Abstract
The fusion of lipid bilayers is studied with dissipative particle dynamics simulations. First, to achieve control over membrane properties, the effects of individual simulation parameters are studied and optimized. Then, a large number of fusion events for a vesicle and a planar bilayer are simulated using the optimized parameter set. In the observed fusion pathway, configurations of individual lipids play an important role. Fusion starts with individual lipids assuming a splayed tail configuration with one tail inserted in each membrane. To determine the corresponding energy barrier, we measure the average work for interbilayer flips of a lipid tail, i.e., the average work to displace one lipid tail from one bilayer to the other. This energy barrier is found to depend strongly on a certain dissipative particle dynamics parameter, and, thus, can be adjusted in the simulations. Overall, three subprocesses have been identified in the fusion pathway. Their energy barriers are estimated to lie in the range 8–15 kBT. The fusion probability is found to possess a maximum at intermediate tension values. As one decreases the tension, the fusion probability seems to vanish before the tensionless membrane state is attained. This would imply that the tension has to exceed a certain threshold value to induce fusion.
Introduction
Fusion of biological membranes is an essential process in many areas of cell biology, ranging from vesicular trafficking and synaptic transmission to cell-cell fusion or viral fusion. Biological membranes are complex systems composed of many different lipids and proteins. For a better understanding of the fundamental processes involved, lipid vesicles are often used as simplified model systems (1). Even in the absence of proteins, such model membranes can be induced to fuse experimentally by a variety of methods.
For a fusion pore to form, drastic topological rearrangement of the two membranes and a destruction of their bilayer structure is necessary at least locally. On the other hand, lipid bilayer membranes in water are very stable structures that do not easily form holes. This makes the fusion process and its energetics an interesting problem, which has received much attention in recent years.
The initial fusion pore is believed to be a necklike connection with an initial size of ∼10 nm. The corresponding timescale has not been directly measured, but both patch-clamp methods applied to synaptic membranes (2) and ultrafast optical microscopy of giant vesicles (3) suggest that the fusion pore can be formed in <100 μs. Since it is currently not possible to resolve these length- and timescales experimentally, theoretical or computational models are employed to gain insight into the process of fusion pore formation.
Theoretical descriptions are based on elastic theories for membrane sheets, which postulate intermediate configurations and try to find the lowest energy transition states. However, these lowest energy states usually correspond to relatively high-energy barriers. Despite modifications of the assumed intermediates to lower the energy barriers, these barriers are still estimated to be ∼40 kBT.
Computer simulations such as Brownian dynamics (4), Monte Carlo simulations (5), coarse-grained molecular dynamics (MD) (6–8), dissipative particle dynamics (DPD) (9,10), and atomistic MD (11,12), on the other hand, give a molecular picture of the process and are not restricted with respect to the structure of intermediate configurations. These simulation studies observe different fusion pathways and highlight the importance of lipid conformations in the process, but they do not usually allow us to measure the energy barriers between states. One exception is a Markovian state model based on coarse-grained MD (13,14) that has managed to deduce the energy difference between the initial state and several intermediates from the transition rates.
In this study, DPD simulations have been used to probe the statistics of many fusion attempts, while still being able to simulate the relevant length- and timescales. From the statistics of the fusion time in combination with separate simulations of enforced interbilayer flips, in which one tail of a lipid molecule is moved from one bilayer to the other leading to a splayed conformation of this lipid, the energy barriers for the fusion observed in these simulations could be estimated as already outlined in Grafmüller et al. (15).
We focus on the presumably simplest way to induce lipid bilayer fusion, i.e., via a global membrane tension, which is coupled to hydrodynamics and can be directly controlled in MD (16) and DPD (9,17) simulations with explicit water. The experimentally observed frequency of fusion events increases with osmotic inflation of the vesicles (18), which indicates that the energy barriers for fusion can be reduced by increasing the membrane tension. The simulations reported here attempt to elucidate the fusion pathway of this mechanism and its dependence on specific lipid properties as well as the membrane tension, and to reveal the energy barriers between the intermediate states. Unlike experimental suggestions, previous DPD simulations (9) did not indicate any tension-dependent energy barriers. However, as shown in this article, membranes built from the parameter set used in Shillcock and Lipowsky (9) are characterized by fast exchange of lipids between adhering membranes. Investigation of the effects of individual parameters on the membrane properties have allowed us to carefully adjust the simulation parameters to 1), obtain bilayers with improved stretching behavior; and 2), create bilayers that exhibit an energy barrier to those interbilayer exchange of lipids. In the following, we report a systematic variation of the simulation parameters and discuss the fusion pathway for the chosen parameter set with all intermediate states. In particular, one of the simulation parameters that determines the energy barrier of a relevant subprocess for fusion can be identified and thus can be tuned by comparison with available data on this barrier.
Our fusion geometry consists of a vesicle with a diameter of 14 or 28 nm in contact with a planar bilayer. To obtain sufficient statistics, the time evolution of >160 fusion attempts of a vesicle to a planar bilayer patch is monitored. In those simulations, the initial projected area per molecule, A, is varied systematically and serves as a control parameter.
Since we study tension-induced fusion of membranes and vesicles, a few general remarks about membrane tension are appropriate. At first sight, membrane tension seems to be analogous to the interfacial tension between two fluid phases. The latter tension can be defined in two ways:
-
1.
Thermodynamically, via an expansion of the system's free energy in powers of the system size.
-
2.
Mechanically, via the work expended to increase the area of the interface. This work can be expressed in terms of the pressure or stress tensor of the fluid system.
Both definitions turn out to be equivalent even though this equivalence is far from obvious (see, e.g., (19)).
Compared to interfaces, membranes have more configurational freedom. Indeed, in contrast to interfaces, membranes consist of thin molecular bilayers that can form unilamellar or multilamellar vesicles and a variety of thermodynamic phases. Furthermore, a single membrane can rupture, fold back on itself, or undergo other types of morphological transitions. In fact, a sufficiently large membrane segment that is stretched and, thus, under mechanical tension can always lower its free energy by rupture or poration. Therefore, a thermodynamic definition of membrane tension, which necessarily involves the limit of large membrane area, is beset with conceptual difficulties. On the other hand, the mechanical definition of tension via the stress tensor can also be applied to relatively small membrane segments as studied in computer simulations (16). Thus, when we use the term “membrane tension,” we always mean the mechanically defined tension.
The article is organized as follows. In Methods, the simulation method and the model systems are summarized. Then follows a description of the material properties of the simulated membranes, which emphasizes the improved stretching behavior of the membranes and introduces a simplified implementation of the tension measurements. The dependence of these properties and the stretching behavior on the individual simulation parameters is discussed in Parameter Dependence of Bilayer Equilibrium Properties. Two other subsections, The Fusion Pathway, and Other Pathways and Fusion Statistics, give a detailed description of possible time evolutions and outcomes. The intermediate stages of the fusion process are analyzed in detail and the statistics of these are discussed. Finally, in Fusion Statistics and Energy Barriers, the fusion time distributions and their tension dependence are analyzed to estimate the tension-dependent energy barriers of the fusion pathway.
Methods
Simulation method and parameters
Dissipative particle dynamics (DPD) is a coarse-grained, particle-based simulation technique that explicitly includes water and reproduces hydrodynamic behavior (20,21). The DPD particles or beads represent small volumes of fluid rather than single atoms so that their interactions are softly repulsive and short-ranged (see Supporting Material, Data S1, for details). All interaction potentials have the same range r0, but their amplitudes aij differ for different bead species. Both self-assembly and phase behavior of lipids have been reproduced with DPD (17,22,23).
The systems considered here are built up from three bead species: lipid head (H), lipid chain (C), and water (W) beads. The more complex architecture of the lipid molecules is constructed by connecting adjacent beads with spring potentials. In addition, the hydrocarbon chains are stiffened by a bending potential for two consecutive bonds.
The model lipids have a headgroup consisting of three H beads and two hydrophobic tails, each of which is made up from four C beads (shown in Fig. 1). This architecture was introduced in the context of molecular dynamics simulations (16) and also used in previous DPD simulations (9,17).
Figure 1.
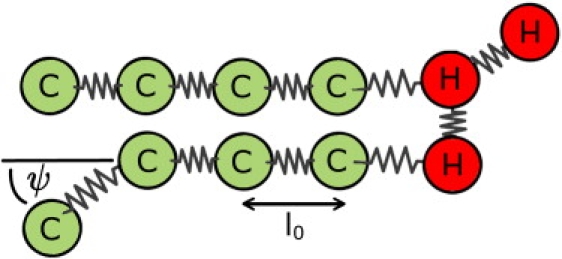
A coarse-grained model dimyristoyl-phosphatidylcholine (DMPC) with a H3(C4)2 architecture consisting of three head (H) beads and two hydrocarbon chains each consisting of chain (C) beads. Each chain bead C represents 3.5 CH2 groups. Consecutive beads are connected by springs of unstretched length l0. The hydrophobic chains are stiffened by a three-body potential constraining the angle ψ between two consecutive bonds.
In general, the simulation parameters are chosen to reproduce the mesoscopic behavior of the system. Thus, the overlap and interaction strength of the water beads used here reproduce the compressibility and density fluctuations of water. The remaining force amplitudes aij are fine-tuned to match the properties of lipid bilayers, by carefully determining the effects of changes to each parameter on the bilayer properties. There are some important constraints for a reasonable bilayer model such as a well-defined bilayer structure without interdigitation of the two monolayers, lateral fluidity, and bending flexibility. In addition, there are inherent limits to the range of reasonable values of aij (24). Our choice of the force parameters also reproduces the bilayer thickness relative to the molecular area A/N, the area expansion modulus and, in addition, a reasonable barrier against lipid exchange between contacting bilayers.
This strategy to obtain the simulation parameters by adjusting them to yield the correct mesoscopic behavior is rather similar to the one used to obtain appropriate interaction parameters in atomistic simulations. Atomistic force fields must also be optimized for different situations to reproduce experimental results, as demonstrated, e.g., by the large number of atomistic water models (see (25)).
As the simulated membranes in our model are relatively thin compared to the area per molecule, dimyristoyl-phosphatidylcholine (DMPC), which is a common, membrane-forming lipid with a comparatively short chain length of C14, is used as reference molecule for comparisons with experimental data. To represent DMPC, one tail bead must correspond to 3.5 CH2 groups.
The DPD simulations reported here use the set of force amplitudes aij as given in Table 1 (b). These represent an improved set compared to those in Table 1 (a) used in previous simulations (9,17). The tail chains have a bending stiffness k3 = 15 compared to k3 = 20 used before, but all other parameters are the same as in Shillcock and Lipowsky (9). The new set of force amplitudes improves the properties of the resulting membranes in two respects: 1), in their overall stretching behavior (see Stretching Behavior, below); and 2), in the stability of two adhering membranes against lipid exchange between those membranes. The previous parameter set in Table 1 (a) shows no observable energy barrier to interbilayer exchange of lipids in adhering bilayers, making this state highly unstable. Lipid exchange begins immediately and the two bilayers intermix. This does not reflect a realistic situation, as hydration of the lipid headgroups will present a considerable barrier for the hydrophobic tails and have a stabilizing effect on adhesion. Note that all interaction strengths aij satisfy aij ≥ 10, ensuring correct diffusive behavior (24).
