestion in 2% SDS. Results shown in Figure 3 indicate that a delay in adding proteinase K and 2% SDS by centrifugation caused an equal increase of oh8-dG (∼3 oh8-dG/105 dG) in DNA from both normal and ischemic cortices (Fig. 3). This delay caused an elevation in the oh8-dG content of 25–35%, with p values of 0.04 and 0.05 by t test for normal and ischemic samples, respectively. Therefore, in subsequent studies, homogenates were not centrifuged, and proteinase K and SDS were added as soon as they were made. We found that the oh8-dG content (5/105dG) in the normal and sham-operated controls did not differ significantly (Fig. 4). This value was comparable with that found in nuclear DNA (2 × 10−5 dG), but was significantly lower than that of the mtDNA (22 × 10−5 dG) from the cerebral cortex of the human brain (Mecocci et al., 1993). Most importantly, we observed a significant increase (fourfold) in the oh8-dG contents (an average of 20 ± 1/105 dG) in the ischemic cerebral cortex during 10–240 min of reperfusion (Fig. 4). No significant difference in the level of DNA lesions was noted between samples from transgenic Big Blue mice and nontransgenic litter mates (our unpublished observations).
Fig. 3.
The effect of centrifugation on sample preparation in the measurement of DNA lesions after forebrain ischemia–reperfusion. DNA lesions were measured by the HPLC–EC method. Data are expressed as mean ± SEM. The number of animals (n) is indicated at the bottom ofeach bar. The statistical significance (ttest) between total DNA (without centrifugation) and nuclear DNA (with centrifugation) was indicated by the p values within the control (Normal) and ischemic groups.
Fig. 4.
Measurement of damage to total cellular DNA after forebrain ischemia–reperfusion by the HPLC–EC method. Each time point contained samples from at least four animals, except samples from the 360 min reperfusion time point (n = 2), which were not included in statistical analysis but are presented with a bar representing the range of data from two animals. (* denotes significance level at p ≤ 0.05 as compared with the control groups using normal and sham-operated animals).
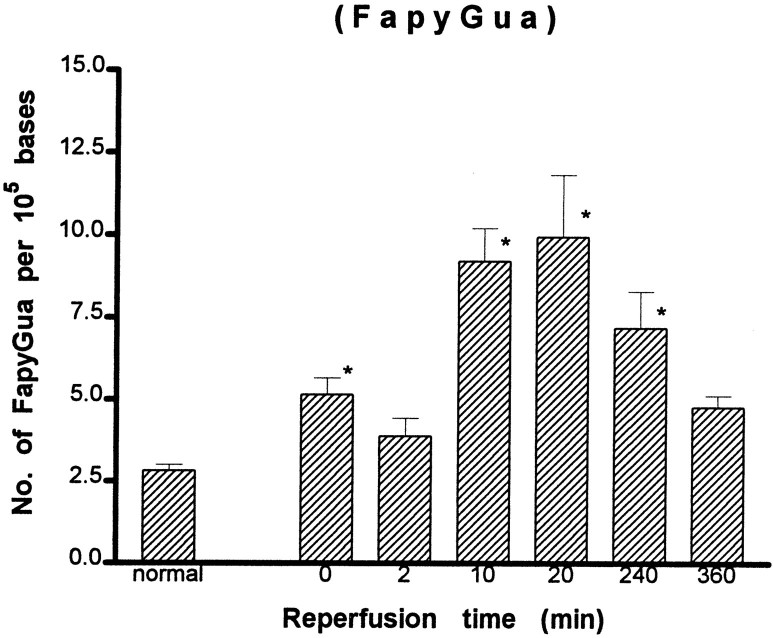
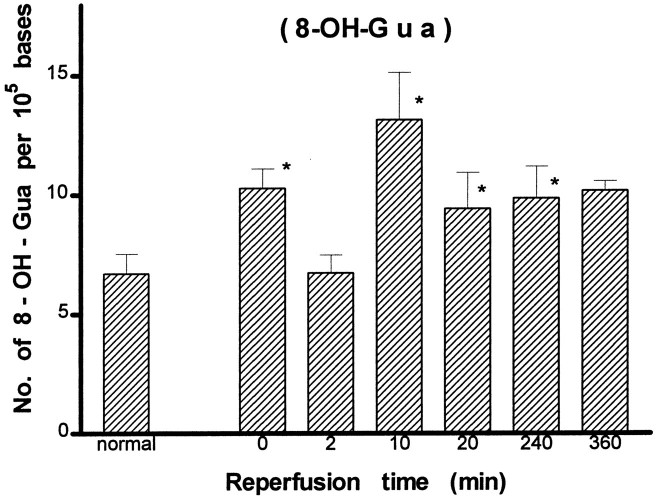
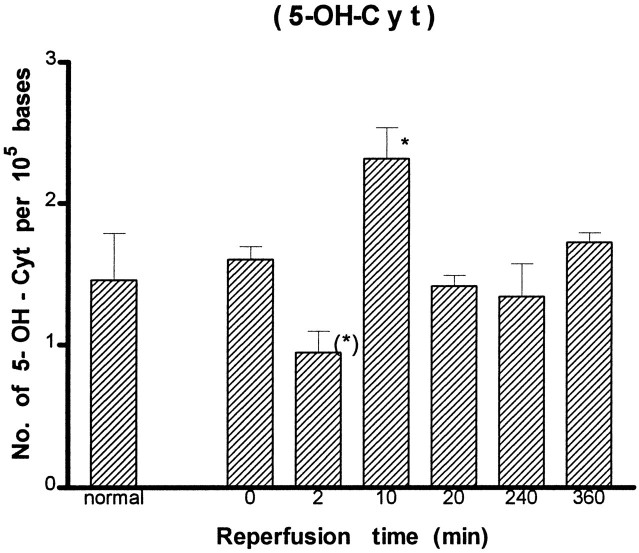
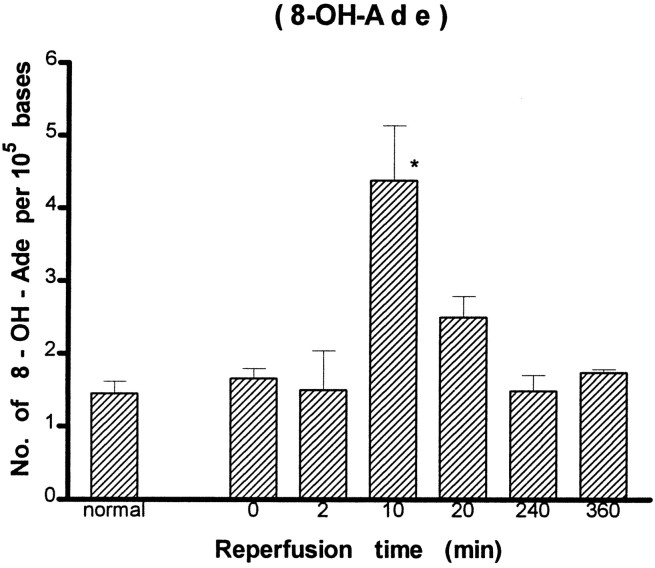
A detailed analysis of modified bases was carried out further using the GC/MS-SIM method. We identified eight DNA lesions from cellular DNA of the cortex: 8-OH-Ade, FapyAde, 2-OH-Ade, 5-OH-Cyt, FapyGua, 8-OH-Gua, 5-OH-5-MeHyd, and 5-OH-Hyd. The number of DNA lesions in the normal animals in this study (Figs. 5-8) was similar to or less than those reported in chromatin preparations from mouse liver (Mori et al., 1993), rat kidney (Kasprzak et al., 1992), or human cells (Dizdaroglu, 1992). The content of FapyGua and 8-OH-Gua increased significantly (p < 0.05) at the end of 30 min of ischemia (Figs. 5, 6). There was no increase within 2 min of reperfusion, but the lesions increased significantly again (two- or fourfold) at 10 min of reperfusion and remained significantly elevated for at least 240 min. The results based on the GC/MS-SIM method indicate cerebral oxidative stress in this model and agree with the results using the HPLC–EC method shown in Figure 4. A significant increase was observed in 5-OH-Cyt and 8-OH-Ade lesions at 10 min of reperfusion after 30 min of ischemia (Figs. 7, 8). The level of 5-OH-Cyt at 2 min of reperfusion was consistently the lowest level among all lesions in ischemic samples (Fig. 7); nevertheless, the reduction was small (≤1 × 10−5 base). The total level of DNA damage involving three measurable G and C lesions (FapyGua, 8-OH-Gua, and 5-OH-Cyt) increased from 10/105bases in the normal sample to 25/105 bases in the sample with 10 min of reperfusion. We estimated that each cell contains ∼1.5 × 106 G/C lesions at 10 min of reperfusion after 30 min of ischemia.
