Abstract
In the plant blue-light sensor phototropin, illumination of the chromophoric LOV domains causes activation of the serine/threonine kinase domain. Flavin mononucleotide (FMN) is a chromophore molecule in the two LOV domains (LOV1 and LOV2), but only LOV2 is responsible for kinase activation. Previous studies reported an important role of an additional helix connected to the C-terminal of LOV2 (Jα helix) for the function of phototropin; however, it remains unclear how the Jα helix affects light-induced structural changes in LOV2. In this study we compared light-induced protein structural changes of the LOV2 domain of Arabidopsis phot1 in the absence (LOV2-core) and presence (LOV2-Jα) of the Jα helix by Fourier-transform infrared spectroscopy. Prominent peaks were observed only in the amide-I region (1650 (−)/1625 (+) cm−1) of LOV2-Jα at physiological temperatures (≥260 K), corresponding to structural perturbation of the α-helix. The peaks were diminished by point mutation of functionally important amino acids such as Phe-556 between FMN and the β-sheet, Gln-575 being hydrogen-bonded with FMN, and Ile-608 on the Jα helix. We thus conclude that a light signal is relayed from FMN through these amino acids and eventually changes the interaction between LOV2-core and the Jα helix in Arabidopsis phot1.
Introduction
Plants have developed many sensory systems to adapt to environmental variations in light, gravity, temperature, and chemical substrates. Light is one of the most important environmental factors for plants because they use it as an energy source. Plants have three major photoreceptor proteins to sense the intensity, direction, and quality of the light environment. Phytochrome (1) senses red/far-red light and acts as a photoreversible molecular switch. Other photoreceptor proteins, such as cryptochrome (2) and phototropin (3–6), respond to blue light; however, a new class of blue light receptors, FKF1 (flavin-binding Kelch repeat F-box 1) family members (including FKF1 (7), ZTL (8) and LKP2 (9)), respond to UV-A/blue light (320–500 nm). Phototropin (phot), which was originally identified as a photoreceptor for phototropism in Arabidopsis (10), has also been found to regulate chloroplast relocation movements (11–13), light-induced stomatal opening (14), cotyledon expansion (15), leaf expansion (16), and rapid inhibition of hypocotyl elongation (17). These functions are deeply involved in optimizing the efficiency of photosynthesis. Most higher plants have two isoforms of phot: phot1 and phot2 (6). Stomatal opening is mediated redundantly by both phot1 and phot2 (14). In contrast, phot1 and phot2 share tropic responses and chloroplast accumulation, depending on the fluence rate of light in Arabidopsis (12), whereas chloroplast avoidance is regulated by only phot2 (11).
Phototropins are composed of ∼1000 amino acid residues and two prosthetic flavin mononucleotide (FMN) molecules (Fig. 1). The N-terminal half has two chromophoric domains, each of which binds an FMN molecule noncovalently, and the C-terminal half has a serine/threonine kinase domain. The photochemical reaction of FMN yields kinase activation through the domain-domain interaction change, although the mechanism is not yet clear. The chromophoric domains (∼100 residues) are named LOV1 and LOV2 because they have primary (18) and tertiary (19) structures highly homologous to bacterial light-sensor photoactive yellow protein (PYP), oxygen-sensor FixL, and voltage sensor HERG of a channel protein (thus the domain is called the LOV (light, oxygen, and voltage sensing) domain). The protein fold belongs to the Per-Arnt-Sim (PAS) superfamily. Previous x-ray crystallography analyses of the LOV2 domain of Adiantum neochrome1 (neo1) (19) and the LOV1 domain of Chlamydomonas phot (20) showed that the structures of the two LOV domains in phototropin are very similar.
Figure 1.
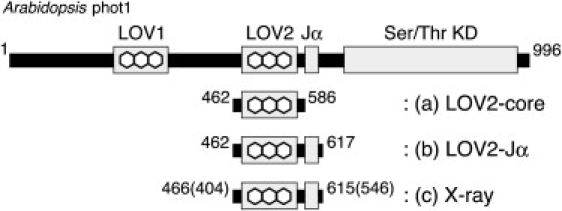
Schematic illustration of the LOV2 constructs of phototropin used in this study. Images a and b correspond to the LOV2 domain without (LOV2-core) and with an extra helix region (LOV2-Jα) at the C-terminal side, respectively. The LOV2 construct of oat phot1 used in the x-ray study (39) is also shown in c, where the number in parentheses represents the amino acid numbering in oat phot1. Note that seven amino acids are deleted between LOV2 and Jα in oat compared with Arabidopsis.
The primary photoreaction in the LOV domain is an adduct formation between FMN and a nearby cysteine (21–25). After light absorption by FMN, intersystem crossing leads to the formation of a triplet-excited state that absorbs at 660 nm (L660) and appears with a time constant of 3 ns in Adiantum neo1-LOV2 and oat phot1-LOV2 (26). Then, adduct formation accompanies the appearance of the S390 intermediate with time constants of 4 μs in oat phot1-LOV2 (23) and 0.9 and 4 μs in Chlamydomonas phot-LOV1 (27). Although various models have been proposed for the reactive cysteine, previous FTIR studies revealed that cysteine is protonated in both the ground (28–31) and triplet-excited (32) states of FMN. A dynamical protein motion probably plays an important role in adduct formation (33,34).