Table 1.
Values of aij
| aij | j=H | j=C | j=W | |
|---|---|---|---|---|
| (a) | i = H | 25 | 50 | 35 |
| i = C | 50 | 25 | 75 | |
| i = W | 35 | 75 | 25 | |
| (b) | i = H | 30 | 35 | 30 |
| i = C | 35 | 10 | 75 | |
| i = W | 30 | 75 | 25 | |
(a) Old parameter set for the conservative force in DPD. All aii values are chosen to reproduce the compressibility of water. (b) New parameter set as used here. All values of aij satisfy aij ≥ 10 to ensure correct diffusive behavior (24).
The large value of the chain-water interaction, aCW = 75, was kept from the original parameter set. It leads to the strong effective adhesion between membranes that come into close proximity. Presumably, it will also lead to a relatively large hydration energy of the lipids. However, since the fusion pathway discussed below does not involve direct contact of chain and water beads, this overestimation of the hydration energy is not expected to change the results.
The head-tail interaction, aHT, is optimized to reproduce the energy barrier presented by the hydrated headgroups against interbilayer flips. On the other hand, this interaction is not optimized with respect to flip-flops within one bilayer, which occur more frequently than in experimental membranes. In simulations of a single bilayer containing 1640 lipids, 12 flip-flops from one monolayer to the other are observed within 1 μs. This would correspond to an average dwell time of 0.1 ms of a lipid in one monolayer. This is faster than observed experimentally, where the dwell time is ∼1 h (26,27).
Another set of force parameters has been used in Gao et al. (28). The latter set uses different force amplitudes aij 1), for pairs of C beads that belong to the same or to different chains; and 2), for the end beads Ce of the chains, which leads to even more parameters that need to be adjusted. In contrast, for the parameter set used here, all pairs of C beads are governed by the same force amplitudes as given in Table 1 (b). In Gao et al. (28), the repulsion between different tail chains is much smaller than between the beads along one chain, which introduces an effective attraction between tail chains. The resulting bilayers are much stiffer than those in our model and the tail chains are much more ordered and aligned. This leads to a different fusion pathway as discussed at the end of Energy Barrier for Interbilayer Flips.
Implementation of the stress tensor
The calculation of the mechanical membrane tension Σ in the simulations is based on the macroscopic stress tensor Σαβ. From its components, the stress profile s(z) = 0.5(Σxx + Σyy) − Σzz can be calculated with the method introduced by Schofield and Henderson (29) and extended to m-body potentials by Goetz and Lipowsky (16). The membrane tension Σ is the z integral over the stress profile s(z).
To obtain the stress profile s(z) in a simulation, the simulation box is subdivided into thin slices and the slice integration is expressed by multiplying the expressions with two Heaviside step functions as described in Goetz and Lipowsky (16). The membrane tension, on the other hand, depends only on the z-integrated components of this averaged stress tensor. It can therefore be calculated by directly integrating the components of the stress tensor in the z direction. This integration avoids the use of step functions and leads to expressions that are easy to implement.
In the calculations, the contributions to the microscopic stress tensor σαβ are separated into contributions from interaction clusters of size m, i.e., into two-body, three-body, etc., interactions with corresponding interaction potentials U(2), U(3) etc., as described in Data S2. For the model considered here, only two-body interactions from the conservative interaction and the bond potentials between two beads, and three-body interactions from the bond-pair potentials, contribute to the stress tensor Σαβ. The z-integrated contributions from the two-body interactions, I2αβ, and from the three-body interactions, I3αβ, are given by
| (1) |
and
| (2) |
where L|| is the lateral size of the membrane and the superscripts α and β can take the values x, y, or z, and specify the respective vector components. The sum over 〈j〉 is over all m clusters, in this case over all triplets, and 〈k, l〉 denotes all particle pairs within the cluster with positions and , so that is the vector connecting the two particles.
These expressions correspond to the generalized second and third virials. The sum over the contributions from I2αβ and I3αβ give the αβ-components of the z-integrated total stress tensor. Using α = β = x and α = β = z, the tangential components of the macroscopic stress tensor, Σxx = (I2xx + I3xx) and Σyy = I2yy + I3yy, and the normal component Σzz = (I2zz + I3zz), which define the membrane tension, are found. As the membrane is fluid, all components with α ≠ β should vanish. This is fulfilled in the simulations within ±0.002 kBT/r03. The tension calculated from this direct integration is plotted in Fig. 2, together with the tension calculated from the detailed stress profile. Clearly, the two methods give the same result.
Figure 2.

Bilayer tension as a function of area per molecule calculated from the stress profile s(z) for the old DPD parameter set (9,17) (circles) and the new parameter set with improved stretching behavior (diamonds). For the new parameter set, has also been measured by direct z integration of the microscopic stress tensor (crosses). Error bars represent the standard error. The two methods are found to yield the same results. The two parameter sets are given in Table 1.
Rescaled parameters in dissipative particle dynamics
The bead diameter r0 and bead mass m0 represent natural units of length and mass in the simulation. Energies are measured in units of kBT. The basic unit of time constructed from these quantities is . It is convenient for the analysis of the simulation results to introduce dimensionless quantities, which will be indicated by a bar. Thus, we define the dimensionless area per molecule ≡ A/r20 and the dimensionless tension ≡ Σr20/kBT with corresponding compressibility modulus ≡ KAr20/kBT.
To obtain physical units, the length- and timescale r0 and τ of the system must be chosen in an appropriate manner from characteristic properties of the system. For studies of fluid membranes, the area per molecule A of a tension-free bilayer and the diffusion coefficient D|| for lateral diffusion of lipids in the bilayer provide natural length- and timescales. The area per molecule in the simulations is 1.25 r02, whereas the experimentally measured value is 0.596 nm2 (30), so that the basic length scale is r0 = 0.69 nm. The in-plane diffusion coefficient of DMPC is D|| = 5 μm2/s (31). In the simulations, the in-plane diffusion coefficient of the lipids is D|| = 0.016r02/τ. Thus, for the correct diffusive behavior, a mapping of τ = 1.6 ns is required. Accordingly, a Δt = 0.02 τ timestep in the simulations corresponds to Δt = 0.0314 ns.
The simulated systems
All simulations are done in the NVT ensemble with a density ρ = 3/r03.
The choice of NVT or NPT depends on the system that is simulated. If the membranes represent a small piece of a much larger membrane, a small fusion site with a vesicle will have little effect on the membrane tension and NPT would be an appropriate ensemble. For two membranes of roughly the same size, a pore or fusion site will reduce the membrane tension considerably and the effects are better represented by an NVT ensemble. The membranes in our simulations reach experimentally feasible sizes and the NVT ensemble has the advantage that the results are comparable to simulations of two fusing vesicles with the same diameter, which behave very similar to the fusion to a planar bilayer described here.
In this ensemble, the projected membrane area remains constant, whereas the bilayer tension fluctuates around its average value. The standard deviation of the tension values decreases strongly with increasing system size L|| up to L|| ≃ 30 r0, and then seems to approach a constant value (see Supporting Material, Fig. S1).
In this study, four types of simulations have been performed (see Data S3):
-
1.
A random mixture of lipids and solvent to test whether the lipids self-assemble to a bilayer structure.
-
2.
A preassembled planar bilayer for characterization of the membrane properties.
-
3.
A vesicle in close proximity to a planar bilayer patch to observe fusion.
-
4.
Two adhering planar bilayers, which are used to measure the average work required to enforce the flipping of one-lipid tails from one bilayer to the other.
Bilayers and vesicles are assembled with a prescribed area per molecule, A. The planar bilayer is built up from 6700 to 8300 lipid molecules, the 28-nm vesicle from 6800 to 7500 lipids, and the 14-nm vesicle from 1400 to 1600 lipids. The membranes are equilibrated with a smaller timestep of Δt = 0.005 τ for 10 ns. For fusion simulations, a vesicle is placed in close proximity to a planar bilayer patch, separated by a thin layer of water beads. The thickness of this initial water layer is ∼1.5 nm. For such a separation, the two membranes usually come into contact by diffusion within <150 ns. No external forces are applied to bring the membranes into contact. Those simulations for which the vesicle diffuses away from the planar bilayer are discarded.
Fusion is induced by applying a lateral tension to the planar bilayer. This is achieved by increasing the value of A, which is directly related to the bilayer tension. Most fusion simulations have been done for two different vesicle diameters, 20 r0 and 40 r0, corresponding to ∼14 and 28 nm, respectively, in a simulation box with an area of (72 r0)2 ≃ (50 nm)2, and a height of 52 r0 and 72 r0 for the 20 r0 and the 40 r0 vesicle, respectively. Additional simulations of a vesicle with diameter 20 r0 in a smaller box of (36 r0)3 were performed to further explore the observed dependence of the results on the vesicle size. To obtain relevant fusion statistics, >160 fusion simulations have been monitored. For each data point corresponding to a particular value of the control parameter, an average over at least 18 independent simulations is taken.
To measure the flipping energy, the average work required for an interbilayer flip, a single lipid from the lower bilayer, is selected and a slowly moving, external harmonic potential is applied to one of its tail beads as in Fig. 3 a. The potential starts centered on the bead's z coordinate and moves slowly toward the other bilayer at a constant speed of 0.009 nm/ns, until the tail has flipped into the other bilayer and the lipid has assumed a splayed conformation as in Fig. 3 b. At each timestep, both the displacement of the bead from its original position, zbead, and the spring force required to hold it at that position, are recorded.
Figure 3.

Simulations enforcing interbilayer flips of lipid tails are used to measure the energy barrier for interbilayer flips. (a) From two adhering bilayers (head beads are blue/green, tail beads are omitted for clarity), a single lipid is selected (several red and one yellow tail beads), and a slowly moving external harmonic force F applied to one of its tail beads (yellow), until the tail has flipped to the other bilayer, so that the lipid has assumed a splayed configuration with one tail inserted in each bilayer as shown in panel c. (b) The energy landscape Eα for the bead is sketched as a function of the displacement z of the yellow bead. It has a high barrier in the center corresponding to the repulsive headgroups and increases to the sides reflecting displacement of the headgroup into the hydrophobic region.
To simulate quasistatic pulling, the motion has to be sufficiently slow. The appropriate velocity can be determined from simulations pulling a lipid through the flat energy landscape of a uniform solvent. Fast pulling leads to a nonzero spring force, caused by the friction of the surrounding fluid. The friction coefficient found in these simulations is ∼2 × 10−10 ns/m. At sufficiently low pulling speeds, the average position of the bead is always close to the position of the harmonic potential.