Fig. 5.
Measurement of DNA lesions in total cellular DNA after forebrain ischemia–reperfusion by the GC/MS-SIM method (see Fig.4 for details). (* denotes significance level at p ≤ 0.05 as compared with the control groups.)
Fig. 6.
Measurement of DNA lesions in total cellular DNA after forebrain ischemia–reperfusion by the GC/MS-SIM method (see Fig.4 for details). (* denotes significance level at p ≤ 0.05 as compared with the control groups.)
Fig. 7.
Measurement of DNA lesions in total cellular DNA after forebrain ischemia–reperfusion by the GC/MS-SIM method (see Fig.4 for details). (* denotes significance level at p ≤ 0.05 as compared with the control groups.)
Fig. 8.
Measurement of DNA lesions in total cellular DNA after forebrain ischemia–reperfusion by the GC/MS-SIM method (see Fig.4 for details). (* denotes significance level at p ≤ 0.05 as compared with the control groups.)
Oxidative damage in nuclear genes
Because cellular DNA that was used may contain mtDNA in addition to nuclear DNA, the increase in DNA lesions of all samples shown above may be attributed to DNA damage in both mtDNA and nuclear DNA. To demonstrate that nuclear genes were also targets of ROS and indeed were damaged after ischemia–reperfusion, we examined the presence of DNA lesions in nuclear genes that are sensitive to digestion by Fpg protein. Fpg protein of E. coli removes FapyGua, 8-OH-Gua, and 5-OH-Cyt lesions using its glycosylase activities, followed by removal of the DNA without base [apurine/apyrimidine (AP) sites] using its AP endonuclease activities (Chetsanga and Lindahl, 1979; Tchou et al., 1991; Boiteux et al., 1992;Hatahet et al., 1994). This assay detects the presence of FPGSS, i.e., DNA lesions that are susceptible to digestion by Fpg protein. With the Fpg protein treatment in test tubes, specific lesions are removed and strand breaks are generated. DNA is converted to smaller fragments than that without treatments. As a result, high molecular weight DNA seems lost in the agarose gel. This is illustrated with two DNA fragments in the agarose gel (Fig. 9,bottom): one is a fragment of ≥4 × 104 bp located at 15 mm, the other is a fragment of ≤1200 bp at 93 mm from gel wells. The first fragment in lanes N, S, and 1–9 (Fig. 9,bottom) was expected to have a minimum of four FPGSS according to Figures 4, 5, 6, 7. After Fpg protein digestion, this fragment was no longer present (see the adjacent lanes in the same location). On the other hand, the second DNA fragment of ≤1200 bp, which is expected to have less than one FPGSS in the DNA, was relatively resistant to Fpg protein as compared with the first fragment shown in Figure 9. The uneven appearance in the gel was most likely related to differences in degrees of Fpg digestion. Variations in ethidium bromide stain caused by DNA gel loading appeared to be negligible.
Fig. 9.
DNA damage in nuclear genes after forebrain ischemia–reperfusion. EcoRI-digested DNA with or without additional Fpg protein digestion was resolved in 0.8% neutral agarose gel using field-inversion electrophoresis (see Materials and Methods) and then stained with ethidium bromide (bottom). One of two gels is shown. Three molecular weight markers (lane m) that represent 19, 12, and 8 kb are visible in the bottom panel. A ruler with increments of 10 mm is shown on the left side of the gel. The samples without added Fpg protein were normal (lane N), sham operation (lane S), and 30 min with reperfusion of 0 min (lane 1), 2 min (lane 3), 10 min (lane 5), 20 min (lane 7), 240 min (lane 9), and 360 min (lane 11). The samples with added Fpg protein (thelanes without label, bottom) were on the adjacent lanes to the right of each individual sample. The DNA in the gel was then transferred to a membrane. The membrane was hybridized to32P-labeled, gel-purified single-stranded cDNA probe. A 2 kb fragment of γ-actin genes was detected in the autoradiogram and presented in the top panel. The radioactivities in the blot were stripped (see Materials and Methods), and the blot was hybridized to a second probe of DNA polymerase-β; a fragment of 6–7 kb was detected (center panel). The label on each lane was the same as in agarose gel except that the DNA sample with Fpg protein treatments was identified with +.
