Abstract
In mammalian circadian rhythms, the transcriptional-translational feedback loop (TTFL) consisting of a set of clock genes is believed to elicit the circadian clock oscillation. The TTFL model explains that the accumulation and degradation of mPER and mCRY proteins control the period-length (tau) of the circadian clock. Although recent studies revealed that the Casein Kinase Iεδ (CKIεδ) regurates the phosphorylation of mPER proteins and the circadian period-length, other kinases are also likely to contribute the phosphorylation of mPER. Here, we performed small scale screening using 84 chemical compounds known as kinase inhibitors to identify candidates possibly affecting the circadian period-length in mammalian cells. Screening by this high-throughput real-time bioluminescence monitoring system revealed that the several chemical compounds apparently lengthened the cellular circadian clock oscillation. These compounds are known as inhibitors against kinases such as Casein Kinase II (CKII), PI3-kinase (PI3K) and c-Jun N-terminal Kinase (JNK) in addition to CKIεδ. Although these kinase inhibitors may have some non-specific effects on other factors, our mini screening identified new candidates contributing to period-length control in mammalian cells.
Keywords: circadian rhythm, clock genes, imaging, real-time monitor, kinase inhibitors
I. Introduction
The mammalian circadian clock system generates daily fluctuations in the several gene expressions to regulate many aspects of near-24-hour rhythmicity such as behavior, physiology, and metabolic processes, thereby allowing mammals to anticipate the momentum of the day [4, 7, 12].
The molecular oscillation of circadian clock consists of an interlocked positive and negative transcription/translation feedback loop (TTFL) involving a set of clock genes and clock-controlled output genes that link the oscillator to clock-controlled processes [14]. CLOCK and BMAL1 are basic-helix-loop-helix (bHLH) PAS transcription factors that heterodimerize and transactivate the core clock components such as Period (Per1, 2, 3), Cryptochrome (Cry1 and Cry2) and Rev-Erbα [7, 12, 14]. PER and CRY proteins thus suppress the activity of the CLOCK/BMAL1, whereas REV-ERBα suppresses Bmal1 gene expression.
It has been thought that speed at which negative factors such as mPERs and mCRYs accumulate in the cellular nuclei control the period-length of circadian clock oscillator. This nuclear accumulation of those proteins is affected by their gene expression levels and nuclear translocation efficiency, and the mPER2 and mCRY1 shuttle between nucleus and cytoplasm we previously reported may also contribute to the nuclear accumulation process [18]. Among the various steps of the circadian molecular oscillator, the phosphorylation of clock proteins are believed to be important for the regulation of period-length. In this study, we performed a screening analysis using a kinase inhibitor library containing 84 compounds, focusing on the period-length of the mammalian cellular circadian clock.
II. Materials and Methods
Cell culture and cell line establishment
Rat-1 fibroblast (HSRRB, Osaka, Japan) and C6 cells were cultured in DMEM with 10% FBS and penicillin-streptomycin. When cells were analyzed in our high-throughput real time monitor system, the medium was changed to HEPES-buffered phenol-red free DMEM, 24 hr after transfection.
Real-time circadian rhythm monitoring
The mechanics of the bioluminescence detection system used to analyze the circadian rhythm have been described in previous reports [6]. Rat-1 or C6 cells were cultured in 10% FBS and penicillin-streptomycin containing medium. Cells were plated in 35 mm dishes (2×105 cells/dish), the medium was changed to luciferine (0.2 mM) and HEPES (15 mM) containing DMEM without phenol-red. Cells were synchronized by treatment with 100 nM dexamethasone and set on the turntable of our real-time monitoring system.
Kinase inhibitor screening assay
For screening assay, we used a kinase inhibitor library purchased from BIOMOL containing 84 compounds. These kinase inhibitors were resolved in DMSO as 10 mM concentration. For 24-well base screening using C6 cells, cells were cultured in DMEM with 10% FBS for two days before dexamethasone (Dex) treatment for synchronization. After medium change to luciferin containing recording medium as described above, cells were synchronized by 100 nM Dex. Just after synchronization (within 10 min), the cells were treated with the kinase inhibitors (see Fig. 1). The final concentration of all inhibitors was 30 µM. All inhibitors were applied to three wells for each compound. Circadian clock oscillation was analyzed by 24-well based real-time monitoring machines [6]. For detailed studies of the candidate kinase inhibitors after the screening, we also used compounds purchased from Sigma and Calbiochem.