S390 is the only ground-state intermediate during the photocycle of LOV domains. Therefore, it is believed that S390 is the active state for the light-sensing function of phototropin. Although the crystal structures of the unphotolyzed (19) and S390 (25) states of neo1-LOV2 show identical surfaces, our FTIR studies indicate a highly temperature-dependent nature of the amide-I vibrations of neo1-LOV2. Structural perturbation of the loop region is prominent at low temperatures (<250 K), whereas those of the α-helix and β-sheet appear at higher temperatures (>250 K), suggesting the presence of progressive protein structural changes (35). Other FTIR (36) and circular dichroism (CD) studies (37) have also demonstrated that photoactivation of purified LOV2 includes structural changes in the LOV-domain apoprotein. NMR and x-ray crystallographic studies of extended LOV2 fragments from oat phot1 revealed that an extra α-helix (named the Jα helix) associates with the surface of LOV2 in the dark state (38,39). The Jα helix is located at the C-terminus of LOV2 and is amphipathic in nature, consisting of polar and apolar sides, the latter of which docks onto the β-sheet strands of the LOV2 core. The interaction between Jα and LOV2 is disrupted upon cysteinyl adduct formation. Recently, light-induced conformational changes of Jα helix were also observed by means of a transient grating method (40–43). However, it is still unclear how the conformational changes in the LOV2 core by the adduct formation are transmitted to the Jα helix.
In this study we examined the role of Jα in the photoactivation of LOV2 by means of FTIR spectroscopy, a powerful tool for investigating protein structural changes. To that end, we prepared the LOV2 domains from Arabidopsis phot1 in the absence (LOV2-core) and presence (LOV2-Jα) of the Jα helix. We observed a significant spectral difference in the helical structure (1660–1640 cm−1) of the amide-I region between LOV2-core and LOV2-Jα, which suggests that they originate from conformational changes of the Jα helix. To put this observation on a firm basis, we also measured three mutants: I608E, Q575L, and F556L. Harper et al. (44) reported that the I608E mutant exhibits constitutive kinase activity. Since Ile-608 faces the β-sheet on the Jα helix, an Ile-to-Glu mutation probably disrupts the interaction between the LOV-core and the Jα helix. The other two mutants were the replacement of amino acids in the LOV2-core. Previous x-ray and FTIR studies invoked the importance of Gln-575 (Gln-1029 in the case of Adiantum neo1) on the Iβ strand, whose hydrogen bond with FMN is switched upon formation of S390 (45). It was found that kinase activation is significantly decreased for the Q575L mutant of Arabidopsis phot1 (46). On the other hand, our recent FTIR study (47) showed that Phe-556 (Phe-1010 in the case of Adiantum neo1) on the Hβ strand is a determinant in characterizing the LOV2 domain, whereas the corresponding amino acid is leucine for LOV1. These three amino acids (Ile-608, Gln-575, and Phe-556) are the candidates which are responsible for intramolecular signal relay. In fact, the observed prominent bands of the α-helix in the wild-type almost disappeared in these mutants. We propose an activation mechanism of the LOV2 domain in Arabidopsis phot1 on the basis of our previously reported results and the FTIR analysis presented here.
Materials and Methods
Construction of expression vectors
With the use of cDNA of Arabidopsis phot1 as a template, the required DNA fragments with appropriate restriction sites were synthesized by polymerase chain reaction and oligonucleotide primers as described previously (48,49). Amplified DNA was isolated, digested, and cloned into a pGEX4T1 bacterial expression vector (GE Healthcare UK Ltd., Buckinghamshire, UK) as a translational fusion to glutathione S-transferase (GST). The following two polypeptides, fused with GST in the N-terminus, were prepared: GST-LOV2-core (Lys-462-Arg-586) and GST-LOV2-Jα (Lys-462-Asp-617). In addition to the wild-type polypeptides, the F556L and Q575L mutants were prepared as LOV-core side mutants in each construct, and the I608E mutant was prepared as a Jα-helix side mutant in the LOV2-Jα construct. The amino acid substitutions were introduced with the use of a Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, CA), in accordance with the supplier's instructions. These amino acid substitutions were confirmed by DNA sequencing.
Expression and purification of recombinant proteins
GST fusion proteins were prepared by overexpression systems with Escherichia coli as described previously (48,49). The purified proteins were concentrated to give a final concentration of 2.5 mg/mL by using an Amicon Ultra PL-10 (Millipore, Billerica, MA) and dialyzed against 1 mM K/phosphate buffer (pH 7). 4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride, which works as an inhibitor of thrombin protease, was added to give a final concentration of 0.1 mM. Then 70–80 μL of the solution were placed on a BaF2 window and the dry films were hydrated by dropping H2O next to the film on the window. The hydration conditions for each sample were identical. The hydrated films were stable, as judged from their spectral integrity after storage for a few months at −80°C.