Unlike steered MD simulations that usually pull along a given direction vector, which can introduce a bias if the system cannot adapt its orientation on the MD timescale, in our simulations the potential is applied only to the z coordinate of the bead, which is free to move in the xy plane. Furthermore, the low pulling speed and small size of the molecule make it unlikely that the direction will bias the resulting work, as molecules can easily adapt.
The simulation code used for these simulations has been developed and tested by our group. More information about this simulation code may be obtained from the authors. Simulations were run on single processors (Intel Xenon 3.6 GHz), and needed ∼7 days/μs.
Results and Discussion
Material properties of the simulated bilayers
Simulations starting from a random mixture of lipids and solvent show that the molecules with a H3(C4)2 architecture, as shown in Fig. 1, and the interaction parameter set in Table 1 (b), self-assemble into bilayer vesicles (Fig. S2). With the length and timescale of the system determined from the area per molecule A of the tension-free bilayer and in-plane diffusion coefficient D||, other equilibrium properties of the bilayer can be measured and compared to experimental findings.
The density profiles of the individual components show that the lipids form proper bilayer structures with water completely excluded from the interior and with the head beads accumulated at the interface between hydrophobic tails and water (Fig. S3). The two monolayers are well separated and there is no interdigitation. The weak repulsion aCC = 10 leads to relatively narrow bilayers with a high density of the hydrophobic core. As a consequence, when mapping the amphiphiles to real molecules, a lower mass is associated with the tail beads as demonstrated by Ortiz et al. (32). When the bilayers are stretched, the volume per lipid remains approximately constant, as is observed experimentally for the incompressible hydrocarbon chains.
Stretching behavior
In the fusion simulations, fusion is induced by controlling the area per molecule in the bilayers, which effectively puts the membranes under tension. To study tension-dependent behavior, a knowledge of how the tension Σ, in the bilayer depends on the molecular area is required.
The stress profile across the membrane has similar characteristics to those found in other coarse-grained simulations (16,17), with positive peaks from interactions with water at the lipid-solvent interface and negative contributions from the bond and bond-angle potentials in the bilayer center (see Fig. S4).
The membrane tension Σ is the z integral over the stress profile s(z). To find the dependence of Σ on the molecular area, a series of simulations at different values of is performed. Fig. 2 shows the membrane tension as a function of the area per molecule for the parameter set introduced here in comparison to the previously used set. The comparison shows that whereas the area per molecule of the tensionless state is the same ( = 1.25) in both cases, the stretching behavior of the two membranes is rather different. The old parameter set leads to very stable membranes, whose area per molecule can be increased by ∼60% without rupturing the membrane within a few microseconds. The dimensionless tension ≡ Σr20/kBT is a nonlinear function of this molecular area, increasing steeply from the tensionless state but then growing much more slowly. Experimental studies of large vesicles show that these cannot sustain extensions of >3–5% for lipid vesicles (33) and ∼20% for polymersomes (34). The new parameter set reduces the stretchability of the simulated membrane to ∼20% and leads to an essentially linear relation between tension and molecular area , as is also observed experimentally. The very large stretchability is a common property of simulated bilayers and should be attributed to the small length- and timescales. For example, at the limit of metastability for the tense planar membrane, corresponding to the maximum tension value in Fig. 2, the bilayer remains stable over the 1.5-μs simulation performed to obtain this data, whereas a pore forms in a longer simulation of 9 μs. In addition, this stability limit for the stretched membrane should also depend on the box size. In Tolpekina et al. (35), the free energies of a stretched and a porated membrane, both with the same base area and containing the same number of molecules, have been compared. The analysis leads to a stability limit of the porated membrane, and a coexistence point for the two states. The corresponding stretch scales as L||−2/3 with the linear size L|| of the base area. The stability limit of the stretched membrane, on the other hand, lies at infinite stretch. If we assumed that these scaling laws can also be applied to the rupture of a membrane, a 100-μm2 DPD membrane would only be stretchable by 0.7%.
The linear relation between and implies that the molecular area, which is used as a control parameter, can easily be converted to membrane tension to describe tension-dependent processes. The area compressibility modulus KA is given by the slope of the tension as a function of at the tensionless state with . The corresponding dimensionless modulus ≡ KAr20/kBT can be deduced directly from the plot in Fig. 2 and has the value = 22.75. Converted back to physical units, this leads to a value of 200 dyn/cm, which is slightly lower, but of the same order of magnitude as the experimentally measured values for phospholipids, which range from 234 dyn/cm for DMPC to 265 dyn/cm for DOPC (30). Thus, the agreement with experiment is greatly improved compared to the previous DPD parameter set, which leads to = 700–1000 dyn/cm.
Parameter dependence of bilayer equilibrium properties
To improve the macroscopic properties of the simulated bilayers, the various DPD parameters have been varied systematically. In this study, the stretching behavior, i.e., the membrane tension as a function of molecular area , is of particular interest, because membrane tension is used to induce fusion.
Here we give a brief overview of the effects of different parameter changes. Their effects on the membrane's stretching behavior are summarized in Fig. S5(a).
Changing the relative magnitudes of the head-head and tail-tail force amplitudes, aHH and aCC, changes the effective size of the headgroups or tail chains, respectively, and alters their compressibility and thus magnitudes in the density profile: lower aii values lead to higher densities and vice versa. If the values become too large or too small the spontaneous curvature of the monolayers becomes too large and the bilayer state becomes unstable. These effective size changes of the beads also affect the membrane stretching behavior. Larger headgroups, corresponding to larger aHH values, shield the hydrophobic tails more effectively from water, so that the bilayer becomes more stable. The functional dependence of membrane tension on molecular area becomes approximately linear and the molecular area of the tension-free bilayer increases, as the larger headgroups need more space. Smaller values of aCC, on the other hand, corresponding to smaller, more compressible tail chains, decrease the membrane stretchability and also lead to more linear tension curves. In the final parameter set of Table 1 (b), the combination of the relatively strong head-bead interactions and the closer packing of the chains due to their reduced repulsion, generate a monolayer curvature that facilitates lipid rearrangements to cover the rim of a forming pore.
The tail-water force amplitude aCW represents the hydrophobic effect. It has to be strong enough to drive self-assembly, but otherwise has little effect on the tension's dependence on the molecular area. A slight increase in the values of appears as the tail-water contributions to the stress profile increase, but this effect is small if the hydrophobic region is well shielded by the headgroups.
The effect of chain length on the membrane properties has been subject to both experimental and simulation studies (17,34,36,37). The simulation studies have found that with longer tails, the packing of lipids becomes denser and that both the area stretch modulus KA and the tensionless value of the reduced molecular area depend sensitively on the chain length. The experiments, on the other hand, found that the area stretch modulus is independent of chain length over a wide range (from 13 to 22 carbons/chain). The simulations performed here in the context of the parameter optimization show that the chain-length dependence of the bilayer behavior varies with the parameter set used in the simulations. Fig. 4, a and b, shows the membrane tension as a function of reduced molecular area for lipids with different chain lengths for the two parameter sets in Table 1, (a) and (b). In both cases, the stability of the tense membrane increases with chain length. However, in the original parameter set (Table 1 (a)), the bump in the tension curve becomes more pronounced as the chain length increases, dividing the stretching process into two regimes with a high and a low tension gradient. At the same time, the compressibility modulus KA increases with chain-length. For the new parameter set used here as given in Table 1 (b), the curves also start to show a slight deviation from the linear dependence. In contrast to the original parameter set, the compressibility modulus now decreases slightly with chain-length. However, both effects are much less pronounced than in the original case, with a deviation in the compressibility modulus KA of ∼1% for an increase in chain length from three to five beads.
Figure 4.

Membrane tension as a function of molecular area for three different chain lengths of 3, 4, and 5 per tail chain, in the old (a) and new (b) parameter set. There is no state with zero tension for bilayers formed from lipids with three beads per chain in the old parameter set (a), since at low values the bilayer structure is not stable.
Finally, the parameter ratio aHC/aCC, also plays an important role. This ratio represents a measure of how strongly headgroups and hydrocarbon chains repel each other. The old parameter set is characterized by aHC/aCC = 2, which does not suffice to stabilize two adhering bilayers. In the latter situation, a relatively large number of lipid tails starts immediately to undergo interbilayer flips, leading to a strongly perturbed adhesion zone on the timescale of 1 μs. If the ratio aHC/aCC is increased, interbilayer flips become noticeably slower. For the larger ratio of aHC/aCC = 3.5 as used here, the barrier for interbilayer flips is measured to be ∼8 kBT.
The fusion pathway
To shed light on the molecular mechanism of membrane fusion, >160 DPD simulations of fusion attempts between a vesicle and a tense planar bilayer patch have been monitored. Successful fusion attempts in these simulations all involve the same sequence of events. Here we present a detailed description of the observed fusion mechanism.
The simulations start with the vesicle in close proximity to the planar bilayer. The evolution of the system is monitored from the first contact between vesicle and planar bilayer until the fusion pore has opened. If fusion has not occurred within 20 μs, the attempt is counted as unsuccessful.
Fig. 5 shows snapshots of the development of one successful fusion event between a planar bilayer (red heads/green tails) and a vesicle (orange heads, yellow tails) with a diameter of 30 nm. The system is shown from two perspectives, where the z axis is taken to be normal to the planar bilayer: cross sections viewed from the side and cuts through the planar membrane, viewed from above. For the top views, the cuts are performed through the midplane of the planar membrane. In these top views, all the green tail beads, which originally belonged to the planar membrane, are made transparent. In the upper top views, one sees the appearance of the yellow tail beads that enter the planar membrane by interbilayer flips from the vesicle. In the lower top views, the yellow tail beads are transparent as well, revealing the orange head beads of the vesicle lipids. The white areas in the lower top views correspond to hydrophobic regions containing only tail beads. A few water particles can be seen outside the vesicle. These have leaked through the membrane during the initial equilibration period, where the lipids are linear and arranged on a lattice. There is no leakage at later times.
Figure 5.