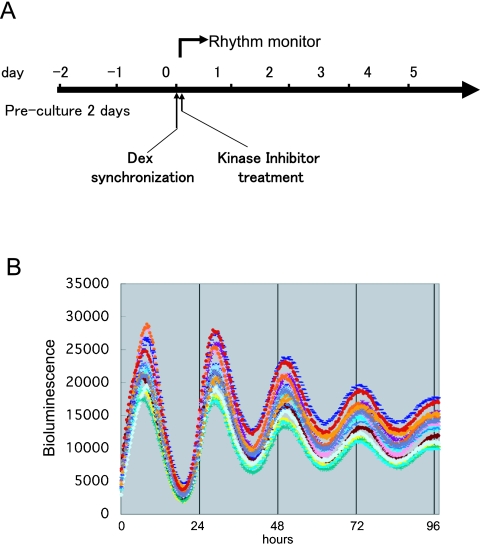
Fig. 1.
Experimental design of the screening. (A) Experimental design of screening examining the effect of kinase inhibitors on the circadian period-length in C6 and rat-1 cells. Before the dexamethasone synchronization of cellular circadian clock, cells were precultured for two days. Just after Dex stimulation, kinase inhibitors were applied to the cells. For this screening, all kinase inhibitors were applied at a final concentration of with 30 µM. At synchronization, the cells were confluent. (B) Control observation of mPer2:luc stably transfected C6 cell line using our 24-well based high-throughput real-time bioluminescence monitoring system. Bioluminescence oscillation from all wells showed almost the same phase and period-length.
III. Results
Previous studies revealed that post-translational modification PER2 protein by phosphorylation is closely related with period-length (tau) in mice and human [16, 17]. These studies suggest that the phosphorylation of PER2 by CKIεδ is the key step of the regulation of protein stability, and then contributes the period-length control of clock oscillation. These findings made us hypothesize that the phosphorylation step of clock components may control the circadian period-length in mammals. In addition to the control of mPER stability, recent reports revealed that F-box protein FBXL3 was essential for mCRY ubiquitination, and that the dysfunction of FBXL3 resulted in the severe period-length change of circadian rhythm [1, 5, 15]. In general, F-box proteins target the phosphorylation of substrates to start poly-ubiquitination. The expression of FBXL3 level is constitutive in all tissues examined. Moreover, we recently reported that only faint changes of circadian period-length were observed during the constitutive over-expression of mCRY1 in rat-1 cells [20]. A recent in vivo study using mCry1 in constitutively over-expressed transgenic mice supports this result [11]. These findings suggest that the control of phosphorylation in clock proteins can be the key process to maintain the stable period-length of the mammalian circadian clock. To address this hypothesis, we next planned a screening analysis using a kinase inhibitor library focusing on the period length control of the circadian clock.
Then we performed screening analysis using kinase inhibitor library including 84 compounds to investigate their effect on period length signals (Fig. 1A). For the first screening, C6 glioma cells were used, since this cell line has been frequently used to investigate the various signaling pathways in the neuroscience field in addition to exhibiting clear circadian oscillation [3].
Before the screening, to evaluate the reproducibility of bioluminescence rhythms, mPer2:luc reporter integrated C6 glioma cell lines were tested using the our 24-well plate based high-throughput real-time monitor system. After synchronization by dexamethasone treatment, the bioluminescence rhythms detected from each well showed almost the same period lengths and phases (Fig. 1B).
We made a set of certain criteria to select candidate inhibitors focused on period-length change: 1) changed period-length, 2) arrhythmic, 3) unchanged period-length (see Fig. S1, S2 and S3, respectively). Among the criteria we chose only inhibitors showing period-length change, since inhibitors showing arrhythmic phenotype in clock oscillation must be appearing through a mixed effect, such as toxic effect, although these may also contain some important candidate for circadian clock regulation.