FTIR spectroscopy
Infrared spectra of the hydrated films were measured with an FTS-40 (Bio-Rad) spectrophotometer as described previously (31,34,35,45). Low-temperature spectra were measured by using a cryostat (Optistat DN; Oxford Instruments, Oxfordshire, UK) and a temperature controller (ITC 4; Oxford Instruments, Oxfordshire, UK) with liquid nitrogen as the coolant. Hydrated films were illuminated by a >400 nm light for 1 min, which was supplied by a combination of a halogen-tungsten lamp (1000 W) and a long-pass filter (L42; AGC Techno Glass, Chiba, Japan).
Results
Light-induced FTIR spectral changes in LOV2-core and LOV2-Jα at various temperatures
Fig. 2 shows the light minus dark difference FTIR spectra of LOV2-core (solid lines) and LOV2-Jα (dotted lines) in the 1800–1000 cm−1 region at several temperatures. It is obvious that there are few spectral differences at 150 and 250 K, indicating that protein structural changes of the LOV2 domain are not influenced by the presence of the Jα helix. In contrast, a clear difference between LOV2-core and LOV2-Jα was observed at 260, 273, and 295 K. This observation can be interpreted in terms of structural changes in either Jα itself or the LOV2-core affected by Jα. It should be noted that the spectra of the LOV2-core (solid lines) are almost identical from 250 to 295 K, whereas there are some spectral alterations at 150 K. This is clearly seen in Fig. S1 in the Supporting Material, where the spectra at 1750–1600 cm−1 involving the amide-I region overlap well with each other except at 150 K.
Figure 2.
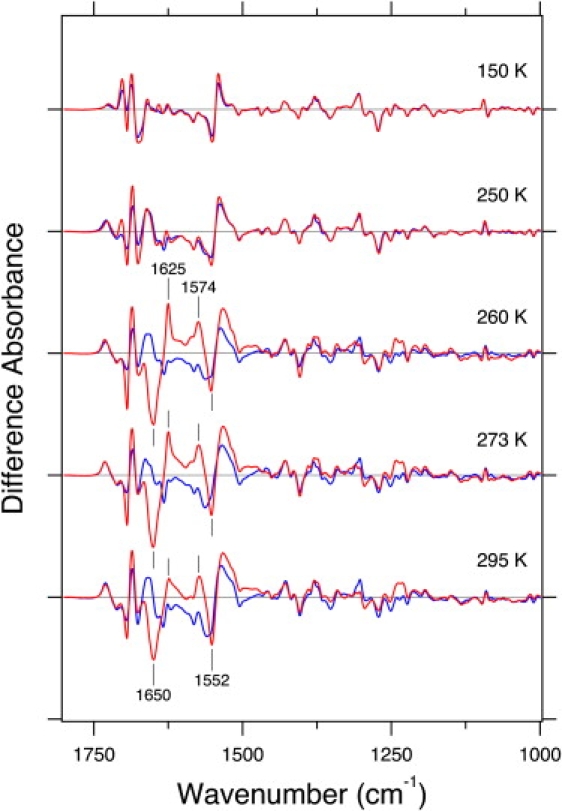
Light minus dark difference infrared spectra for LOV2-core (solid lines) and LOV2-Jα (dotted lines) in the 1800–1000 cm−1 region. Spectra are measured at 150, 250, 260, 273, and 295 K. One division of the y axis corresponds to 0.02 absorbance units.
Although there are various spectral differences at 1500–1000 cm−1 at higher temperatures (260–295 K), the most characteristic for LOV2-Jα are the amide-I bands at 1650 (−) and 1625 (+) cm−1, and the amide-II band at 1574 (+) cm−1. The negative amide-I frequency at 1650 cm−1 is typical for the α-helix, and the sharp negative peak in the amide-II region at 1552 cm−1 is also typical for the α-helix. Thus, it is likely that structural changes in the α-helix take place in LOV2-Jα at 260-295 K, but not at lower temperatures or in the LOV2-core. In general, protein structures around the FMN chromophore only change at low temperatures. These results strongly suggest that the observed structural perturbation of the α-helix originates from Jα, which is further examined by measuring mutant proteins. Below, we highlight the amide-I region in more detail.
Fig. 3 shows the light minus dark difference infrared spectra of LOV2-Jα in the 1750–1600 cm−1 region, where amide-I vibration appears. The C2=O and C4=O stretching vibrations of FMN also appear in this frequency region, and a previous study of neo1-LOV2 (50) identified these frequencies at 1677 and 1711 cm−1 by use of 13C2=O- and 13C4=O-labeled FMN, respectively. Therefore, it is likely that the bands at 1676 and 1712 cm−1 in Fig. 3 b originate from C2=O and C4=O stretching vibrations, respectively, in the unphotolyzed state of LOV2-Jα. The corresponding bands in the S390 intermediate are possibly at 1686 and 1730 cm−1 for C2=O and C4=O stretching vibrations, respectively (Fig. 3 b). Higher-frequency shifts of the C2=O and C4=O stretches commonly occur upon formation of S390 in Arabidopsis phot1-LOV2 and neo1-LOV2.