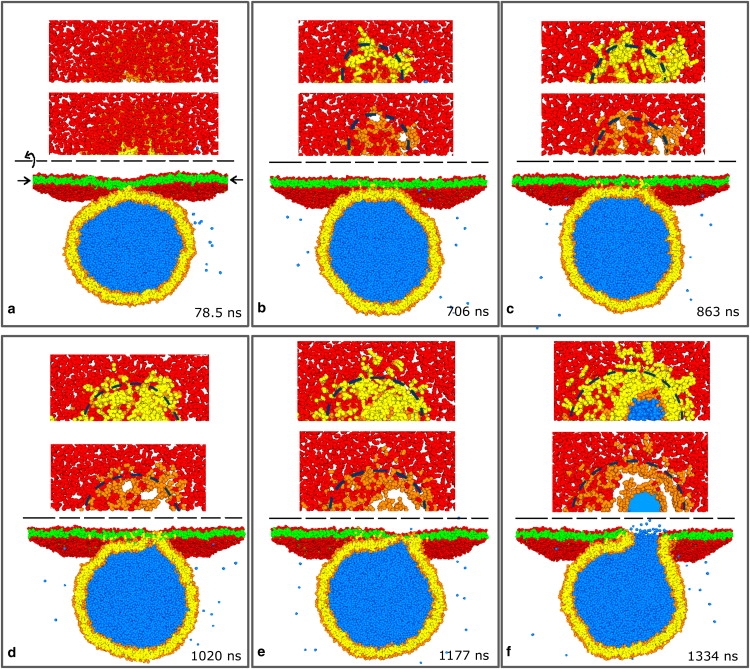
Fusion of a vesicle with a diameter of 28 nm to a planar membrane with a projected area of (50 nm)2. The vesicle consists of 6869 lipids (orange heads; yellow chains) while the planar membrane contains 6911 lipids (red heads; green chains). The water beads originally inside the vesicle are blue, those outside are not shown for clarity. Six snapshots illustrating the development of the fusion event from 78.5 ns after the first contact until opening of the fusion pore after 1334 ns. For each time, the system is shown from two perspectives: cross sections cut through the center of the vesicle viewed from the side, and two cross sections through the midplane of the planar membrane, as indicated by the arrows in panel a viewed from above. In the upper top views, the green hydrophobic beads from the planar bilayer are made transparent, revealing the yellow hydrophobic chains of vesicle lipids that have flipped into the planar bilayer. In the lower top views, all hydrophobic beads are set transparent, so that white areas in the headgroup plane indicate purely hydrophobic areas. Lipid tails start to undergo interbilayer flips after 78.5 ns. The growth of the contact area enhances these at the contact line, indicated by the blue broken line in panel b, creating a bean-shaped, disordered hydrophobic contact that nucleates into a hemifused diaphragm after 1177 ns.
Upon first contact, the vesicle adheres to the planar membrane patch. The contact area grows and the vesicle membrane spreads onto the planar membrane, forming a relatively sharp contact angle at the contact line, i.e., the boundary of the contact area. As the contact area grows, lipid tails start to move from the vesicle into the planar bilayer. These exchange processes or interbilayer flips take place mainly at the contact line, see Fig. 5 b, because
-
1.
There the vesicle membrane has a pronounced kink of high curvature with increased strain and more strongly compressed lipid tails, a situation that has been previously discussed in another context (38).
-
2.
The vesicle lipids are already tilted with respect to the planar bilayer. Both effects tend to lower the energy cost of interbilayer flips along the contact line.
The interbilayer flipping of the lipid tails disturbs the local double-bilayer structure and leads to the formation of a disordered membrane domain within the contact zone. The hydrophobic tails moving through the headgroups bring the hydrophobic centers of the two bilayers into direct contact. This hydrophobic contact expands not radially symmetric, but rather following the contact line, assuming a beanlike shape, which can be seen as the open areas in Fig. 5 b.
Finally, within this disordered hydrophobic-contact region, lipids reorder to form a small patch of a single, hemifused bilayer with a diameter of a few nanometers. The hemifused bilayer leads to a more favorable area per molecule and thus reduces the bilayer tension. It expands for a short time and finally ruptures at the rim to form the fusion pore. In the example shown in Fig. 5, this takes place 1334 ns after the onset of the process. All successful fusion events observed in our simulations involve the same sequence of membrane conformations described here.
In addition, several fusion events between two vesicles with a diameter of 28 nm have been simulated. The fusion of the two vesicles follows the same pathway as the fusion between a vesicle and a planar membrane.
Adhesion and interbilayer flips
In our DPD simulations, the adhesion of bilayers arises because of two effectively attractive interactions. First, the repulsive interaction aWC between the water beads and the chain beads is larger than the repulsive interaction aHC between the head beads of one membrane and the chain beads of another membrane. Therefore, the interaction energy is reduced if the water beads adjacent to one membrane are replaced by another membrane. Second, the small water beads push the large membranes together by depletion interactions of entropic origin (39).
Experimentally, the adhesion of DMPC membranes has been somewhat controversial. Multilayer stacks of DMPC membranes in excess water exhibit an equilibrium spacing of ∼2.8 nm (40). Such stacks have also been observed to form spontaneously at the air-water interface (41). When two DMPC membranes are immobilized on mica surfaces, their adhesion energy W was estimated to be W ≃ 0.1 mJ/m2 as deduced from surface force measurements (42). On the other hand, the adhesion of DMPC vesicles as studied by micropipette aspiration led to the smaller estimate W ≃ 0.01 mJ/m2 (43). Furthermore, the group of Helfrich reported that large DMPC membranes do not adhere in distilled water unless they experience some tension (44).
These different observations can be reconciled by the following theoretical considerations (45). The van der Waals interaction between two planar membranes that have the same lipid composition is always attractive as follows from the general Lifshitz theory for these forces (see, e.g., (46) and references therein). This van der Waals interaction is, however, renormalized by the shape fluctuations or undulations of the membranes provided the membrane tension is sufficiently low (45,47). This effect is stronger for larger membrane segments, since the number of undulation modes is proportional to the membrane area. Thus, for sufficiently large membrane segments, the undulations can lead to a renormalized interaction that is purely repulsive.
In the surface force experiments, all membrane undulations will be suppressed and one should, thus, measure the bare van der Waals attraction not affected by undulations. In the micropipette experiments, membrane undulations are also suppressed but only down to a certain minimal wavelength that depends on the applied tension (45). The remaining fluctuations still act to reduce the effective van der Waals attraction, which explains the smaller value of the adhesion energy as estimated from the micropipette experiments.
In the system considered here, membrane undulations are suppressed for two different reasons. First, because of the small size of the vesicles, the number of possible undulation modes is rather limited even in the absence of tension. Second, these remaining undulation modes are further suppressed by the tension Σ. Therefore, the planar membrane and the vesicle should exhibit an adhesion energy of W ≃ 0.1 mJ/m2 or 2.5 × 10−2 kBT/nm2 at room temperature. As shown in the following, the DPD membranes studied here have an adhesion energy, which is rather similar to this experimentally determined value even though the DPD parameters were not optimized with respect to this energy.
In our simulations, the system remains in the adhered state for a long time if the initial membrane tension is relatively small. The contact area grows until the system reaches a mechanically stable state, in which the energy gain from the effective adhesion energy density |W| is balanced by the cost of deforming the membranes.
Because the membranes attain a relatively large tension during the adhesion, the membrane shapes are tension dominated and approach spherical caps. The contact curvature radius Rco observed in the snapshots is small compared to the linear size of the vesicle Rve. Thus, as Rco = (κc/2|W|) >> Rve, the bending energy is negligible and an effective contact angle θeff can be defined as in Seifert and Lipowsky (48). These simple system geometries can be mechanically stable and thus yield estimates of further mechanical properties of the system from simulation snapshots.
A balance of the forces on the contact line in the directions parallel and perpendicular to the planar bilayer leads to two independent Neumann equations for the tensions in the planar bilayer, Σpl and the vesicle membrane, Σve (Data S4), as given by
| (3) |
where θ1 and θ2 are the angles formed by the membrane segments of the vesicle with the bilayer plane as sketched in Fig. 6.
Figure 6.
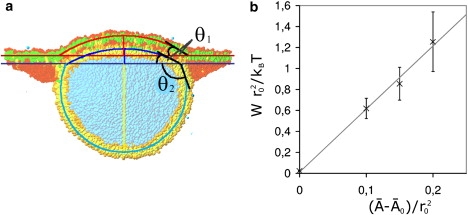
(a) Example of a vesicle that adheres to a planar bilayer patch. Both the vesicle and the deformed segment of the planar membrane are well fitted by spherical caps, which define the two contact angles θ1 and θ2. (b) The dependence of the adhesion energy density W on the difference of the area per molecule from its tensionless value. Each data point represents the average over data from 10 to 20 different snapshots. Error bars are mean ±1 SD and the solid line is the best linear fit.
The geometry of the adhering membranes consists of three membrane segments: two spherical cap regions for the vesicle and the contact area, and a planar segment. For this geometry, the area per lipid in the planar membrane can be calculated and the corresponding tension Σpl determined from the tension-area plot in Fig. 2. If this tension value is inserted into the Neumann equations (Eq. 3), these two equations can be solved for |W| and Σve. In Fig. 6 b, the result for the average adhesion energy |W| is displayed as a function of the initial molecular area in the planar membrane. Inspection of this figure shows that |W| is linearly related to with the best fit given by
| (4) |
For , the strength of the attractive interaction between the membranes is thus ∼2 × 10−2 kBT/nm2, which is of the same order of magnitude as the values of the van der Waals attraction between PC membranes obtained from surface force apparatus measurements (see, e.g., (42)). The tensionless molecular area of the vesicle is found to be ≃ 1.05 r20. These estimates represent a lower bound for |W| and , as, especially at low tensions, other factors such as the bending energy may also contribute to the force balance.
This mechanically stable adhered state is characterized by a low rate of interbilayer flips. Fig. 7 a illustrates such interbilayer flips at low tension. The two images are constructed in the same way as the upper top views in Fig. 5: All beads of the planar membrane except the head beads of the proximal monolayer are made transparent. Thus one sees the appearance of the yellow tail beads that enter the planar membrane by interbilayer flips from the vesicle. Inspection of Fig. 7 a reveals that these interbilayer flips preferentially occur along the circular contact line. In addition, this figure shows an example for in-plane lipid diffusion, as indicated by the green arrow: after both tails of a lipid molecule have undergone an interbilayer flip, the lipid molecule is free to diffuse away from the contact area.
Figure 7.
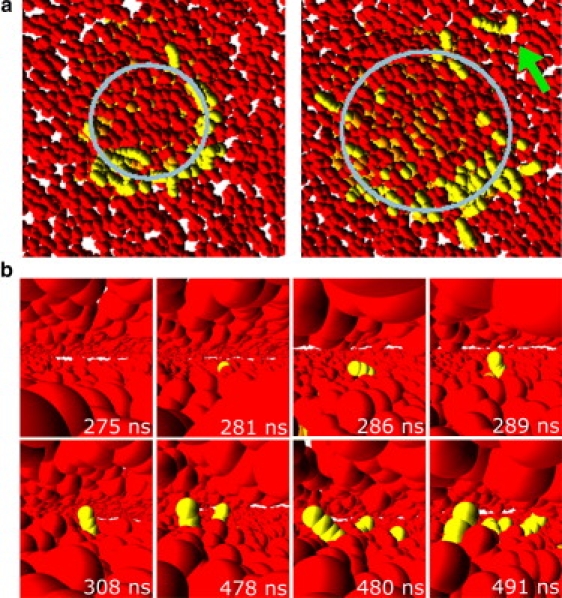
The first interbilayer flips of the vesicle's hydrophobic chains (yellow) into the planar bilayer (head beads are red, chain beads not shown). (a) Top view of the planar membrane, constructed in the same way as the upper top views in Fig. 5. The hydrophobic chains and the upper monolayer of the planar bilayer are transparent, revealing the yellow hydrophobic beads of vesicle lipids that have flipped into the planar membrane. The rate of interbilayer flips is low, so that the influence of the contact line (gray circle) on the probability for interbilayer flips becomes clearly visible. The arrow indicates a flipped lipid that has diffused away from the contact area. At slow flipping rates this diffusion competes with the flipping. (b) Snapshots of the center of the planar membrane. These snapshots show the details of the first hydrophobic chains belonging to vesicle lipids (yellow) moving into the planar bilayer. At first only one tail flips, so that the lipid assumes a splayed conformation. Further lipids undergo the same transition in the vicinity, presumably because the splayed lipid sufficiently disturbs the bilayer structure. In the final snapshot, one lipid has flipped both of its tails into the other bilayer.