Through the screening assay, we could obtain seven inhibitors showing apparent period-length change: IC261 (CKIε,δ inhibitor), DRB (5,6-dichloro-1-β-D-ribofuranosyl-benzimidazole; CKII inhibitor), LY294002 (PI3K inhibitor), SP600125 (JNK inhibitor), BML-297 (Akt inhibitor), SB203580 and SB202190 (p38 MAPK inhibitors) (Fig. 1). Among them, although p38 MAPK inhibitors showed prolonged period length, the bioluminescence intensity after treatment by these compounds were severely decreased. Also the effect of Akt inhibitor on the cellular circadian period-length seemed to be weak. Thus, for detailed analysis, we here chose the IC261, DRB, LY294002 and SP600125 inhibitors investigate the regulation mechanisms of circadian period-length in mammalian cells.
Next, to avoid the possibility that the effects of these kinase inhibitors were C6 cell specific phenomena, we investigated the effect of these compounds also using rat-1 fibroblast cells. The treatment of these kinase inhibitors resulted in the longer period length of mPer2:luc driven bioluminescence oscillation in rat-1 cells dose dependently (Fig. 2A and B). In these confirmation experiments, we used DMAT (2-dimethylamino-4,5,6,7-tetrabromo-benzimidazole) instead of DRB, since DMAT is a specific CK2 inhibitor and DRB is known to affect RNA polymelase II activity as well [13]. Using Bmal1:luc reporter stably transfected cells, we confirmed similar changes in their circadian period lengths (data not shown). These results suggest that various kinase and intracellular signaling pathways may contribute to maintaining the stable period length of the mammalian circadian clock.
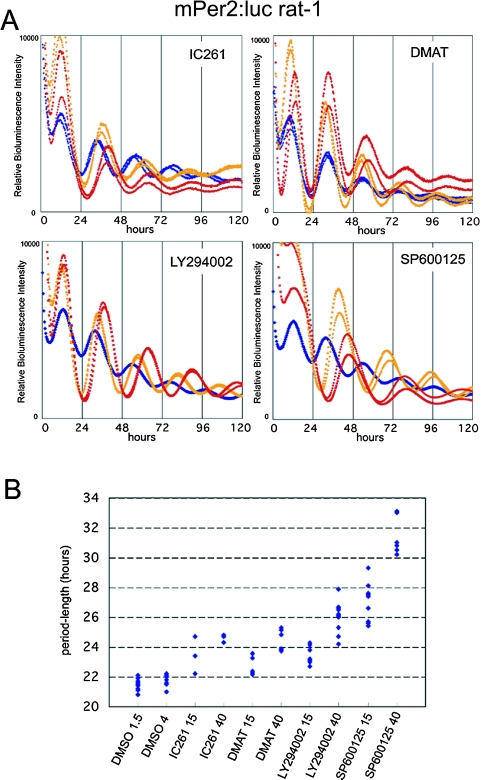
Fig. 2.
Inhibitors against CKIεδ, CKII, PI3-K amd JNK dose-dependently extend the period-length of mPer2 promoter driven molecular oscillation in rat-1 cells. (A) Kinase inhibitor screening assay revealed that the inhibitors against CKIεδ (IC261), CKII (DMAT), PI3-K (LY294002) and JNK (SP600125) dose-dependently extend the period-length of mPer2 promoter driven molecular oscillation in rat-1 cells. Blue dots represent the DMSO treatment as their control. Yellow and red dots represent 15 µM and 40 µM concentrations of each inhibitor in mPer2:luc single stable transfected rat-1 cells. Two samples were analyzed in each concentration. (B) Quantitative data of period-length in each kinase inhibitor treatment. The numbers 15 and 40 after each inhibitor indicate their final concentration. The numbers 1.5 and 4 after DMSO indicate the amount of DMSO (1.5 µl and 4 µl, respectively) applied in the 2 ml culture medium.
IV. Discussion
Here we suggested the importance of protein phosphorylation by various kinases through various regulation pathways to maintain the period-length of mammalian circadian clock. In this study, we performed a screening assay to search the candidate kinases contributing to circadian period-length control, and we could detect six kinases as candidates. For long time, it has been believed that the TTFL generates the circadian rhythm, and this model is based on the “cyclic accumulation of clock gene products”. In other words, the accumulation levels of core clock components would reflect the time-of-day [10]. However, recent studies including our works suggest that constitutive high-level accumulation of core clock component such as mPer1, mPer2 and mCry1 do not stop the oscillation in mammalian circadian clock [2, 9, 11, 19, 20]. These results strongly suggest that additional unknown mechanisms must reside in the mammalian circadian clock system.