By hybridizing a single-stranded cDNA of γ-actin or DNA polymerase-β, we observed a single fragment of ∼2 kb (actin gene) or 6–7 kb (DNA polymerase-β gene) (McBride et al., 1990) in the autoradiogram (Fig. 9, top two panels). The fragment that remained after Fpg protein treatment represented the gene that was resistant to digestion by Fpg protein. Therefore, the fragment shown was the gene fraction without damage. The ratio of genes with Fpg protein treatment to that without is the zero class. A value of ≥1 in the zero class as shown in DNA from normal and sham-operated samples for both nuclear genes (Table 2) suggests that there was no measurable FPGSS in the genes from animals without ischemia–reperfusion. The result indicates that Fpg protein did not contain a significant amount of nonspecific nuclease activities or that the nuclease activities in Fpg protein preparation did not cause a significant adverse effect on DNA. Most importantly, we observed that there was a reduction in the zero class of both actin and DNA polymerase-β genes in samples with ischemia–reperfusion. The results indicate that there was an increase in DNA lesions in ischemic samples compared with those in normal and sham-operated controls (Table 2). With 30 min of ischemia, the FPGSS frequency calculated from the zero class (Table 2; Mellon et al., 1987) in both genes was 0.3 in samples without reperfusion (0 min). The frequency was increased to 1.2 in actin genes at 20 min of reperfusion. We observed the same for the DNA polymerase-β gene, except that the FPGSS frequency was 0.4 and 0.5 in 10 and 20 min of reperfusion, respectively. Therefore, the frequencies of FPGSS in both genes were the highest at 20 min of reperfusion among all of the time points measured (Table 2). The frequency in both nuclear genes was reduced to ≤0.2 at 4 or 6 hr of reperfusion (Fig. 9). Last, an increase followed by a decrease of FPGSS in nuclear genes suggests the presence of DNA repair after forebrain ischemia–reperfusion in the mouse.
Table 2.
DNA damage of nuclear genes in cerebral cortex after forebrain ischemia
| Pixel values × 10−3 | Zero class R=(+Fpg/−Fpg) | FPGSS frequency (−ln R) | ||
|---|---|---|---|---|
| +Fpg protein | −Fpg protein | |||
| Actin genes | ||||
| Normal (N) | 113 | 86 | ≥1 | 0 |
| Sham operated (S) | 110 | 91 | ≥1 | 0 |
| Ischemic samples with reperfusion of | ||||
| 0 min | 87 | 118 | 0.74 | 0.3 |
| 2 min | 87 | 106 | 0.82 | 0.2 |
| 10 min | 91 | 117 | 0.78 | 0.2 |
| 20 min | 40 | 139 | 0.29 | 1.2 |
| 240 min | 132 | 119 | 0.94 | 0.1 |
| 360 min | 145 | 152 | 0.95 | 0.1 |
| DNA polymerase-β | ||||
| Normal (N) | 415 | 324 | ≥1 | 0 |
| Sham operated (S) | 487 | 399 | ≥1 | 0 |
| Ischemic samples with reperfusion of | ||||
| 0 min | 191 | 269 | 0.71 | 0.3 |
| 2 min | 224 | 260 | 0.86 | 0.2 |
| 10 min | 162 | 243 | 0.67 | 0.4 |
| 20 min | 111 | 178 | 0.62 | 0.5 |
| 240 min | 81 | 94 | 0.86 | 0.2 |
| 360 min | 159 | 170 | 0.94 | 0.1 |
The pixel values are the average of four pixel readings on the same blot presented in Figure 9. FPGSS frequency was calculated according to Mellon et al. (1987).
Brain injury shown as DNA fragmentation in nuclei
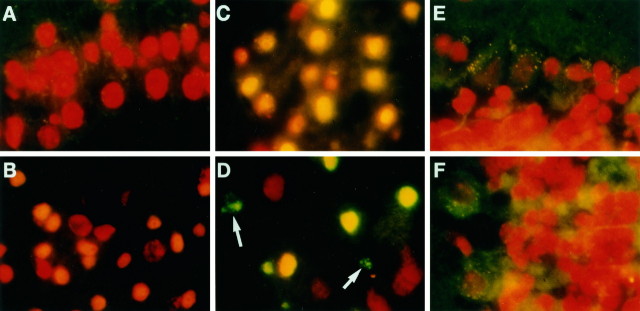
DNA fragmentation as a sign of damage to tissue is observed frequently in rodent stroke models. DNA fragmentation in the ischemic brain can be detected using TUNEL stain and by DNA ladders using gel electrophoresis (Linnik et al., 1993; Dugan et al., 1995; Li et al., 1995a,b; Du et al., 1996). To further delineate the effect of oxidative stress in this stroke model, we explored morphological evidence of damage to brain tissue. We observed that cortical nuclei of normal animals stained bright red because of an intercalation of PI on DNA and did not show TUNEL-positive stain (Fig. 10A). The result indicated minimal DNA fragmentation. After forebrain ischemia–reperfusion, TUNEL-positive nuclei appeared in the caudate putamen, hippocampus, and scattered regions of the cortex. Of the 23 animals that were examined, nuclei of layer II neurons showed bright green TUNEL-positive stain (also double exposure) in none of 10 animals (0%) on day 1, in two of four animals (50%) on day 2, in three of four animals (75%) on day 3, and in all five animals on days 4 and 8 of reperfusion. Although there were no bright green nuclei detected on day 1 (Fig.10B,C), nuclei with yellowish staining perhaps were indicative of a mixture of the red fluorescence from PI dye on intact DNA and a trace of green fluorescence from fluorescein antibody against incorporated dig-dUTP on fragmented DNA, i.e., nuclei that have a few fragmented DNA. Nuclei that have signs of DNA fragmentation appeared to be smaller in size than the adjacent normal nuclei, indicating a possible morphological feature of nuclei shrinkage. Nuclei with condensed aggregates and apoptotic bodies appeared in caudate putamen on day 3 of reperfusion (Fig.10D), and they appeared in the cortex on day 8 of reperfusion (our unpublished observation). The apoptotic bodies were detected among the nuclei with signs of initial fragmentation (yellowish nuclei) in animals with longer reperfusion time (arrow in Fig. 10D), suggesting that late-occurring tissue injury was observed in this model of forebrain ischemia–reperfusion. No TUNEL-positive nuclei were observed in cerebellum from animals without ischemia (normal, Fig.10E) or with ischemia (day 3 of reperfusion, Fig.10F). This observation was consistent with the notion that a lack of oxidative stress resulted in no DNA fragmentation.
Fig. 10.
DNA fragmentation after forebrain ischemia (30 min). Photographs were taken using a mercury light source with filters [dual exposures: a first exposure with Leica (Deerfield, IL) I3 filter (excitation 450–490 nm; barrier at 510 nm) for fluorescein and a second with N2.1 filter (excitation 515–560 nm; barrier at 580 nm) for PI], and with 1000× magnification in one representative animal of selected time point. A, Layer II nuclei of normal cortex from animal without ischemia. B, C, Layer II nuclei of the cortex and CA 3 nuclei of the hippocampus with 1 d of reperfusion, respectively. D, Caudate putamen nuclei with 3 d of reperfusion. E, F, Nuclei from the cerebellum of normal brain and of brain with ischemia and 3 d of reperfusion, respectively. Nuclei with PI stain appeared red, whereas nuclei with anti-dig-fluorescein conjugates and incorporated dig-dUMP on the DNA fragment appearedbright green (D, TUNEL-positive nuclei). Nuclei with the appearance of orange oryellow fluorescence in B, C, andD suggest a blend of red (PI stain) withslight green (conjugates of anti-dig-fluorescein and dig-dUMP).