Figure 3.
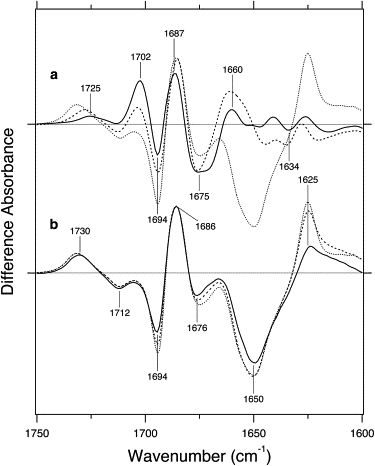
Light minus dark difference infrared spectra of LOV2-Jα in the 1750-1600 cm−1 region measured at 150 (solid line in a), 250 (broken line in a), 260 (dotted lines in a and b), 273 (broken line in b), and 295 (solid line in b) K. One division of the y axis corresponds to 0.017 absorbance units.
All other vibrations in this frequency region originate from those from protein, particularly amide-I mode reflecting C=O stretches of the peptide backbone. It is well known that the amide-I frequencies are characteristic of secondary structures, such as a loop (1670∼1690 cm−1), an α-helix (∼1650 cm−1), and a β-sheet (1620∼1640 cm−1) (51). Since the C2=O stretch of FMN and the amide-I vibration of the loop appear at a similar frequency (1670∼1690 cm−1), isotope labeling is necessary for band assignment of C=O stretches from apoprotein and FMN as was for neo1-LOV2 (50). Here we interpret the vibrational bands based on the comparison with the previous study (50) and the characteristic frequencies of amide-I vibrations. Although there are temperature-dependent spectral changes at >1670 cm−1, prominent spectral alterations are observed at 1650–1620 cm−1. At 150 and 250 K, there are no significant differences at this frequency (Fig. 3 a), implying that there are no structural changes in the α-helix or the β-sheet. On the other hand, prominent peaks newly appear at 1650 (−) and 1625 (+) cm−1 at 260 K. The negative 1650-cm−1 band, characteristic of the α-helix, was not observed for the LOV2-core (Fig. S1). Therefore, disruption of the α-helix structure presumably originates from the Jα helix.
FTIR spectral comparison of the wild-type and mutant proteins
The data in Figs. 2 and 3 strongly suggest that the observed structural perturbation of the α-helix originates from Jα. We tested three mutants for further clarification. Fig. 4 a compares light minus dark difference infrared spectra among LOV-core (dotted lines), LOV-Jα (thin solid lines), and the I608E LOV2-Jα mutant (thick solid lines). As is clearly seen, the spectrum of I608E LOV2-Jα is similar to that of the LOV2-core, but not to wild-type LOV2-Jα. The prominent bands at 1650 (−)/1625 (+) cm−1 are never observed for the I608E mutant (260–295 K). Similar spectral changes of C4=O and C2=O stretching vibrations between LOV2-core and I608E LOV2-Jα suggest that similar protein structural changes take place inside LOV2-core, but light-induced structural changes of the LOV-core are never transmitted to the Jα helix in I608E LOV2-Jα. Harper et al. (43) reported that the I608E mutation disrupted the interaction between the Jα helix and LOV-core. The constitutive activity of kinase in I608E suggests the importance of the interaction between Ile-608 and its counterpart located in the LOV-core side for transmitting structural changes (44). This observation is fully consistent with the observation by Harper et al. (43), and the prominent negative and positive bands observed in LOV2-Jα at 1650 (−)/1625 (+) cm−1 are closely associated with light-induced kinase activity.
Figure 4.
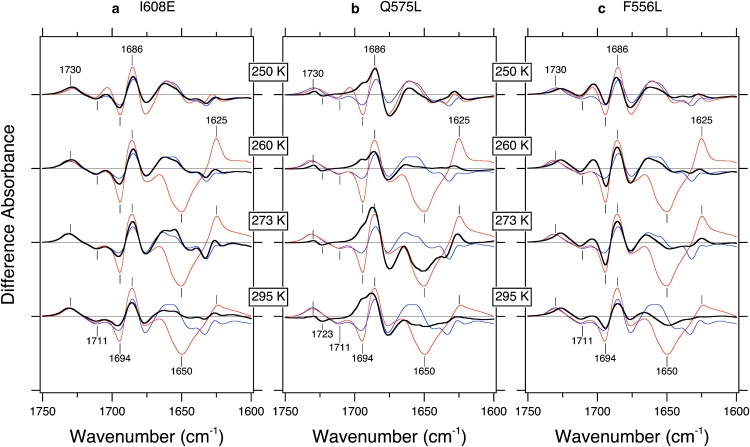
(a) Light minus dark difference infrared spectra for the LOV2-Jα I608E mutant (thick solid lines), LOV2-Jα wild-type (thin solid lines), and LOV2 wild-type (dotted lines). (b) Light minus dark infrared difference spectra for the LOV2-Jα Q575L mutant (thick solid lines), LOV2-Jα wild-type (thin solid lines), and LOV2 wild-type (dotted lines). (c) Light minus dark infrared difference spectra for the LOV2-Jα F556L mutant (thick solid lines), LOV2-Jα wild-type (thin solid lines), and LOV2 wild-type (dotted lines) in the 1750–1600 cm−1. Spectra are measured at 250, 260, 273, and 295 K. One division of the y axis corresponds to 0.02 absorbance units.