In this study, we revealed that the various chemical compounds known as inhibitors for various kinases change the circadian period-length in mammalian cells. Among the targets of these inhibitors, the function of CKIεδ is well investigated in several studies. CKIεδ phosphorylates mPER proteins and control their stability. A recent study reported that CKII phosphorylates mPER2 protein and regulates its stability [8]. This result is also compatible with our data of this study. Gathering all observation in this study, we propose a possible model of mammalian circadian clock system (Fig. 3). This model suggests two mechanistic insights of the mammalian circadian clock: 1) CKIεδ and CKII collaboratively regulate the period-length through controlling the status of mPER2 protein; and 2) multiple pathways control the period-length via multiple clock components in the circadian clock oscillator. Our findings may provide multiple possible therapeutic targets to control circadian rhythm disorder.
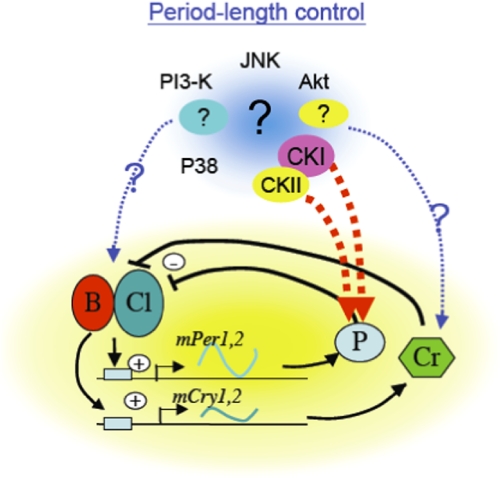
Fig. 3.
Possible model to explain the circadian period-length regulation. Various kinases contribute to control period-length via the various target components comprising the circadian feedback loop. The activity fluctuation of these kinases may be correlated with each other to express a self-sustaining robust circadian oscillation with stable period-length.
V. Acknowledgments
We wish to thank Dr. T. Kondo (Nagoya University) for support and technical advice and discussion. This study was supported by a Grant-in-Aid (K.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
This paper was presented as Young Investigator Award Lecture in 48th Annual Meeting of Japan Society of Histochemistry and Cytochemistry (Yamanashi).
Supplementary Materials
Fig. S1. Results of the first screening examining the effects of kinase inhibitors on circadian period-length in C6 cells. Raw data are presented here. We could detect longer period-length in treatment with DRB (Casein Kinase II), LY294002 (PI3-Kinase), SP600125 (c-Jun N-terminal Kinase), SB203580 (p38 MAPK), SB202190 (p38 MAPK) and BML-257 (Akt). In this screening, we could not detect shorter period-lengths in any kinase inhibitor treatment.
Fig. S2. Several kinase inhibitors resulted in an arrhythmic mPer2:luc-driven bioluminescence. Three representative data are shown. Treatment by some inhibitors showed strong bioluminescence intensity but no apparent oscillation like HA-1077 (upper panel: PKA and PKG inhibitor). Treatment by some inhibitors showed a one cycle up-and-down bioluminescence and then an arrhythmic like H-9 (middle panel: PKA, PKG, MLCK and PKC inhibitor). Treatment by some inhibitors showed very low bioluminescence intensity and no oscillation like KN-93 (lower panel: CaMKII inhibitor).
Fig. S3. Several kinase inhibitors resulted in no apparent change in the phase or period-length in mPer2:luc driven bioluminescence. Three representative data are shown. Tyrphostin51, AG370 and HDBA are inhibitors against EGFRK, PDGFRK and EGFRK, respectively.