A closer look at individual interbilayer flips is shown in Fig. 7 b, which displays a view inside the planar bilayer, with its hydrophobic beads made transparent, so that one can observe how the first vesicle lipid chain (yellow) moves over into the planar bilayer core. In the vicinity of lipids in this splayed tail conformation, the probability of further interbilayer flips is somewhat increased.
A similar onset of fusion with splayed-tail lipids at the rim of the contact area was also reported for other simulation studies (7). In another case, splayed lipids bridge the gap between the vesicles with a stalk forming around them (8). Of course the splayed lipid conformation is a mechanism that can only be observed in simulations using double-tailed amphiphile models.
There is also some experimental support for a splayed tail configuration at the onset of fusion, summarized in Kinnunen and Holopainen (49). Furthermore, it has been argued that the most likely effect of the fusion peptide hemagglutinin is to promote intermembrane flips by dehydration and structural disruption as peptides replace water molecules in the hydration layers (1). Finally it is interesting that, although mediated by fusion proteins, the fusion of vacuoles also takes place at the edge of an extended contact area (50).
The fusion pathway described here provides a direct connection between the onset of fusion and splayed lipid conformations. The latter conformations are more likely to occur for conelike lipids, i.e., for lipid molecules that resemble truncated cones with a relatively small headgroup and relatively bulky tails (sometimes called lipids with “negative spontaneous curvature”). Indeed, when such a conelike lipid is located in the proximal monolayer of the membrane kink along the contact line, its tails are strongly compressed, and the molecule can relax this mechanical strain by flipping one tail into the proximal monolayer of the other membrane. Therefore, according to our simulation results, conelike lipids should enhance the fusion process because their tails are more likely to undergo interbilayer flips and these lipids are more likely to attain splayed conformations. In this way, we provide a new interpretation to experimental observations as reviewed in the literature (51,52) that conelike lipids act to promote membrane fusion.
Hydrophobic contact-disorder at the contact line
Hydrophobic contact occurs if several vesicle lipids flip to the planar bilayer simultaneously and in close proximity. This strongly perturbs the local bilayer structure, where the headgroups are forced apart and brings the hydrophobic cores of the two bilayers into direct contact, so that the tail chains intermix, as in the example in Fig. 8 a. The combined width of the two membranes becomes thinner, since the two intervening layers of headgroups are no longer present. Often small groups of lipid heads remain at their original location, i.e., between the two bilayers. As their tail chains rearrange to form the hydrophobic contact, the headgroups become trapped within this extended hydrophobic region, as they cannot easily move across the hydrophobic material to either side. These sometimes appear later in larger numbers in the distal monolayer of the planar bilayer. Trapped headgroups are also sometimes observed in atomistic simulations (12) and some experimental studies (53,54) observe fast movement of headgroups to the distal monolayer during fusion. An example of this phenomenon is shown in Fig. 8 b.
Figure 8.

(a) Side view: an example of the disordered domain at the contact line. The hydrophobic material of the two bilayers (green/yellow) is no longer separated by a headgroup layer (red/orange) and has mixed. Several orange head beads are trapped in the center of this region. (b) Top view onto the planar bilayer patch. A small region of head beads from vesicle lipids (orange) appears between the planar bilayer lipids (red), indicating a region where several lipids from the vesicle have moved across the planar bilayer.
The hemifused diaphragm and formation of the fusion pore
After some time, a newly ordered hemifused bilayer forms within the perturbed membrane region and expands until it either ruptures or the area per molecule is sufficiently relaxed. If fusion is successful, the size of the hemifused patch usually does not exceed a couple of nanometers and the time until rupture lies between 100 ns and 300 ns. However, as the hemifused patch expands, the membranes gain additional area and the tension relaxes. Therefore, beyond a certain size hemifusion is stable on the timescale of the simulations. In that case, the expansion of the hemifused area continues either until the tension is balanced or until it spans the entire contact area, in which case some tension may remain.
This extended hemifused state is often accompanied by some leakage of solvent beads through small transient pores that form at the contact line (as seen in Fig. 9). The formation of such pores indicates that their line tension is strongly reduced, and confirms that the fusion pore or neck forms at the rim of the hemifused patch. Via these pores, mismatched values of A in the inner and outer monolayer can also be equilibrated.
Figure 9.
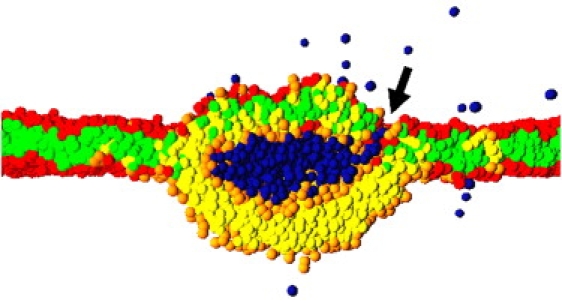
A 14-nm vesicle and part of a 50-nm planar bilayer at , which have formed an extended hemifused contact. Only the central part of the simulation box is shown. A small pore, indicated by the arrow, has formed at the junction of the three bilayers. Such pores allow the pressure of the enclosed water to be reduced and fast lipid flip-flops between the inner and outer monolayers to occur.
A geometric analysis similar to that of the adhered state gives an estimate of the net area per molecule (Data S4). In the cases where the hemifused patch covers only part of the contact area these estimates are very close to the value of A for which the membrane is relaxed. If the contact area is completely hemifused, the tension in all three membrane segments should be balanced. Thus if the tension is not eliminated, the three contact angles should be equal, unless there is a line tension to reduce the length of the three membrane junction line. A study of snapshots of such hemifused systems, where some membrane tension remains, shows that, in fact, the internal angle of the vesicle is larger. This leads to an estimate of ∼2 kBT/r0 for the line tension, as explained in Data S4.
Other pathways and fusion statistics
As the (meta) stable adhesion and hemifusion show, fusion is not the only mechanism of relaxing the tension. Alternatively, tense membranes can rupture or, at low , simulations may still remain in adhesion or hemifusion after 20 μs. Unlike adhesion and hemifusion, bilayer rupture implies a definite failure of the fusion process. For tensions close to the rupture point , bilayer rupture can easily be induced by the area increase due to the deformations caused by the adhering vesicle. This process competes with the fusion process and may thus happen before fusion takes place.
Comparison of the two vesicle sizes shows that 1), the small vesicle can fuse at lower values of than the larger one; and 2), at low tensions, the large vesicle remains in the adhered state, whereas the small vesicle typically hemifuses. These differences are related to the relative size of the two membranes. To have a similar effect on the tension of the planar bilayer, the smaller vesicle has to be more strongly perturbed. Using the adhesion strength obtained from the spherical fits and solving the force balance for stable adhesion, Eq. 3, for the shape of the adhering membranes, with the constraint that the volume of solvent inside the vesicle remains constant, one finds that the tension difference induced by the area increase of the deformation is small compared to the overall tension in the membrane. Therefore, both vesicles would form approximately the same angles with the planar bilayer at a given . As a result the contact area of the 28-nm vesicle is larger by a factor (Rl/Rs)2 = 4, covering ∼20% of the planar bilayer, whereas the 14-nm vesicle can cover only ∼5%. As the adhesion energy is proportional to the adhesion area, the energy reduction for the small vesicle is not sufficient to achieve this at most tensions and the hemifused intermediate is formed, unless the vesicle is practically tension-free. Then, however, the tension is no longer high enough to rupture the hemifused patch and form the fusion pore. As a consequence, unsuccessful fusion attempts of this smaller vesicle size are usually trapped in the hemifused state, whereas larger vesicles at the same tension often remain in the adhered state.
Fusion statistics and energy barriers
Fusion time distribution
The tension determines not only the success rates, but also the timescale of fusion. Each successful fusion event has a corresponding fusion time, tfu, defined as the time elapsed from first contact between the vesicle and the planar membrane patch until the formation of the fusion pore has been completed. The distribution of these fusion times depends on the membrane tension, as already described in Grafmüller et al. (15). Each fusion time distribution, corresponding to a certain , can be characterized by the average value 〈tfu〉 and the width . Both the average fusion times obtained from the fusion time distributions of the simulations, 〈tfu〉a and the width of the distributions are shown as functions of the molecular area in Fig. 10. Inspection of this figure shows that both quantities appear to grow exponentially with decreasing . It becomes, therefore, increasingly difficult to determine the timescale of fusion from computer simulations as the tensionless state is approached. To obtain an accurate estimate, not only do the longer average fusion times themselves have to be accessible to the simulations, but they also have to sample the increasingly broad distribution.
Figure 10.
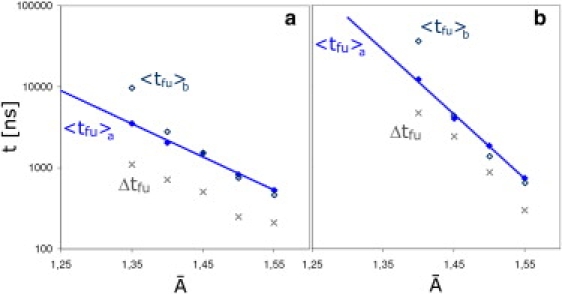
The average fusion times 〈tfu〉a (solid diamonds) and 〈tfu〉b (open diamonds) as functions of the area per molecule displayed together with the widths Δtfu of the fusion time distributions (crosses) (a) for the 14-nm and (b) for the 28-nm vesicle. The two averages 〈tfu〉a and 〈tfu〉b represent a lower and upper bound for the average fusion time 〈tfu〉. Both 〈tfu〉a and Δtfu seem to decrease exponentially with .
The different outcomes and overlapping time distributions observed in these simulations demonstrate clearly that the results of individual trajectories should not be overinterpreted. To obtain reliable results, or quantitative relations, such as the tension dependence of the fusion times, it is necessary to perform a large number of runs.
Because these distributions only include successful fusion events and disregard final states of adhering or hemifused vesicles, which might still go on to fuse after longer waiting times, the average values obtained from these distributions represent lower bounds for the average fusion times. An upper bound can also be obtained from a second estimate that averages over the fusion rates and includes the adhering and hemifused final states as . The resulting upper bound of the average fusion time, , is also presented in Fig. 10. At high bilayer tensions, the data points for 〈tfu〉b more or less coincide with those for 〈tfu〉a, but at low tensions, 〈tfu〉b deviates from 〈tfu〉a toward longer fusion times, and diverges when no successful fusion events are observed. The true dependence of the average fusion time 〈tfu〉 on the molecular area lies between the two data sets for 〈tfu〉a and 〈tfu〉b.