Fig. 4 b compares light minus dark difference infrared spectra among LOV-core (dotted lines), LOV-Jα (thin solid lines), and the Q575L LOV2-Jα mutant (thick solid lines) in the 1750–1600 cm−1 region. The prominent bands at 1650 (−)/1625 (+) cm−1 are also not observed at 260 and 295 K. Of interest, Q575L LOV2-Jα exhibits the 1650 (−)/1625 (+) cm−1 bands only at 273 K, suggesting that activation of the Jα helix is not completely lost in this mutant. The interaction between LOV-core and Jα presumably is normal for Q575L LOV2-Jα, but protein structural changes inside LOV-core must be perturbed. In fact, there are many spectral differences from the wild-type in the 1750–1660 cm−1 region. The lack of bands at 1730 (+)/1711 (−) cm−1 for Q575L LOV2-Jα supports the assignment as the C4=O stretching vibration, because the side chain of Gln-575 forms a hydrogen bond with the C4=O group of FMN. Newly appeared small bands at 1730 (+)/1723 (−) cm−1 for Q575L LOV2-Jα presumably originate from the C4=O stretching vibration of FMN, where the hydrogen bond of the C4=O group is weakened. We previously reported a similar observation for the neo1-LOV2 domain in Adiantum (45). In addition, the negative band at 1694 cm−1 disappeared for Q575L LOV2-Jα, though it is observed for both LOV-core and LOV2-Jα. It is likely that the loop structure is significantly modified in this mutant. A physiologically important role of glutamine at this position has been also reported (46). The results presented here show that the rearrangement of the hydrogen-bonding network between Gln and FMN is an eventual prerequisite for the signal relay to be transferred to the Jα helix.
Fig. 4 c compares light minus dark difference infrared spectra among LOV-core (dotted lines), LOV-Jα (thin solid lines), and the F556L LOV2-Jα mutant (thick solid lines) in the 1750–1600 cm−1 region. Phenylalanine at this position is conserved for LOV2, whereas the corresponding amino acid for LOV1 is leucine in almost all plant phototropins. The Jα helix is believed to exist only at the C-terminal side of LOV2, not LOV1. We proposed that Phe and Leu are one functional determinant between LOV2 and LOV1, respectively, and the recent Phe-to-Leu mutation of neo1-LOV2 indeed converted LOV2 to LOV1-like (47). Therefore, we expected that F556L LOV2-Jα in Arabidopsis phot1 would impair normal protein structural changes. As expected, the prominent negative and positive bands at 1650 (−)/1625 (+) cm−1 disappeared for F556L LOV-Jα. In addition, the positive band at 1730 cm−1 in LOV2-Jα wild-type is downshifted at higher temperatures. Thus, rearrangement of the hydrogen-bonding network around FMN and successive secondary structural changes in the LOV2 core are influenced by this mutation.
Double difference FTIR spectra
To discern the signal from the Jα helix, Fig. 5 shows double difference spectra for the wild-type: LOV2-Jα wild-type minus LOV2-core wild-type (thick solid lines); I608E mutant: LOV2-Jα I608E mutant minus LOV2-core wild-type (thin solid lines); Q575L mutant: LOV2-Jα Q575L mutant minus LOV2-core Q575L (broken lines); and F556L mutant: LOV2-Jα F556L mutant minus LOV2-core F556L (dotted lines) in the 1800–1000 cm−1 region measured at 250 and 260 K. There are only small differences in wild-type at 250 K, indicating that the Jα helix does not undergo structural changes at this step. On the other hand, prominent negative and positive bands appeared at 1650 (−)/1625(+) cm−1 in wild-type at 260 K, indicating that the Jα helix undergoes a structural change at this step. Thus, a transition phase exists between 250 and 260 K. The prominent positive and negative bands at 1650 (−)/1625 (+) cm−1 disappeared in each mutant. This result also strongly suggests that the structural change of the Jα helix does not occur in these mutants.
Figure 5.