VI. References
- 1.Busino L., Bassermann F., Maiolica A., Lee C., Nolan P. M., Godinho S. I., Draetta G. F., Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome protein. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 2.Fan Y., Hida A., Anderson D. A., Izumo M., Johnson C. H. Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr. Biol. 2007;17:1091–1100. doi: 10.1016/j.cub.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujioka A., Takashima N., Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochem. Biophys. Res. Commun. 2006;346:169–174. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 4.Gachon F., Nagoshi E., Brown S. A., Ripperger J., Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 5.Godinho S. I., Maywood E. S., Shaw L., Tucci V., Barnard A. R., Busino L., Pagano M., Kendall R., Quwailid M. M., Romero M. R., O’neill J., Chesham J. E., Brooker D., Lalanne Z., Hastings M. H., Nolan P. M. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 6.Kiyohara Y. B., Tagao S., Tamanini F., Morita A., Sugisawa Y., Yasuda M., Yamanaka I., Ueda H. R., van der Horst G. T., Kondo T., Yagita K. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. U S A. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowrey P. L., Takahashi J. S. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maier B., Wendt S., Vanselow J., Wallach T., Reischl S., Oehmke S., Schlosser A., Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Numano R., Yamazaki S., Umeda N., Samura T., Sujino M., Takahashi R., Menaker M., Tei H. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl. Acad. Sci. U S A. 2006;103:3716–3721. doi: 10.1073/pnas.0600060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura H., Yamaguchi S., Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res. 2002;309:47–56. doi: 10.1007/s00441-002-0572-5. [DOI] [PubMed] [Google Scholar]
- 11.Okano S., Akashi M., Hayasaka K., Nakajima O. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neurosci. Lett. 2009;451:246–251. doi: 10.1016/j.neulet.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Reppert S. M., Weaver D. R. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 13.Sarno S., Ruzzene M., Frascella P., Pagano M. A., Meggio F., Zambon A., Mazzorana M., Di Maira G., Lucchini V., Pinna L. A. Development and exploitation of CK2 inhibitors. Mol. Cell. Biochem. 2005;274:69–76. doi: 10.1007/s11010-005-3079-z. [DOI] [PubMed] [Google Scholar]
- 14.Schibler U., Naef F. Cellular oscillators: rhythmic gene expression and metabolism. Curr. Opin. Cell Biol. 2005;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Siepka S. M., Yoo S.-H., Park J., Song W., Kumar V., Hu Y., Lee C., Takahashi J. S. Circadian mutant overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanselow K., Vanselow J. T., Westermark P. O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y., Toh K., Jones C., Shin J.-Y., Fu Y.-H., Ptacek L. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yagita K., Tamanini F., Yasuda M., Hoijmakers J. H. J., van der Horst G. T. J., Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto Y., Yagita K., Okamura H. Role of cyclic mPer2 expression in the mammalian cellular clock. Mol. Cell. Biol. 2005;25:1912–1921. doi: 10.1128/MCB.25.5.1912-1921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka I., Koinuma S., Shigeyoshi Y., Uchiyama Y., Yagita K. Presence of robust circadian clock oscillation under constitutive over-expression of mCry1 in rat-1 fibroblasts. FEBS Lett. 2007;581:4098–4102. doi: 10.1016/j.febslet.2007.07.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Results of the first screening examining the effects of kinase inhibitors on circadian period-length in C6 cells. Raw data are presented here. We could detect longer period-length in treatment with DRB (Casein Kinase II), LY294002 (PI3-Kinase), SP600125 (c-Jun N-terminal Kinase), SB203580 (p38 MAPK), SB202190 (p38 MAPK) and BML-257 (Akt). In this screening, we could not detect shorter period-lengths in any kinase inhibitor treatment.
Fig. S2. Several kinase inhibitors resulted in an arrhythmic mPer2:luc-driven bioluminescence. Three representative data are shown. Treatment by some inhibitors showed strong bioluminescence intensity but no apparent oscillation like HA-1077 (upper panel: PKA and PKG inhibitor). Treatment by some inhibitors showed a one cycle up-and-down bioluminescence and then an arrhythmic like H-9 (middle panel: PKA, PKG, MLCK and PKC inhibitor). Treatment by some inhibitors showed very low bioluminescence intensity and no oscillation like KN-93 (lower panel: CaMKII inhibitor).
Fig. S3. Several kinase inhibitors resulted in no apparent change in the phase or period-length in mPer2:luc driven bioluminescence. Three representative data are shown. Tyrphostin51, AG370 and HDBA are inhibitors against EGFRK, PDGFRK and EGFRK, respectively.