Overall energy barrier
The tension-dependent fusion times indicate a tension-dependent energy barrier for fusion. In an attempt to identify states that may constitute such a barrier, the fusion process has been decomposed into three subprocesses:
-
1.
Subprocess α starts with the first contact between the two membranes and represents the adhesion and spreading of the vesicle onto the planar bilayer up to the time when the first interbilayer flip of a lipid tail has taken place. The duration of this process defines the first flipping time tα.
-
2.
Subprocess β consists of the intermixing and partial fusion of the two bilayers, starting from the first interbilayer flip until the nucleation of the hemifused patch. As above, the duration of process β defines the reordering time tβ.
-
3.
Subprocess γ corresponds to the rupture of the hemifused patch, which leads to the opening of the fusion pore and defines the rupture time tγ.
By definition, the total fusion time is given by the sum tfu = tα + tβ + tγ. The average duration of the subprocesses α and β, 〈tα〉 and 〈tβ〉, are displayed together with 〈tfu〉a as a function of the molecular area in Fig. 11. Clearly, both timescales decay exponentially with increasing and thus with increasing tension. The duration of subprocess γ, on the other hand, appears to be relatively independent of both tension and vesicle size. The hemifused diaphragms in all fusion events that do not involve a (meta)stable hemifused state rupture within a time interval of 150–300 ns.
Figure 11.
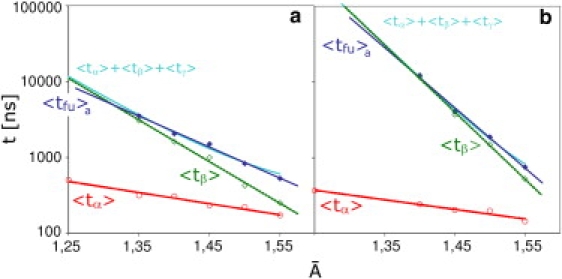
The average duration of the tension-dependent subprocesses 〈tα〉 (red circles) and 〈tβ〉 (green open diamonds) displayed together with 〈tfu〉a (blue solid diamonds) as a function of the area per molecule (a) for the 14-nm and (b) for the 28-nm vesicle. Both 〈tα〉 and 〈tβ〉 show an exponential dependence on . The light blue curve represents a new fit of the fusion time based on the sum 〈tα〉 + 〈tβ〉 + 〈tγ〉, where 〈tγ〉 is the rupture time of the hemifused diaphragm.
An improved fit of the average fusion time 〈tfu〉a thus consists of two superimposed exponentials plus a constant for the rupture time (as also shown in Fig. 11). While at high tensions it agrees with an exponential fit to the fusion time, at low tensions it deviates toward longer times and coincides with 〈tβ〉.
The time dependence of the flipping and the reordering process implies that the corresponding energy barriers governing the flipping rate and the nucleation of the hemifused diaphragm should depend linearly on the membrane tension as and . Here and are the respective barriers for a tension-free membrane and and are characteristic areas. The tension dependence of the corresponding timescales is described by and for process α and β, respectively. The exponential fits to the simulation data lead to values of the characteristic areas and for the 14-nm and and for the 28-nm vesicle. Numerical values for the energy barriers and will be discussed in the next subsections. The dependence of the flipping time on the tension is much weaker than that of the reordering time. Consequently, as tension is lowered, the fusion time is more and more dominated by the reordering time.
These simulations show that the fusion process has at least three subprocesses with corresponding energy barriers, two of which depend on the membrane tension. To find the magnitude of these energy barriers for relaxed membranes, the exponential fits of Fig. 11 can be extrapolated to zero tension. However, to find the values of and , knowledge of the preexponential scaling factor, tsc, is also required.
The flipping subprocess
Subprocess α is the process of interbilayer flips of single lipid tails. This local process, which changes the conformation of a single lipid relative to its surroundings, is accessible to direct simulation. The energy barrier for this process is provided by the (partially) hydrated polar headgroups of the proximal monolayers. It is intuitively clear that this barrier should decrease with increasing tension Σ, as the tension causes the headgroups in the planar membrane to move further apart and thus makes it easier for the hydrophobic chains to cross from one bilayer to the other.
To measure this energy barrier, simulations enforcing such interbilayer flips have been performed. In two adhering membranes, a single lipid tail is pulled slowly from its original position into the other bilayer, so that the lipid has one tail in each bilayer as observed in the fusion simulations. The average work required for this process in 20 independent simulations was found to be 〈W〉 = 9 ± 2 kBT. This value constitutes an upper bound for the energy barrier and should correspond to the barrier itself for very slow pulling.
Another estimate for the energy barrier comes from the Jarzynski relation (55) as given by
| (5) |
This equality should hold irrespective of how fast the process happens, if a sufficiently large number of trajectories is sampled. The average value from the enforced lipid flip simulations gives a barrier height of 8 kBT.
The distribution of the expended work W is relatively wide (see Fig. S7), because both the bead in the potential and the barrier itself fluctuate. Therefore, it is hard to draw any definite conclusion about the shape of the work distribution from the data.
To check the consistency of our previous estimate for the average expended work, we now use a cumulant expansion of the quantity 〈exp[W/kBT]〉. Truncating this expansion at second order, we obtain the average work
| (6) |
which leads to 〈W〉/kBT ≈ 8.9. This estimate is rather close to the previously mentioned value 〈W〉/kBT = 9 ± 2 as obtained by directly averaging the work, which shows that the higher order contributions of the cumulant expansion are small. For the very slow pulling speeds as used here, one would expect an essentially Gaussian distribution, for which the cumulants Cn = 0 for n > 2, and the results of the cumulant expansion agree very well with this expectation.
Finally, the decrease of the flipping times 〈tα〉 with increasing tension as observed in the fusion simulations indicates that the energy barrier for the flipping process is tension-dependent. Additional simulations of enforced interbilayer flips, in which the area per molecule is varied systematically, reproduce this tension dependence (as shown in Fig. 12) for two different values of the force amplitude aHC.
Figure 12.
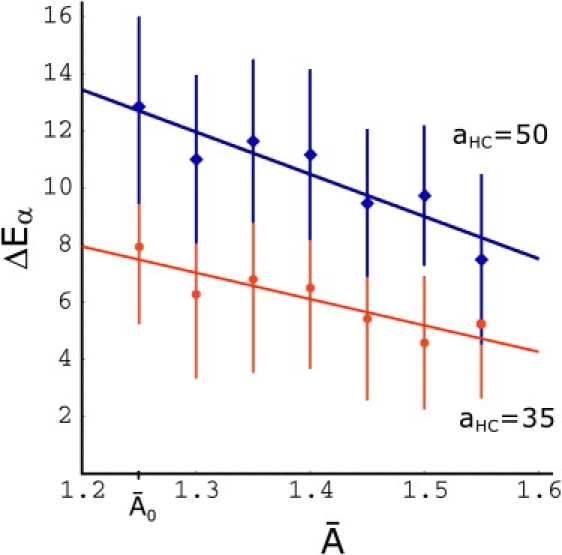
The energy barrier for the interbilayer flip of a lipid tail as a function of the area per molecule for two values of the head-tail force amplitude aHC, aHC = 50 (blue diamonds), and aHC = 35 (red circles). Each point is the average of 20 independent enforced interbilayer flips and the error bars represent mean ±1 SD. In the fusion simulations, the parameter value aHC = 35 has been used. The area corresponds to the tensionless membrane.
Energy barrier for interbilayer flips
The energy barrier for the flipping process, which is due to hydration in real membranes, is implemented in the coarse-grained simulations in the form of the stronger force amplitude aHC between head (H) and tail (C) beads. Therefore, for a given lipid architecture and parameter set, the height of the barrier can be expected to be determined by the value of the aHC.
A series of simulations of enforced interbilayer flips with different values of the aHC parameter in the range aHC = 35 to aHC = 50 confirms that expectation. Fig. 13 shows both the expectation value 〈W〉 (blue) and the energy barriers resulting from the Jarzynski relation (red) determined in these simulations as a function of aHC. The bars on the Jarzynski points indicate the addition of the second cumulant and show that the higher orders are small for all points. The plots clearly show that the height of the energy barrier increases with increasing strength of aHC.
Figure 13.
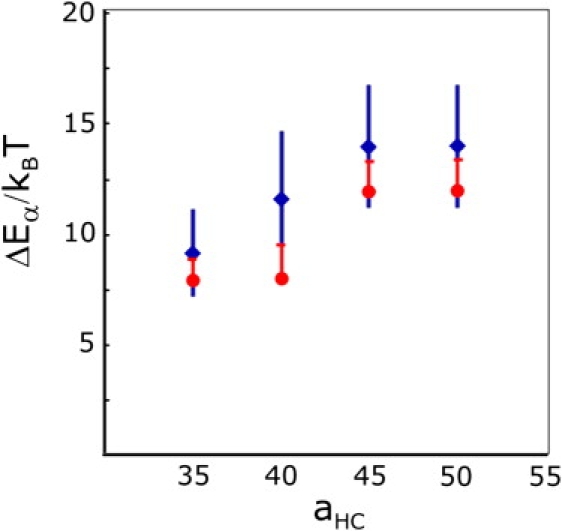
(Diamonds) The average energy barrier height plotted against the strength of the head-tail force amplitude aHC. Each point is determined from 20 independent enforced interbilayer flips. The error bars represent mean ±1 SD. (Circles) The energy determined by the Jarzynski relation 5. The end of the upward bar indicates the average expended work up to second order of the cumulant expansion. The average barrier height clearly increases with larger values of aHC. Therefore, the latter parameter can be used to fine-tune the energy barrier for interbilayer flips.
Since the flipping barrier depends on aHC, its magnitude can be tuned in such a way that the energy barrier is consistent with available reference data. A possible experimental estimate of the barrier height can be deduced from the hydration energy of the hydrocarbon chains, which can be estimated from the critical micelle concentration. The hydration energy of DMPC is ∼23.4 kBT, thus the hydration energy for only one of the hydrocarbon tails, roughly half of this estimate, should be ∼10 kBT. Pulling on two tails simultaneously in the simulations confirms that this costs approximately twice as much energy as to flip a single one.
Potential of mean force calculations in atomistic MD (56) and experimental studies (57,58) of the partitioning of hexane, i.e., parts of lipid tails with six carbon atoms, between water and the hydrophobic interior of DOPC bilayers also estimate the transfer energy to be of ∼10 kBT. In many fusion experiments the membranes are strongly dehydrated (see, e.g., (59,60)) and this barrier for interbilayer flips might be even lower.
Fusion simulations using the old DPD parameter set ((9), and see Table 1 (a)) gave different results both for the pathway and statistics:
-
1.
Fusion starts not at the contact line, but somewhere within the contact area.
-
2.
Successful fusion events occur very fast, usually within 300 ns and no tension dependence of the fusion time distributions could be deduced.
-
3.