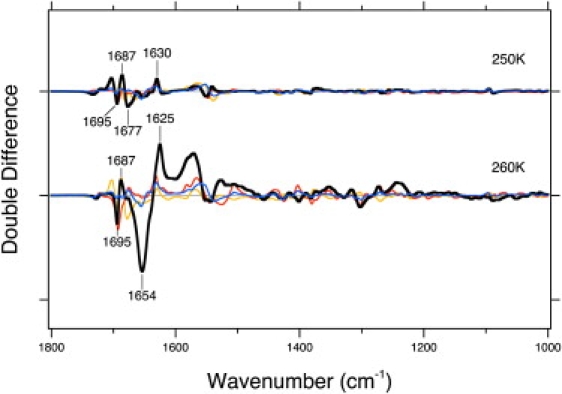
Double difference spectra for the wild-type: LOV2-Jα wild-type minus LOV2 wild-type (thick solid lines); I608E mutant: LOV2-Jα I608E mutant minus LOV2 wild-type (thin solid lines); Q575L mutant: LOV2-Jα Q575L mutant minus LOV2 Q575L (broken lines); and F556L mutant: LOV2-Jα F556L mutant minus LOV2 F556L (dotted lines). One division of the y axis corresponds to 0.02 absorbance units.
Discussion
In this study we aimed to reveal the role of the Jα helix at the C-terminus of the LOV2 domain in light-induced activation of phototropin. To achieve this, we compared light-induced FTIR spectra between LOV2-core and LOV2-Jα in Arabidopsis phot1. In the case of LOV2-core, there is no prominent band in the 1660–1600 cm−1 region, suggesting that the secondary structure of the α-helix and the β-sheet hardly change their conformation. In contrast, LOV2-Jα exhibits a secondary structural alteration of the α-helix at ≥260 K, where the observed helix probably originates from the Jα helix. The importance of the Jα helix as a switch was first indicated by NMR studies of extended LOV2 fragments from oat phot1, where the Jα helix associates or dissociates with LOV2-core in the dark or light state, respectively (38). The Jα helix is located at the C-terminus of LOV2 and is amphipathic in nature, consisting of polar and apolar sides, the latter of which docks onto the β-sheet strands of LOV2-core. Light-induced conformational changes of the Jα helix have also been observed by means of a transient grating method (40–43). In addition, a recent x-ray crystallography study of oat phot1-LOV2 showed an association of LOV-core and the Jα helix (39). The FTIR study presented here clearly provides additional structural information regarding the role of the Jα helix in structural changes for activation of phototropin.
The secondary structures corresponding to the 1650 (−)/1625(+) cm−1 bands are noteworthy. The negative band at 1650 cm−1 can be assigned to the Jα helix, which is supported by the spectral comparison between LOV2-core and LOV2-Jα (Fig. 2) and the mutation study (Fig. 4). The frequency of the positive band at 1625 cm−1 is characteristic of a β-sheet. Therefore, one possibility is that the Jα region is rearranged into a β-sheet to maintain a hydrophobic interaction between the LOV2-core surface and the Jα region. Another possibility is suggested by the less-intense positive peak at 1625 cm−1 compared with the negative 1650-cm−1 band. It is known that the intensity of amide-I vibrations of the α-helix is enhanced because of collective motion of the C=O stretching vibrations in the same phase (51); therefore, molecular coefficients of amide-I vibrations can be higher in the α-helix than in the loop and random structures. The amide-I frequency of a random structure has been reported at 1660–1640 cm−1, which is close to that of the α-helix (51). The random coil structure presumably is not stable in the unphotolyzed state, which is not seen in the crystal structure, but is possibly involved in transient intermediate states. Thus, if the conversion of an α-helix to a random structure takes place, observation of an only negative band at ∼1650 cm−1 may be expected. The less-intense positive peak at 1625 cm−1 compared with the negative 1650-cm−1 band suggests that this is the case. A positive 1625-cm−1 band may originate from aggregates of unfolded structures. The conversion from an α-helix to a random coil and unfolded structures is consistent with findings from the transient grating studies (40–43).
By using mutant proteins, we were able to gain insight into the intramolecular signal relay mechanism. Fig. 6 illustrates the location of the amino acids and scheme of the activation of the LOV2 domain. Ile-608 is located in the middle of the Jα helix and faces toward the Iβ strand of LOV2-core (Fig. 6, left). It was shown that the I608E mutant has no interaction with the LOV-core, yielding constitutive activation (38,44). The FTIR study provided difference spectra of I608E LOV2-Jα similar to those of LOV2-core, which are fully consistent with the previous reports. These results strongly suggest that the Jα helix does not fold successfully or that it folds into a helical structure, but does not interact with LOV-core. Thus, the spectral change at 1650 (−)/1625 (+) cm−1 seems to be a prerequisite for the LOV2 domain to activate the kinase domain.
Figure 6.
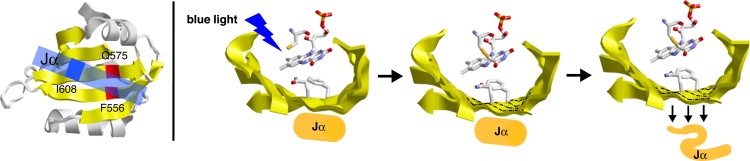
Schematic illustration of signal transduction from LOV-core to the Jα helix in phot1-LOV2.