DISCUSSION
We have described here the detection of nuclear gene mutation in a forebrain ischemia–reperfusion model of transgenic mice containing a reporter lacI gene. The mutation in the nuclei of the brain is most likely initiated by DNA damage caused by oxidative stress. This conclusion is supported by an elevation of mutagenic lesions in cellular DNA (Figs. 4, 5, 6, 7, 8) and the presence of FPGSS in nuclear genes (Fig. 9). The mutagenic lesions in cellular DNA that we detected are characteristic of products resulting from the interaction between free radicals and DNA (Jaruga and Dizdaroglu, 1996), suggesting free radical-mediated damage in this model. The damage in DNA is repaired with time. Nevertheless, mutations in the reporter gene lacI located in nuclei were detected. Our observations are consistent with the notion that this model of forebrain ischemia–reperfusion induces oxidative stress, which is mutagenic to brain cells.
Different DNA lesions are formed by different pathways. Four DNA lesions were elevated in this model. The lesion detected after oxidative stress reflects a net result of the formation and removal (repair) of all lesions. Under normal conditions, one oxygen molecule is reduced to two H2O molecules with four electrons. Incomplete reduction of oxygen molecules results in the formation of ROS when either of the two molecules is unbalanced (Lehninger, 1982). At least two separate mechanisms may account for the formation of ROS in this model. Ischemia produces ROS when the reduction of oxygen molecules occurs within a condition starting with balanced oxygen and electrons in transit to a condition of diminishing oxygen molecules but increasing hydrogen ions. The formation of ROS depends on the available oxygen molecules. On the other hand, a reverse condition may be in effect during ischemia–reperfusion. It is predicted that the level of ROS is elevated gradually until oxygen molecules and electrons are balanced. Among these ROS, hydroxyl radicals are believed to be genotoxic. Hydroxyl radicals have a very short half-life; therefore, they have to be generated at the site near nuclear DNA. One way this is accomplished is from a homolytic cleavage of peroxynitrites (Lipton et al., 1993; Oury et al., 1993; Crow et al., 1994). Peroxynitrites are formed from superoxide ions and nitric oxide (NO) at a rate constant of 6.7 × 109m/sec (Beckman et al., 1990;Huie and Padmaja, 1993). The stability of peroxynitrite ions may enable them to travel a distance equivalent to several cells.
Each individual lesion in an organ such as the brain is possibly repaired via complex repair pathways or pathways that are at least as complex as those in single-cell systems. Recent reports indicate that the half-life of 8-OH-Ade and 5-OH-Cyt is 12 min, whereas the half-lives of FapyGua and 8-OH-Gua are 34 and 55 min, respectively (Jaruga and Dizdaroglu, 1996). The half-life of these lesions predicts that 8-OH-Ade and 5-OH-Cyt should “disappear” earlier than FapyGua and 8-OH-Gua. The levels of four DNA lesions that were detected after 30 min of ischemia are consistent with the half-lives of these lesions. Our results using whole animal studies as shown in Figures 5, 6, 7, 8 confirm these predictions. Malinski et al. (1993) reported an elevation of NO and related free radicals in the first 15 min of a 1 hr cerebral ischemia. The elevation returns to that of the pre-ischemia level for the remainder of the ischemia period, and then a second elevation of NO and related free radicals starts gradually (not immediately) on reperfusion. The gradual increase in the free radicals indicates that oxidative stress in the brain during the first few minutes of reperfusion may be low but will gradually increase with reperfusion time. Therefore, no detectable DNA damage was observed in our model at 2 min of reperfusion, whereas there was an increase in DNA damage at 10 min for all four lesions. Again, because of the half-life, the guanine lesions remain elevated, whereas 8-OH-Ade and 5-OH-Cyt levels return to the control level after 20 min of reperfusion. Alternatively, other factors may affect the rates at which the lesions are removed. For example, the repair of FapyGua and 8-OH-Gua lesions may consist of two phases: the initial component is fast and occurs within 2 min of reperfusion as in the case of 5-OH-Cyt, and the second phase is slow and may last longer than 20 min. This biphasic repair agrees with the removal of guanine adducts in the brain (Gaubatz and Tan, 1994). The observation of a late burst of NO and related radicals along with a possible inhibition of Fpg protein (or its mammalian equivalents) by the presence of NO (Wink and Laval, 1994) could increase lesion formation with a concurrent reduction of the repair of FapyGua and 8-OH-Gua lesions during reperfusion of >20 min (Figs. 4, 5, 6). Our data, therefore, support a possible role of NO and related free radicals in mediating neurotoxicity after ischemia–reperfusion (Wink et al., 1991; Nguyen et al., 1992; Malinski et al., 1993; Marragos et al., 1993; Huang et al.,1994; Schmutte et al., 1994; Zhang et al., 1994).
An elevation in lacI mutants 8–24 hr after forebrain ischemia using the Big Blue transgenic system is consistent with oxidative stress that may damage nuclear DNA. DNA damage is repaired efficiently with minimal risk of error if the damage level is low or the repair function is accurate. Conversely, if the repair is less accurate or the damage is severe, mutation may increase. Although a detailed evaluation is needed to determine the mechanism of repair in the brain, three lines of supporting evidence suggest that nuclear DNA damage in this model is repaired: (1) an increase of DNA repair synthesis by fourfold after 30 min of cerebral ischemia (Tobita et al., 1991), (2) an increase by two- to fourfold followed by a decrease of FPGSS in two nuclear genes during reperfusion (Fig. 9), and (3) a correlation between the majority of altered sequences in thelacI mutant (Table 1) and the DNA lesions in G or C bases (Figs. 4, 5, 6, 7, 8). Mutations initiated by ROS damage may depend on neighboring nucleotides on the same strand of DNA and the type of DNA polymerase performing the repair synthesis (Tkeshelashvili et al., 1991; Feig et al., 1994). Mutations detected in this study most likely arise from either a deficit in DNA repair or a replication error of DNA polymerase-β, an enzyme that is the predominant DNA polymerase in the adult brain and is devoid of exonuclease activities for proofreading (Loeb et al., 1986). It is known that DNA synthesis mediated by DNA polymerase-β is less accurate than that with exonuclease (Feig and Loeb, 1993). Approximately 80% of the lacI mutation by forebrain ischemia in Table 1 occurred at the sequence of G or C bases. Approximately half of the mutation was found to contain deletions and insertions, and nine of them resulted in frame shifts (deletion and/or insertion in 3 ± 1 bases). This type of mutation spectrum needs further confirmation in additional mutant isolates. On the other hand, the spectrum of mutation by forebrain ischemia–reperfusion is different from that observed in spontaneous lacI mutation of Big Blue mice (Kohler et al., 1991). The base changes in our study also bear a striking similarity to ROS-induced mutagenesis in cell culture conditions (Kasai et al., 1986; Kuchino et al., 1987; Moraes et al., 1990; Wood et al., 1990; Akman et al., 1991; McBride et al., 1991;Shibutani et al., 1991; Tkeshelashvili et al., 1991; Feig et al., 1994) and to the mutation spectrum that resulted from replication of templates that contain oxidative damage in G bases using DNA polymerase with no proofreading capacity (Kunkel, 1990; Shibutani and Grollman, 1993; Feig and Loeb, 1994). Such a mechanism would occur in the brain during repair synthesis by DNA polymerase-β on a transcribed strand using a damage-infested nontranscribed strand as a template. More studies are required to confirm this hypothesis.