The success rates are comparatively low, with hemifusion as the most likely result over a large range of tensions.
Simulations of enforced interbilayer flips show that there is no appreciable barrier for interbilayer flips for the old DPD parameter set (data not shown). The adhered state is unstable and membranes start to intermix upon contact. Furthermore, the membranes are very stable against pore formation and can be stretched by 60% before rupture.
Comparison of the two parameter sets shows that the aHC parameter of the old set is much higher than that of the new set used here, which leads to the conclusion that it is not the aHC parameter alone, but rather the ratio aHC/aCC, that determines the flipping barrier. For the old and new parameter set, this ratio is aHC/aCC = 2 and aHC/aCC = 3.5, respectively. In the absence of this barrier, lipids can intermix freely, so that the kink at the contact line that serves to lower the flipping energy has no influence on the initiation of fusion. Hemifusion can form rapidly and relax the tension. Fusion then only succeeds for very high tensions or when it is rapid enough to proceed before hemifusion has completely relaxed the membrane tension. Combined with the great stability against pore formation, this serves to stabilize the hemifused state.
If the aHC parameter of the old parameter set is raised by only five, i.e., aHC = 55 with all other parameters unchanged as in Table 1(b), the fusion pathway no longer proceeds without a barrier. Fusion starts at the contact line as observed in the simulations here and the fusion time increases from 250 to 600 ns. To determine the tension dependence, more simulations at different tensions would be required.
The parameter set used by Gao et al. (28) to study the fusion of two vesicles represents much stiffer bilayers where the tail chains tend to align and stick together. This makes splayed lipid conformations much more unlikely and thus leads to different fusion mechanisms in which the tail chains remain more aligned.
Additional energy barriers
Since the energy barrier for one of the processes of fusion could be measured in independent simulations, the scale factor tsc has become accessible. From the waiting time 〈tα〉 at and the estimate of ≃ 8 kBT obtained from the enforced interbilayer flips, the prefactor is found to be tsc = 0.12 ns and tsc = 0.16 ns for the 28-nm and 14-nm vesicle, respectively.
Using these values, we can now estimate the energy barriers for subprocess β corresponding to the nucleation of the hemifused patch, and for subprocess γ describing the rupture of this patch. The energy barrier for subprocess β is found to be = 11.1 ± 2 kBT and = 14.4 ± 2 kBT for the 14-nm and 28-nm vesicles, respectively, and the barrier for subprocess γ is estimated as 8 kBT.
At low tensions the total fusion time tfu is dominated by the reordering time, 〈tβ〉 (see Fig. 11), so that the energy barrier for fusion at low membrane tensions will also be dominated by the barrier for the reordering process. Thus the simulation statistics presented here suggest that the main energy barrier for fusion of tensionless bilayers is size-dependent and of ∼11 kBT and 14 kBT for fusion of the 14-nm and 28-nm vesicles, respectively.
Dependence of fusion times on vesicle radius
Similar to the success rates, there is a difference between the statistics for the two vesicle sizes. A comparison of the two graphics shows that the fusion times for the 30-nm vesicle are much longer than those for the 14-nm vesicle. This size dependence originates from the reordering process. The characteristic area for the 28 nm vesicle is almost a factor of two larger than that for the 14-nm vesicle, while the flipping time 〈tα〉 and the rupture time 〈tγ〉 appear to be independent of vesicle size.
We now want to argue that the different fusion behavior of the two vesicle sizes is again related to the ratio Rve/L|| of the vesicle radius to the size of the planar membrane, or more precisely to the area difference, , i.e., the difference between the actual area of the stretched membrane and that of a relaxed membrane with the same number of lipids.
Similar to the analysis by Tolpekina et al. (35), one would have to add lipid molecules to relax the tension of the planar membrane. In our system, these lipids can be added via interbilayer flips from the vesicle, and the subsequent formation of a hemifused patch.
If the area difference is large compared to the vesicle area, the number of lipids required to reduce the tension in the planar bilayer represents a considerable part of the vesicle membrane and will perturb the vesicle strongly. For a larger vesicle, this number represents a much smaller fraction of the vesicle's lipids and thus requires a much smaller perturbation of the membrane. This dependence indicates that the fusion probability and time presumably depend on the ratio Rve/L||.
To explicitly confirm this conclusion, additional simulations of a small vesicle with diameter 14 nm in a smaller simulation box with base length L|| = 36 nm have been performed. For this system, the ratio Rve/L|| is the same as for the system with the 28-nm vesicle. The corresponding data for the two systems are displayed in Fig. S8. By comparing these data with those of Fig. 11, we conclude that the different fusion times observed for the two vesicle sizes are primarily determined by the ratio Rve/L|| rather than the vesicle size Rve alone. The difference that can be seen in Fig. S8 between the data for (L||, Rve) = (37,21) and (L||, Rve) = (72,41) is likely to stem from finite size effects.
Tension-dependence of fusion probability
In the previous subsections, we have focused on the successful fusion events and analyzed the corresponding fusion times. These times decrease exponentially with increasing membrane tension (see Figs. 10 and 11). Extrapolation to small tensions then leads to the quoted energy barriers for tensionless membranes. It is important to note, however, that these barriers apply only to the fusion pathway and do not take any other pathway into account as observed during the unsuccessful fusion attempts. As mentioned before, these alternative pathways consist primarily of rupture at large membrane tensions and of adhesion and hemifusion at small tensions.
The fraction of successful fusion events, which defines the fusion probability, is shown in Fig. 14 as a function of the molecular area , which is directly related to the tension . Inspection of this figure shows that the fusion probability exhibits a maximum at intermediate tension values and there has a value close to one. For larger tensions, the fusion probability decreases because of rupture; for smaller tensions, it also decreases because of adhesion and hemifusion. Linear extrapolation of the simulation data to smaller values of the molecular area suggests that the fusion probability may vanish before the membrane reaches its tensionless state. Indeed, no spontaneous fusion events of the smaller vesicle are observed for runs up to 20 μs, i.e., twice as long as predicted from the extrapolation in Fig. 11.
Figure 14.
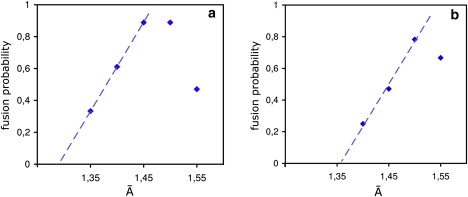
Fusion probability as a function of molecular area for (a) the 14-nm and (b) the 28-nm vesicles. In both cases, the fusion probability, which represents the fraction of fusion attempts that lead to fusion within 20 μs, exhibits a maximum at with 1.45 < < 1.5 in panel a and ≃ 1.5 in panel b corresponding to the tensions ≃ 3.36 and ≃ 4.25, respectively. At higher tensions, fusion becomes less likely because of membrane rupture; at lower tensions, fusion is more and more replaced by adhesion or hemifusion. A linear extrapolation of the data to smaller values of indicates a molecular area threshold for fusion at = 1.29 for the 14-nm and = 1.36 for the 28-nm vesicle. This corresponds to a tension threshold ≃ 0.56 for the 14-nm vesicle and ≃ 1.79 for the 28-nm vesicle.
Thus one may speculate that the system exhibits a threshold value, , for the membrane stretch, which also implies a threshold tension : for , or , the unfused state is the globally stable one. Such a threshold is not implausible: If a relaxed bilayer takes up lipids from the vesicle or forms a hemifused patch, it does not lower its energy, but rather has to be compressed.
Conclusions
From a detailed study of the effects of individual simulation parameters on the properties of simulated bilayers, it was possible to construct a coarse-grained membrane with more realistic properties. In particular, the improved stretching behavior and the introduction of an energy barrier against interbilayer flips between bilayers have led to a different fusion pathway, and a realistic dependence of the fusion time on the membrane tension.
A large number of fusion simulations show a common fusion pathway, which involves much more disordered and less symmetric intermediate states than is typically assumed. In this process, lipids in a splayed tail conformation, with one tail inserted in each membrane, play an important role during the onset of fusion.
The timescales of the fusion events suggest that the fusion process consists of at least three consecutive subprocesses: Interbilayer flips of lipid tails; nucleation of a small hemifused area; and pore formation. The first two of the timescales of the simulated membranes depend exponentially on the tension, revealing two tension-dependent energy barriers. Using simulations that enforce interbilayer flips of individual lipid tails and utilizing two different methods, one of which makes use of the Jarzynski relation, the energy scale for these barriers could be determined. Furthermore, the simulation parameter aHC was shown to be closely related to the energy scale of the interbilayer flips (see Fig. 13) and can be used to tune this energy barrier to a desired size.
These simulations reveal that the conformations of individual molecules play a crucial role in membrane processes on such small scales and cannot easily be neglected. Furthermore the study demonstrates that the fusion process has a stochastic character reflecting the thermal fluctuations of the lipid and water molecules. Therefore, to obtain reliable data on the average fusion times and other observable quantities, it is necessary to study a large number of fusion events. Indeed, both the average fusion time and the width of the fusion time distribution grow exponentially as one lowers the membrane tension, see Fig. 10. Because of the large width of the distributions, a single trajectory does not suffice to reveal the dependence of the average behavior.
For further studies of this kind it would be highly desirable to obtain experimental data on intermembrane flips and the corresponding energy barrier. It will then be useful to extend these simulations to more complex systems containing mixtures of lipids with realistic properties and including possible effects of proteins or other fusogens.
Acknowledgments
MEMPHYS is supported by the Danish National Research Council.