Gln-575 is located on the Iβ strand and donates a hydrogen bond to the C4=O group of FMN (Fig. 6, left). The functional importance was first suggested by x-ray crystallographic structures of the dark and light states of neo1-LOV2 (25), and subsequently by FTIR and biochemical studies (45,46). In the study presented here, the Q575L mutant showed that the positive and negative bands at 1650 (−)/1625 (+)cm−1 are not completely observed at 260 K; these bands appeared slightly at 273 K, and then almost disappeared again at 295 K. These results suggest that the Jα helix is still affected by the conformational change of the LOV-core side in the Q575L mutant, although this perturbation is considerably smaller than that in the wild-type. These perturbations may not be sufficient for kinase regulation. Jones et al. (46) recently reported that the kinase activation level of the Q575L mutant was significant reduced, and that Q575L plays an important role in light-induced kinase activation.
Phe-556 is located on the Hβ strand and sandwiches FMN with a reactive cysteine (Fig. 6, left). We recently reported the important role of Phe at this position as one of the determinants of the LOV2 domain for Adiantum neo1 (47). In the study presented here, we observed no helical perturbation for F556L LOV2-Jα. Therefore, we infer that the F556L mutant has no kinase activation in a light-dependent manner. Since this amino acid is conserved only for the LOV2 domain that possesses an additional Jα helix, the introduction of Phe at this position might lead to the creation of a signal transduction system using the Jα helix.
Fig. 6 (right) shows that the LOV2 domain is composed of a U-shaped β-strand that contains the FMN chromophore. Gln-575 and Phe-556 on the β-sheet are similarly in contact with FMN, but in a different manner. Light-induced adduct formation between FMN and Cys causes environmental changes for these residues, presumably leading to structural perturbations on the β-sheet region. Such structural perturbations on the β-sheet region consequently yields interaction changes between the β-sheet and the Jα helix, eventually releasing the Jα helix from the LOV-core. This scenario of light-signal transduction in the LOV2 domain is based on the FTIR results and other observations presented here.
Acknowledgments
This work was supported in part by grants in aid from the Ministry of Education, Science, Sports and Culture in Japan (19370067 to H.K. and 17084008 to S.T.).
Footnotes
D. Matsuoka's present address is Research Center for Environmental Genomics, Kobe University, Kobe, Japan.
Supporting Material
References
- 1.Quail P.H. Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell Biol. 2002;14:180–188. doi: 10.1016/s0955-0674(02)00309-5. [DOI] [PubMed] [Google Scholar]
- 2.Lin C., Shalitin D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 3.Briggs W.R., Christie J.M. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- 4.Crosson S., Rajagopal S., Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 5.Celaya R.B., Liscum E. Phototropins and associated signaling: providing the power of movement in higher plants. Photochem. Photobiol. 2005;81:73–80. doi: 10.1562/2004-08-22-IR-282. [DOI] [PubMed] [Google Scholar]
- 6.Briggs W.R., Beck C.F., Cashmore A.R., Christie J.M., Hughes J. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 8.Mas P., Kim W.Y., Somers D.E., Kay S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 9.Fukamatsu Y., Mitsui S., Yasuhara M., Tokioka Y., Ihara N. Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 2005;46:1340–1349. doi: 10.1093/pcp/pci144. [DOI] [PubMed] [Google Scholar]
- 10.Christie J.M., Reymond P., Powell G.K., Bernasconi P., Raibekas A.A. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 11.Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- 12.Sakai T., Kagawa T., Kasahara M., Swartz T.E., Christie J.M. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarillo J.A., Gabrys H., Capel J., Alonso J.M., Ecker J.R. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- 15.Ohgishi M., Saji K., Okada K., Sakai T. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101:2223–2228. doi: 10.1073/pnas.0305984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto K., Briggs W.R. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folta K.M., Spalding E.P. Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 2001;26:471–478. doi: 10.1046/j.1365-313x.2001.01038.x. [DOI] [PubMed] [Google Scholar]
- 18.Huala E., Oeller P.W., Liscum E., Han I.S., Larsen E. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 19.Crosson S., Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedorov R., Schlichting I., Hartmann E., Domratcheva T., Fuhrmann M. Crystal structures and molecular mechanism of a light-induced signaling switch: the Phot-LOV1 domain from. Chlamydomonas reinhardtii. Biophys. J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salomon M., Christie J.M., Knieb E., Lempert U., Briggs W.R. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 22.Miller S.M., Massey V., Ballou D., Williams C.H., Jr., Distefano M.D. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry. 1990;29:2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- 23.Swartz T.E., Corchnoy S.B., Christie J.M., Lewis J.W., Szundi I. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 24.Salomon M., Eisenreich W., Durr H., Schleicher E., Knieb E. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. USA. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crosson S., Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennis J.T., Crosson S., Gauden M., van Stokkum I.H., Moffat K. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 27.Kottke T., Heberle J., Hehn D., Dick B., Hegemann P. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys. J. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwata T., Tokutomi S., Kandori H. Photoreaction of the cysteine S-H group in the LOV2 domain of Adiantum phytochrome3. J. Am. Chem. Soc. 2002;124:11840–11841. doi: 10.1021/ja020834c. [DOI] [PubMed] [Google Scholar]