Increases in fragmentation of genetic material as detected by chromosomal aberrations and by cell death after exposures to chemical mutagens or ionizing radiation have been described using cells from patients with neurological disorders (Moshell et al., 1980; Scudiero et al., 1981; 1986; Chamberlain and Lewis, 1982; McGovern and Webb, 1982;Robbins et al., 1983, 1984, 1985; Tarone et al., 1983, 1984; Otsuka et al., 1985; Lavin et al., 1989; Tobi et al., 1990; Boerrigter and Vijg, 1993). Patients in whom a defect occurs during removal of a class of oxygen free radical-induced base lesions may have an increased probability of developing a neurodegenerative disorder, suggesting accumulative effects of oxidative DNA damage (Mazzarello et al., 1992;Satoh et al., 1993; Kruk et al., 1995). In the present study, four lesions have been detected in DNA samples using a forebrain ischemia–reperfusion model, and these lesions are among those found by oxidative damage in human cells (Jaruga and Dizdaroglu, 1996). The mechanisms that initiate mutagenesis and cell death in this model may not be the same. On the other hand, frame-shift mutation, as described in our model and by others in postmitotic neurons (Evans et al., 1994), may inactivate critical genes in certain cell populations in the ischemic brain. The altered gene expression after overt DNA damage may be responsible for cell death at a later time (Zhang et al., 1994). Genetic data supporting this notion have been reported in mutant cell lines that carry lower than normal contents of superoxide dismutase (SOD) and catalase. These mutants are more sensitive to the cytotoxic effect of ROS than the cell line with normal levels of the enzymes (Cerutti et al., 1989). Recent successes in developing transgenic mice overexpressing SOD (Kinouchi et al., 1991; Yang et al., 1994), with NO synthase knockout (Huang et al., 1994) and the neuroprotective effects of scavenger enzymes that neutralize ROS (Chan et al., 1987; Liu et al., 1989; Imaizumi et al., 1990; Matsumiya et al., 1991; He et al., 1993) confirmed that ROS play an important role in brain damage caused by cerebral ischemia–reperfusion. ROS have been implicated in apoptotic cell death (Beauchamp and Fridovich, 1970; Imlay and Linn, 1988; Moraes et al., 1989; Floyd and Carney, 1992; Grammas et al., 1993; Andrus et al., 1994). Therefore, morphological features suggestive of apoptosis that are observed at a later time after ischemia in our model could also be mediated by ROS. Whether mutations of nuclear genes altered the expression of genes that control cell cycles (Liu et al., 1996), resulting in delayed neuronal death, remains to be determined. Our studies present one additional mechanism of DNA damage in the brain, an organ that is well protected from exposure to environmental mutagens. Mutagenesis in this ischemia–reperfusion model raises the possibility of DNA damage and gene dysfunction as the consequence of oxidative stress.
Footnotes
This work was supported by the Vivian L. Smith Foundation for Restorative Neurology and completed during the term of a Grant-in-Aid from the American Heart Association (94012700 to P.K.L.), by grants from National Institutes of Health (NS34810 to P.K.L.; NS25545, NS28995, and NS32636 to C.Y.H.; AG09690 to R.A.F.; GM 37216 to Y.W.K.), and by an Office of Naval Research grant (C4114503-01 to C.Y.H.). Certain commercial equipment or materials are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose. We thank Dr. Y. Y. He (Washington University, St. Louis, MO), Mr. L. L. Maidt (Oklahoma Medical Research Foundation, Oklahoma City, OK), Ms. Jun Wei (Emory University, Atlanta, GA), and Dr. S. Provost (Stratagene, La Jolla, CA) for technical assistance; Mr. W. Eaton (Baylor College of Medicine) for assistance in computer programming; and Ms. Gay Horelica for word processing.
Correspondence should be addressed to Philip K. Liu, Division of Restorative Neurology and Human Neurobiology, Baylor College of Medicine, One Baylor Plaza, Room S815, Houston, TX 77030.
REFERENCES
- 1.Akman SA, Forrest GP, Doroshow JH, Dizdaroglu M. Mutation of potassium permanganate- and hydrogen peroxide-treated plasmid pZ189 replicating in CV-1 monkey kidney cells. Mutat Res. 1991;261:123–130. doi: 10.1016/0165-1218(91)90058-t. [DOI] [PubMed] [Google Scholar]
- 2.Althaus JS, Andrus PK, Williams CM, VonVoigtlaner PF, Cazers AR, Hall ED. The use of salicylate hydroxylation to detect hydroxyl radical generation in ischemic and traumatic brain injury. Mol Chem Neuropathol. 1993;20:147–162. doi: 10.1007/BF02815368. [DOI] [PubMed] [Google Scholar]
- 3.Andrus PK, Taylor BM, Sun FF, Hall ED. Effects of the lipid peroxidation inhibitor tirilazad mesylate (U-74006F) on gerbil brain eicosanoid levels following ischemia and reperfusion. Brain Res. 1994;659:126–132. doi: 10.1016/0006-8993(94)90871-0. [DOI] [PubMed] [Google Scholar]
- 4.Barone FC, Knudsen DJ, Nelson AH, Feuerstein GZ, Willette RN. Mouse strain differences in susceptibility to cerebral ischemia are related to cerebral vascular anatomy. J Cereb Blood Flow Metab. 1993;13:683–692. doi: 10.1038/jcbfm.1993.87. [DOI] [PubMed] [Google Scholar]
- 5.Beauchamp C, Fridovich I. A mechanism for the production of ethylene from methional. J Biol Chem. 1970;245:4641–4646. [PubMed] [Google Scholar]
- 6.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerrigter METI, Vijg J. Studies on DNA repair defects in degenerative brain disease. Age Ageing. 1993;22:S44–S52. doi: 10.1093/ageing/22.suppl_1.s44. [DOI] [PubMed] [Google Scholar]
- 8.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 9.Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J. Homogenous Escherichia coli Fpg protein: a DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidine sites. J Biol Chem. 1990;165:3918–3922. [PubMed] [Google Scholar]
- 10.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 11.Cerutti P, Larsson R, Krupitza G, Muehlematter D, Crawford D, Amstad P. Pathophysiological mechanisms of active oxygen. Mutat Res. 1989;214:81–88. doi: 10.1016/0027-5107(89)90200-5. [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain S, Lewis PD. Studies of cellular hypersensitivity to ionising radiation in Friedreich’s ataxia. J Neurol Neurosurg Psychiatry. 1982;45:1136–1138. doi: 10.1136/jnnp.45.12.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan PH, Longar S, Fishman RA. Protective effects of liposome-entrapped superoxide dismutase on posttraumatic brain edema. Ann Neurol. 1987;21:540–547. doi: 10.1002/ana.410210604. [DOI] [PubMed] [Google Scholar]
- 14.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 15.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colotta F, Polentarutti N, Sironi M, Mantovani A. Expression and involvement of c-fos and c-jun protooncogenes in programmed cell death induced by growth factor deprivation in lymphoid cell lines. J Biol Chem. 1992;267:18278–18283. [PubMed] [Google Scholar]
- 17.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nature Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 18.Cortopassi GA, Shibata D, Soong N-W, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci USA. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crow JP, Spruell C, Chen J, Gunn C, Ischiropoulos H, Tsai M, Smith CD, Radi R, Koppenol WH, Beckman JS. On the pH-dependent yield of hydroxyl radical products from peroxynitrite. Free Radic Biol Med. 1994;16:331–338. doi: 10.1016/0891-5849(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 20.Danenberg PV, Horikoshi T, Volkenandt M, Danenberg K, Lenz H-J, Shea LCC, Dicker AP, Simoneau A, Jones PA, Bertino JR. Detection of point mutations in human DNA by analysis of RNA conformation polymorphism(s). Nucleic Acids Res. 1992;20:573–579. doi: 10.1093/nar/20.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. Chemical determination of oxidative DNA damage by gas chromatography–mass spectrometry. Methods Enzymol. 1994;234:3–16. doi: 10.1016/0076-6879(94)34072-2. [DOI] [PubMed] [Google Scholar]
- 23.Du C, Hu R, Csernansky CA, Hsu CY, Choi DW. Very delayed infarction after mild focal cerebral ischemia: a role for apoptosis? J Cereb Blood Flow Metab. 1996;16:195–201. doi: 10.1097/00004647-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Dugan LL, Choi DW. Excitotoxicity, free radicals, and cell membrane changes. Ann Neurol. 1994;35:S17–S21. doi: 10.1002/ana.410350707. [DOI] [PubMed] [Google Scholar]
- 25.Dugan LL, Lin TS, He YY, Hsu CY, Choi DW. Detection of free radicals by microdialysis/spin trapping EPR following focal cerebral ischemia and reperfusion and a cautionary note on the stability of 5,5-dimethyl-1-pyrroline N-oxide (DMPO). Free Radic Res. 1995;23:27–32. doi: 10.3109/10715769509064016. [DOI] [PubMed] [Google Scholar]
- 26.Evans DAP, Van Der Kleij AAM, Sonnemans MAF, Burbach JPH, Van Leeuwen FW. Frameshift mutations at two hotspots in vasopressin transcripts in post-mitotic neurons. Proc Natl Acad Sci USA. 1994;91:6059–6063. doi: 10.1073/pnas.91.13.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feig DI, Loeb LA. Mechanisms of mutation by oxidative DNA damage: reduced fidelity of mammalian DNA polymerase β. Biochemistry. 1993;32:4466–4473. doi: 10.1021/bi00067a040. [DOI] [PubMed] [Google Scholar]
- 28.Feig DI, Loeb LA. Oxygen radical induced mutagenesis is DNA polymerase specific. J Mol Biol. 1994;235:33–41. doi: 10.1016/s0022-2836(05)80009-9. [DOI] [PubMed] [Google Scholar]
- 29.Feig DI, Sowers LC, Loeb LA. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floyd RA, Carney JM. Free radical damage to protein and DNA: mechanisms involved and relevant observations on brain undergoing oxidative stress. Ann Neurol. 1992;32:S22–S27. doi: 10.1002/ana.410320706. [DOI] [PubMed] [Google Scholar]
- 31.Fraga CG, Shigenaga MK, Park J-W, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaubatz JW, Tan BH. Aging affects the levels of DNA damage in postmitotic cells. Ann NY Acad Sci. 1994;719:97–107. doi: 10.1111/j.1749-6632.1994.tb56822.x. [DOI] [PubMed] [Google Scholar]
- 33.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Hanes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses A, Williamson R, Rossor M, Owen M, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 34.Grammas P, Liu G-J, Wood K, Floyd RA. Anoxia/reoxygenation induces hydroxyl free radical formation in brain microvessels. Free Radic Biol Med. 1993;14:553–557. doi: 10.1016/0891-5849(93)90113-9. [DOI] [PubMed] [Google Scholar]
- 35.Halliwell B, Gutteridge JMC. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes: 5-hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 37.Hayakawa M, Hattori K, Sugiyama S, Ozawa T. Age-associated oxygen damage and mutations in mitochondrial DNA in human hearts. Biochem Biophys Res Commun. 1992;189:979–985. doi: 10.1016/0006-291x(92)92300-m. [DOI] [PubMed] [Google Scholar]
- 38.He YY, Hsu CY, Ezrin AM, Miller MS. Polyethylene glycol-conjugated superoxide dismutase in focal cerebral ischemia and reperfusion. Am J Physiol. 1993;265:H252–H256. doi: 10.1152/ajpheart.1993.265.1.H252. [DOI] [PubMed] [Google Scholar]
- 39.Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- 40.Huie RT, Padmaja S. The reaction of NO with superoxide. Free Radic Res. 1993;18:195–199. doi: 10.3109/10715769309145868. [DOI] [PubMed] [Google Scholar]
- 41.Imaizumi S, Woolworth V, Fishman RA, Chan PH. Liposome-entrapped superoxide dismutase reduces cerebral infarction in cerebral ischemia in rats. Stroke. 1990;21:1312–1317. doi: 10.1161/01.str.21.9.1312. [DOI] [PubMed] [Google Scholar]
- 42.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 43.Jaruga P, Dizdaroglu M. Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 1996;24:1386–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 45.Kasprzak KS, Diwan BA, Rice JM, Misra M, Riggs CW, Olinski R, Dizdaroglu M. Nickel (II)-mediated oxidative DNA base damage in renal and hepatic chromatin of pregnant rats and their fetuses: possible relevance to carcinogenesis. Chem Res Toxicol. 1992;5:809–815. doi: 10.1021/tx00030a013. [DOI] [PubMed] [Google Scholar]
- 46.Kinouchi H, Epstein CJ, Mizui T, Carlson E, Chen SF, Chan PH. Attenuation of focal cerebral ischemic injury in transgenic mice overexpressing CuZn superoxide dismutase. Proc Natl Acad Sci USA. 1991;88:11158–11162. doi: 10.1073/pnas.88.24.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruk PA, Rampino NJ, Bohr VA. DNA damage and repair in telomeres: relation to aging. Proc Natl Acad Sci USA. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 50.Kunkel TA. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 51.Lavin MF, Bates P, Le Poidevin P, Chen PC. Normal inhibition of DNA synthesis following γ-irradiation of radiosensitive cell lines from patients with Down’s syndrome and Alzheimer’s disease. Mutat Res. 1989;218:41–47. doi: 10.1016/0921-8777(89)90045-1. [DOI] [PubMed] [Google Scholar]