- 29.Ataka K., Hegemann P., Heberle J. Vibrational spectroscopy of an algal Phot-LOV1 domain probes the molecular changes associated with blue-light reception. Biophys. J. 2003;84:466–474. doi: 10.1016/S0006-3495(03)74866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bednarz T., Losi A., Gartner W., Hegemann P., Heberle J. Functional variations among LOV domains as revealed by FT-IR difference spectroscopy. Photochem. Photobiol. Sci. 2004;3:575–579. doi: 10.1039/b400976b. [DOI] [PubMed] [Google Scholar]
- 31.Iwata T., Nozaki D., Tokutomi S., Kandori H. Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry. 2005;44:7427–7434. doi: 10.1021/bi047281y. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y., Iwata T., Tokutomi S., Kandori H. Reactive cysteine is protonated in the triplet excited state of the LOV2 domain in Adiantum phytochrome3. J. Am. Chem. Soc. 2005;127:1088–1089. doi: 10.1021/ja0436897. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki D., Iwata T., Tokutomi S., Kandori H. Unique temperature dependence in the adduct formation between FMN and cysteine S-H group in the LOV2 domain of Adiantum phytochrome3. Chem. Phys. Lett. 2005;410:59–63. [Google Scholar]
- 34.Sato Y., Nabeno M., Iwata T., Tokutomi S., Sakurai M. Heterogeneous environment of the S-H group of Cys966 near the flavin chromophore in the LOV2 domain of Adiantum neochrome1. Biochemistry. 2007;46:10258–10265. doi: 10.1021/bi701022v. [DOI] [PubMed] [Google Scholar]
- 35.Iwata T., Nozaki D., Tokutomi S., Kagawa T., Wada M. Light-induced structural changes in the LOV2 domain of Adiantum phytochrome3 studied by low-temperature FTIR and UV-visible spectroscopy. Biochemistry. 2003;42:8183–8191. doi: 10.1021/bi0345135. [DOI] [PubMed] [Google Scholar]
- 36.Swartz T.E., Wenzel P.J., Corchnoy S.B., Briggs W.R., Bogomolni R.A. Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry. 2002;41:7183–7189. doi: 10.1021/bi025861u. [DOI] [PubMed] [Google Scholar]
- 37.Corchnoy S.B., Swartz T.E., Lewis J.W., Szundi I., Briggs W.R. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 2003;278:724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- 38.Harper S.M., Neil L.C., Gardner K.H. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 39.Halavaty A.A., Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 40.Eitoku T., Nakasone Y., Matsuoka D., Tokutomi S., Terazima M. Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. J. Am. Chem. Soc. 2005;127:13238–13244. doi: 10.1021/ja052523i. [DOI] [PubMed] [Google Scholar]
- 41.Nakasone Y., Eitoku T., Matsuoka D., Tokutomi S., Terazima M. Kinetic measurement of transient dimerization and dissociation reactions of Arabidopsis phototropin 1 LOV2 domain. Biophys. J. 2006;91:645–653. doi: 10.1529/biophysj.106.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakasone Y., Eitoku T., Matsuoka D., Tokutomi S., Terazima M. Dynamics of conformational changes of Arabidopsis phototropin 1 LOV2 with the linker domain. J. Mol. Biol. 2007;367:432–442. doi: 10.1016/j.jmb.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 43.Eitoku T., Nakasone Y., Zikihara K., Matsuoka D., Tokutomi S. Photochemical intermediates of Arabidopsis phototropin 2 LOV domains associated with conformational changes. J. Mol. Biol. 2007;371:1290–1303. doi: 10.1016/j.jmb.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 44.Harper S.M., Christie J.M., Gardner K.H. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 45.Nozaki D., Iwata T., Ishikawa T., Todo T., Tokutomi S. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 46.Jones M.A., Feeney K.A., Kelly S.M., Christie J.M. Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J. Biol. Chem. 2007;282:6405–6414. doi: 10.1074/jbc.M605969200. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto A., Iwata T., Tokutomi S., Kandori H. Role of Phe1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- 48.Nakasako M., Iwata T., Matsuoka D., Tokutomi S. Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry. 2004;43:14881–14890. doi: 10.1021/bi0485530. [DOI] [PubMed] [Google Scholar]
- 49.Matsuoka D., Tokutomi S. Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. USA. 2005;102:13337–13342. doi: 10.1073/pnas.0506402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwata T., Nozaki D., Sato Y., Sato K., Nishina Y. Identification of the C=O stretching vibrations of FMN and peptide backbone by 13C-labeling of the LOV2 domain of Adiantum phytochrome3. Biochemistry. 2006;45:15384–15391. doi: 10.1021/bi061837v. [DOI] [PubMed] [Google Scholar]
- 51.Krimm S., Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