- 52.Lehninger AL. Electron transport, oxidative phosphorylation, and regulation of ATP production. In: Anderson S, Fox J, editors. Principles of Biochemistry. Worth; New York: 1982. pp. 467–510. [Google Scholar]
- 53.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu C-E, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu Y-H, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995a;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 54.Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KAB, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995b;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Chopp M, Jiang N, Zaloga C. In situ detection of DNA fragmentation after focal cerebral ischemia in mice. Mol Brain Res. 1995a;28:164–168. doi: 10.1016/0169-328x(94)00220-9. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Sharov VG, Jiang N, Zaloga C, Sabbah HN, Chopp M. Ultrastructural and light microscopic evidence of apoptosis after middle cerebral artery occlusion in the rat. Am J Pathol. 1995b;146:1045–1051. [PMC free article] [PubMed] [Google Scholar]
- 57.Linnik MD, Zobrist RH, Hatfield MD. Evidence supporting a role for programmed cell death in focal cerebral ischemia in rats. Stroke. 1993;24:2002–2008. doi: 10.1161/01.str.24.12.2002. [DOI] [PubMed] [Google Scholar]
- 58.Lipton SA, Choi Y-B, Pan Z-H, Lei SZ, Chen H-SV, Sucher NJ, Loscaizo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 59.Liu PK. Enhanced expression of α-type DNA polymerase genes reduces AZT cytotoxicity in hamster tr5 cells. Somatic Cell Mol Genet. 1993;19:211–220. doi: 10.1007/BF01233069. [DOI] [PubMed] [Google Scholar]
- 60.Liu TH, Beckman JS, Freeman BA, Hogan EL, Hsu CY. Polyethylene glycol-conjugated superoxide dismutase and catalase reduce ischemic brain injury. Am J Physiol. 1989;256:H589–H593. doi: 10.1152/ajpheart.1989.256.2.H589. [DOI] [PubMed] [Google Scholar]
- 61.Liu PK, Trujillo JM, Monnat RJ., Jr Spectrum of spontaneous mutation in animal cells containing an aphidicolin-resistant DNA polymerase alpha. Mutat Res. 1993;288:229–236. doi: 10.1016/0027-5107(93)90089-x. [DOI] [PubMed] [Google Scholar]
- 62.Liu PK, Kraus E, Wu TA, Strong LC, Tainsky AM. Analysis of genomic instability in Li-Fraumeni fibroblasts with germline p53 mutations. Oncogene. 1996;12:2267–2278. [PMC free article] [PubMed] [Google Scholar]
- 63.Loeb LA, Liu PK, Fry M. DNA polymerase-α: enzymology, function, fidelity, and mutagenesis. Prog Nucleic Acid Res Mol Biol. 1986;33:57–110. doi: 10.1016/s0079-6603(08)60020-3. [DOI] [PubMed] [Google Scholar]
- 64.McBride OW, Kozak CA, Wilson SH. Mapping of the gene for DNA polymerase β to mouse chromosome 8. Cytogenet Cell Genet. 1990;53:108–111. doi: 10.1159/000132906. [DOI] [PubMed] [Google Scholar]
- 65.McBride TJ, Preston BD, Loeb LA. Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry. 1991;30:207–213. doi: 10.1021/bi00215a030. [DOI] [PubMed] [Google Scholar]
- 66.McGovern D, Webb T. Sensitivity to ionising radiation of lymphocytes from Huntington’s chorea patients compared to controls. J Med Genet. 1982;19:168–174. doi: 10.1136/jmg.19.3.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malinski T, Bailey F, Zhang ZG, Chopp M. Nitric oxide measured by a prophyrinic microsensory in rat brain after transient middle cerebral artery occlusion. J Cereb Blood Flow Metab. 1993;13:355–358. doi: 10.1038/jcbfm.1993.48. [DOI] [PubMed] [Google Scholar]
- 68.Marragos CM, Andrews AW, Keefer LK, Elespuru RK. Mutagenicity of glyceryl trinitrate (nitroglycerin) in Salmonella typhimurium. Mutat Res. 1993;298:187–195. doi: 10.1016/0165-1218(93)90040-k. [DOI] [PubMed] [Google Scholar]
- 69.Martin GM. Genetic and environmental modulations of chromosomal stability: their roles in aging and oncogenesis. Ann NY Acad Sci. 1991;621:401–417. doi: 10.1111/j.1749-6632.1991.tb16995.x. [DOI] [PubMed] [Google Scholar]
- 70.Matsumiya N, Koehler RC, Kirsch JR, Traystman RJ. Conjugated superoxide dismutase reduces extent of caudate injury after transient focal ischemia in cats. Stroke. 1991;22:1193–1200. doi: 10.1161/01.str.22.9.1193. [DOI] [PubMed] [Google Scholar]
- 71.Mazzarello P, Poloni M, Spadari S, Focher F. DNA repair mechanisms in neurological diseases: facts and hypotheses. J Neurol Sci. 1992;112:4–14. doi: 10.1016/0022-510x(92)90125-5. [DOI] [PubMed] [Google Scholar]
- 72.Mecocci P, MacGarvey U, Kaufman AE, Koontz D, Shoffner JM, Wallace DC, Beal MF. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 73.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 74.Moraes EC, Keyse SM, Pidoux M, Tyrrell RM. The spectrum of mutations generated by passage of a hydrogen peroxide damaged shuttle vector plasmid through a mammalian host. Nucleic Acids Res. 1989;17:8301–8312. doi: 10.1093/nar/17.20.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moraes EC, Keyse SM, Tyrrell RM. Mutagenesis by hydrogen peroxide treatment of mammalian cells: a molecular analysis. Carcinogenesis. 1990;11:283–293. doi: 10.1093/carcin/11.2.283. [DOI] [PubMed] [Google Scholar]
- 76.Mori T, Hori Y, Dizdaroglu M. DNA base damage generated in vivo in hepatic chromatin of mice upon whole body γ-irradiation. Int J Radiat Biol. 1993;64:645–650. doi: 10.1080/09553009314551881. [DOI] [PubMed] [Google Scholar]
- 77.Moshell AN, Tarone RE, Barrett SF, Robbins JH (1980) Radiosensitivity in Huntington’s disease: implications for pathogenesis and presymptomatic diagnosis. Lancet i:9–11. [DOI] [PubMed]
- 78.Nguyen T, Brunson D, Crespi CL, Penman BW, Wishnok JS, Tannebaum SR. DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otsuka F, Tarone RE, Seguin LR, Robbins JH. Hypersensitivity to ionizing radiation in cultured cells from Down’s syndrome patients. J Neurol Sci. 1985;69:103–112. doi: 10.1016/0022-510x(85)90011-5. [DOI] [PubMed] [Google Scholar]
- 80.Oury TD, Piantadosi CA, Crapo JD. Cold-induced brain edema in mice. J Biol Chem. 1993;268:15394–15398. [PubMed] [Google Scholar]
- 81.Robbins JH, Otsuka F, Tarone RE, Polinsky RJ, Brumback RA, Moshell AN, Nee LE, Ganges MB, Cayeux SJ (1983) Radiosensitivity in Alzheimer disease and Parkinson disease. Lancet i:468–469. [DOI] [PubMed]
- 82.Robbins JH, Scudiero DA, Otsuka F, Tarone RE, Brumback RA, Wirtschafter JD, Polinsky RJ, Barrett SF, Moshell AN, Scarpinat RG. Hypersensitivity to DNA-damaging agents in cultured cells from patients with Usher’s syndrom and Duchenne muscular dystrophy. J Neurol Neurosurg Psychiatry. 1984;47:391–398. doi: 10.1136/jnnp.47.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robbins JH, Brumback RA, Polinsky RJ, Wirtschafter JD, Tarone RE, Scudiero DA, Otsuka F. Hypersensitivity to DNA-damaging agents in abiotrophies: a new explanation for degeneration of neurons, photoreceptors, and muscle in Alzheimer, Parkinson, and Huntington disease, retinitis pigmentosa, and Duchenne muscular dystrophy. Basic Life Sci. 1985;35:315–344. doi: 10.1007/978-1-4899-2218-2_20. [DOI] [PubMed] [Google Scholar]
- 84.Sakamoto A, Ohnishi ST, Ohnishi T, Ogawa R. Relationship between free radical production and lipid peroxidation during ischemia and reperfusion injury in the rat brain. Brain Res. 1991;554:186–192. doi: 10.1016/0006-8993(91)90187-z. [DOI] [PubMed] [Google Scholar]
- 85.Salminen A, Liu PK, Hsu CY. Alteration of transcription factor binding activities in the ischemic rat brain. Biochem Biophys Res Commun. 1995;212:939–944. doi: 10.1006/bbrc.1995.2060. [DOI] [PubMed] [Google Scholar]
- 86.Satoh MS, Jones CJ, Wood RD, Lindahl T. DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions. Proc Natl Acad Sci USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schellenberg GD, Bird TD, Wijsman EM, Orr HT, Anderson L, Nemens E, White JA, Bonnycastle L, Weber JL, Alonso ME. Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science. 1992;258:668–671. doi: 10.1126/science.1411576. [DOI] [PubMed] [Google Scholar]
- 88.Schmutte C, Rideout WM, III, Shen J-C, Jones PA. Mutagenicity of nitric oxide is not caused by deamination of cytosine or 5-methylcytosine in double-stranded DNA. Carcinogenesis. 1994;15:2899–2903. doi: 10.1093/carcin/15.12.2899. [DOI] [PubMed] [Google Scholar]
- 89.Scudiero DA, Meyer SA, Clatterbuck BE, Tarone RE, Robbins JH. Hypersensitivity to N-methyl-N-nitro-N-nitrosoguanidine in fibroblasts from patients with Huntington disease, familial dysautonomia, and other primary neuronal degenerations. Proc Natl Acad Sci USA. 1981;78:6451–6455. doi: 10.1073/pnas.78.10.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scudiero DA, Polinsky RJ, Brumback RA, Tarone RE, Nee LE, Robbins JH. Alzheimer disease fibroblasts are hypersensitive to the lethal effects of a DNA-damaging chemical. Mutat Res. 1986;159:125–131. doi: 10.1016/0027-5107(86)90121-1. [DOI] [PubMed] [Google Scholar]
- 91.Sen S, Phillis JW. Alpha-phenyl-tert-butyl-nitrone (PBN) attenuates hydroxyl radical production during ischemia and reperfusion injury of rat brain: an EPR study. Free Radic Res. 1993;19:255–265. doi: 10.3109/10715769309056513. [DOI] [PubMed] [Google Scholar]
- 92.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St. George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 93.Shibutani S, Grollman AP. On the mechanism of frameshift (deletion) mutagenesis in vitro. J Biol Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 94.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 95.Suzuki T, Akaike N, Ueno K-I, Tanaka Y, Himori N. MAO inhibitors, clorgyline and lazabemide, prevent hydroxyl radical generation caused by brain ischemia/reperfusion in mice. Pharmacology. 1995;50:357–362. doi: 10.1159/000139304. [DOI] [PubMed] [Google Scholar]
- 96.Tarone RE, Scudiero DA, Robbins JH. Statistical methods for in vitro cell survival assays. Mutat Res. 1983;111:79–96. doi: 10.1016/0027-5107(83)90010-6. [DOI] [PubMed] [Google Scholar]
- 97.Tarone RE, Otsuka F, Robbins JH. A sensitive assay for detecting hypersensitivity to ionizing radiation in lymphoblastoid lines from patients with Duchenne muscular dystrophy and primary neuronal degenerations. J Neurol Sci. 1984;65:367–381. doi: 10.1016/0022-510x(84)90099-6. [DOI] [PubMed] [Google Scholar]
- 98.Tchou J, Kasai H, Shibutani S, Chung M-H, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tkeshelashvili LK, McBride T, Spence K, Loeb LA. Mutation spectrum of copper-induced DNA damage [erratum appears in (1992) J Biol Chem 267:13778]. J Biol Chem. 1991;266:6401–6406. [PubMed] [Google Scholar]
- 100.Tobi SE, Moquet JE, Edwards AA, Lloyd DC, Itzhahi RF. Chromosomal radiosensitivity of lymphocytes from Alzheimer’s disease patients. J Med Genet. 1990;27:437–440. doi: 10.1136/jmg.27.7.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tobita M, Nagano I, Nakamura S, Kogure K (1991) XXI-18. DNA single-strand breaks in postischemic gerbil brain detected by in situ nick translation. J Cereb Blood Flow Metab 11[Suppl 2]:S361. [DOI] [PubMed]
- 102.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS, Keefer LK. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 103.Wink DA, Laval J. The Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenesis. 1994;15:2125–2129. doi: 10.1093/carcin/15.10.2125. [DOI] [PubMed] [Google Scholar]
- 104.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 105.Yamaguchi M, Hirose F, Hayashi Y, Nishimnoto Y, Matsukage A. Murine DNA polymerase β gene: mapping of transcription initiation sites and the nucleotide sequence of the putative promoter region. Mol Cell Biol. 1987;7:2012–2018. doi: 10.1128/mcb.7.5.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang G, Chan PH, Chen J, Carlson E, Chen SF, Weinstein P, Epstein CJ, Kamii H. Human copper-zinc superoxide dismutase transgenic mice are highly resistant to reperfusion injury after focal cerebral ischemia. Stroke. 1994;25:165–170. doi: 10.1161/01.str.25.1.165. [DOI] [PubMed] [Google Scholar]
- 107.Zhang C, Baumer A, Maxwell RJ, Linnane AW, Nagley P. Multiple mitochondrial DNA deletions in an elderly human individual. FEBS Lett. 1992;297:34–38. doi: 10.1016/0014-5793(92)80321-7. [DOI] [PubMed] [Google Scholar]
- 108.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose)synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]