Supporting Material
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Data S1—DPD Simulation Method
Data S2—Stress Tensor Calculation
Data S3—Simulated Systems
Data S4—Geometry of Tense Membranes
References
- 1.Tamm L.K., Crane J., Kiessling V. Membrane fusion: a structural perspective on the interplay of lipids and proteins. Curr. Opin. Struct. Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 2.Lindau M., de Toledo G.A. The fusion pore. Biochim. Biophys. Acta Mol. Cell Res. 2003;1641:167–173. doi: 10.1016/s0167-4889(03)00085-5. [DOI] [PubMed] [Google Scholar]
- 3.Haluska C.K., Riske K.A., Marchi-Artzner V., Lehn J.M., Lipowsky R., Dimova R. Time scales of membrane fusion revealed by direct imaging of vesicle fusion with high temporal resolution. Proc. Natl. Acad. Sci. USA. 2006;103:15841–15846. doi: 10.1073/pnas.0602766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noguchi H., Takasu M. Fusion pathways of vesicles: a Brownian dynamics simulation. J. Chem. Phys. 2001;115:9547–9551. [Google Scholar]
- 5.Müller M., Katsov K., Schick M. New mechanism of membrane fusion. J. Chem. Phys. 2002;116:2342–2345. [Google Scholar]
- 6.Marrink S., Mark A. Mechanism of vesicle fusion as revealed by molecular dynamics simulations. J. Am. Chem. Soc. 2003;125:11144–11145. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 7.Stevens M.J., Hoh J.H., Woolf T.B. Insights into the molecular mechanism of membrane fusion from simulation: evidence for the association of splayed tails. Phys. Rev. Lett. 2003;91:188102. doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 8.Smeijers A.F., Markvoort A.J., Pieterse K., Hilbers A.J. A detailed look at vesicle fusion. J. Phys. Chem. B. 2006;110:13212–13219. doi: 10.1021/jp060824o. [DOI] [PubMed] [Google Scholar]
- 9.Shillcock J., Lipowsky R. Tension-induced fusion of bilayer membranes and vesicles. Nat. Mater. 2005;4:225–228. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 10.Li D.W., Liu X.Y. Examination of membrane fusion by dissipative particle dynamics simulation and comparison with continuum elastic models. J. Chem. Phys. 2005;122:1749091–1749098. doi: 10.1063/1.1889433. [DOI] [PubMed] [Google Scholar]
- 11.Knecht V., Mark A.E., Marrink S.J. Phase behavior of a phospholipid/fatty acid/water mixture studied in atomic detail. J. Am. Chem. Soc. 2006;128:2030–2034. doi: 10.1021/ja056619o. [DOI] [PubMed] [Google Scholar]
- 12.Knecht V., Mark A.E., Marrink S.J. Molecular dynamics simulations of lipid vesicle fusion in atomic detail. Biophys. J. 2007;92:4254–4261. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasson P.M., Kelley N.W., Singhal N., Vrljic M., Brunger A.T. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc. Natl. Acad. Sci. USA. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasson P.M., Zornorodian A., Park S., Singhal N., Guibas L.J. Persistent voids: a new structural metric for membrane fusion. Bioinformatics. 2007;23:1753–1759. doi: 10.1093/bioinformatics/btm250. [DOI] [PubMed] [Google Scholar]
- 15.Grafmüller A., Shillcock J., Lipowsky R. Pathway of membrane fusion with two tension-dependent energy barriers. Phys. Rev. Lett. 2007;98:218101. doi: 10.1103/PhysRevLett.98.218101. [DOI] [PubMed] [Google Scholar]
- 16.Goetz R., Lipowsky R. Computer simulations of bilayer membranes: self-assembly and interfacial tension. J. Chem. Phys. 1998;108:7397–7409. [Google Scholar]
- 17.Shillcock J., Lipowsky R. Equilibrium structure and lateral stress distribution of amphiphilic bilayers from dissipative particle dynamics simulations. J. Chem. Phys. 2002;117:5048–5061. [Google Scholar]
- 18.Finkelstein A., Zimmerberg J., Cohen F.S. Osmotic swelling of vesicles: its role in the fusion of vesicles with planar phospholipid bilayer membranes and its possible role in exocytosis. Annu. Rev. Physiol. 1986;48:163–174. doi: 10.1146/annurev.ph.48.030186.001115. [DOI] [PubMed] [Google Scholar]
- 19.Rowlinson J.S., Widom B. Oxford, Oxford University Press; Oxford, UK: 1982. Molecular Theory of Capillarity. [Google Scholar]
- 20.Groot R.D., Warren P.B. Dissipative particle dynamics: bridging the gap between atomistic and mesoscopic simulation. J. Chem. Phys. 1997;107:4423–4435. [Google Scholar]
- 21.Vattulainen I., Karttunen M., Besold G., Polson J.M. Integration schemes for dissipative particle dynamics simulations: From softly interacting systems towards hybrid models. J. Chem. Phys. 2002;116:3967–3979. [Google Scholar]
- 22.Yamamoto S., Maruyama Y., Hyodo S.-A. Dissipative particle dynamics study of spontaneous vesicle formation of amphiphilic molecules. J. Chem. Phys. 2002;116:5842–5849. [Google Scholar]
- 23.Kranenburg M., Venturoli M., Smit B. Phase behavior and induced interdigitation in bilayers studied with dissipative particle dynamics. J. Phys. Chem. B. 2003;107:11491–11501. [Google Scholar]
- 24.Stoyanov S.D., Groot R.D. From molecular dynamics to hydrodynamics: a novel Galilean invariant thermostat. J. Chem. Phys. 2004;122:114112/1–114112/8. doi: 10.1063/1.1870892. [DOI] [PubMed] [Google Scholar]
- 25.Guillot B. A reappraisal of what we have learnt during three decades of computer simulations on water. J. Mol. Liq. 2002;101:219–260. [Google Scholar]
- 26.Kornberg R.D., McConnell H.M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 27.Wimley W.C., Thompson T.E. Exchange and flip-flop of dimyristoylphosphatidylcholine in liquid-crystalline, gel, and two-component, two-phase large unilamellar vesicles. Biochemistry. 1990;29:1296–1303. doi: 10.1021/bi00457a027. [DOI] [PubMed] [Google Scholar]
- 28.Gao L., Lipowsky R., Shillcock J. Tension-induced vesicle fusion: pathways and pore dynamics. Soft Matter. 2008;4:1208–1214. doi: 10.1039/b801407h. [DOI] [PubMed] [Google Scholar]
- 29.Schofield P., Henderson J.R. Statistical mechanics of inhomogeneous fluids. Proc. R. Soc. Lond. A. 1982;379:231–246. [Google Scholar]
- 30.Nagle J.F., Tristam-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orädd G., Lindblom G., Westermann P.W. Lateral diffusion of cholesterol and dimyristoylphosphatidylcholine in lipid bilayer measured by pulsed field gradient NMR spectroscopy. Biophys. J. 2002;83:2702–2704. doi: 10.1016/S0006-3495(02)75279-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz V., Nielsen S.O., Discher D.E., Klein M.L., Lipowsky R. Dissipative particle dynamics simulations of polymersomes. J. Phys. Chem. B. 2005;109:17708–17714. doi: 10.1021/jp0512762. [DOI] [PubMed] [Google Scholar]
- 33.Evans E., Rawicz W. Entropy driven tension and bending elasticity in condensed-fluid membranes. Phys. Rev. Lett. 1990;64:2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 34.Discher B.M., Won Y.-Y., Ege D.S., Lee J.C.-M., Bates F.S. Polymersomes: tough vesicles made from diblock copolymers. Science. 1999;284:1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 35.Tolpekina T.V., den Otter W.K., Briels W.J. Simulations of stable pores in membranes: System size dependence and line tension. J. Chem. Phys. 2004;121:8014–8020. doi: 10.1063/1.1796254. [DOI] [PubMed] [Google Scholar]
- 36.Illya G., Lipowsky R., Shillcock J. Effect of chain length and asymmetry on material properties of bilayer membranes. J. Chem. Phys. 2005;122:244901. doi: 10.1063/1.1917794. [DOI] [PubMed] [Google Scholar]
- 37.Rawicz W., Olbrich K.C., McIntosh T., Needham D., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor R.S. Lateral pressures in cell membranes: a mechanism for modulation of protein function. J. Phys. Chem. B. 1997;101:1723–1725. [Google Scholar]
- 39.Asakura S., Oosawa F. Interaction of particles suspended in solutions of macromolecules. J. Polym. Sci. [B] 1958;33:183–192. [Google Scholar]
- 40.Lis L.J., McAlister M., Fuller N., Rand R.P., Parsegian V.A. Interactions between neutral phospholipid bilayer membranes. Biophys. J. 1982;37:657–666. [PMC free article] [PubMed] [Google Scholar]
- 41.Cevc G., Fenzl W., Sigl L. Surface-induced x-ray reflection visualization of membrane orientation and fusion into multibilayers. Science. 1990;249:1161–1163. doi: 10.1126/science.249.4973.1161. [DOI] [PubMed] [Google Scholar]
- 42.Marra J., Israelachvili J. Direct measurements of forces between phosphatidylcholine and phosphatidylethanolamine bilayers in aqueous-electrolyte solutions. Biochemistry. 1985;24:4608–4618. doi: 10.1021/bi00338a020. [DOI] [PubMed] [Google Scholar]
- 43.Evans E., Needham D. Physical properties of surfactant bilayer membranes: thermal transitions, elasticity, rigidity, cohesion, and colloidal interactions. J. Phys. Chem. 1987;91:4219–4228. [Google Scholar]
- 44.Helfrich W. Tension-induced mutual adhesion and a conjectured superstructure of lipid membranes. In: Lipowsky R., Sackmann E., editors. Handbook of Biological Physics. Elsevier; Amsterdam, The Netherlands: 1995. [Google Scholar]
- 45.Lipowsky R. Generic interactions of flexible membranes. In: Lipowsky R., Sackmann E., editors. Handbook of Biological Physics. Elsevier; Amsterdam, The Netherlands: 1995. [Google Scholar]
- 46.Lipowsky R. Melting at grain boundaries and surfaces. Phys. Rev. Lett. 1986;57:2876. doi: 10.1103/PhysRevLett.57.2876. [DOI] [PubMed] [Google Scholar]
- 47.Lipowsky R., Leibler S. Unbinding transitions of interacting membranes. Phys. Rev. Lett. 1986;56:2541–2544. doi: 10.1103/PhysRevLett.56.2541. [DOI] [PubMed] [Google Scholar]
- 48.Seifert U., Lipowsky R. Adhesion of vesicles. Phys. Rev. A. 1990;42:4768–4771. doi: 10.1103/physreva.42.4768. [DOI] [PubMed] [Google Scholar]
- 49.Kinnunen P.K.J., Holopainen J.M. Mechanisms of initiation of membrane fusion: role of lipids. Biosci. Rep. 2000;20:465–482. doi: 10.1023/a:1010402819509. [DOI] [PubMed] [Google Scholar]
- 50.Wang L., Seeley E.S., Wickner W., Merz A.J. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 51.Chernomordik L., Kozlov M.M., Zimmerberg J. Lipids in biological membrane-fusion. J. Membr. Biol. 1995;146:1–14. doi: 10.1007/BF00232676. [DOI] [PubMed] [Google Scholar]
- 52.Chernomordik L. Non-bilayer lipids and biological fusion intermediates. Chem. Phys. Lipids. 1996;81:203–213. doi: 10.1016/0009-3084(96)02583-2. [DOI] [PubMed] [Google Scholar]
- 53.Evans K.O., Lentz B.R. Kinetics of lipid rearrangements during poly(ethylene glycol)-mediated fusion of highly curved unilamellar vesicles. Biochemistry. 2002;41:1241–1249. doi: 10.1021/bi011508x. [DOI] [PubMed] [Google Scholar]
- 54.Lentz B.R., Talbot W., Lee J.K., Zheng L.X. Transbilayer lipid redistribution accompanies poly(ethylene glycol) treatment of model membranes but is not induced by fusion. Biochemistry. 1997;36:2076–2083. doi: 10.1021/bi9623340. [DOI] [PubMed] [Google Scholar]
- 55.Jarzynski C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997;78:2690–2693. [Google Scholar]
- 56.MacCallum J.L., Tieleman D.P. Computer simulation of the distribution of hexane in a lipid bilayer: spatially resolved free energy, entropy, and enthalpy profiles. J. Am. Chem. Soc. 2006;128:125–130. doi: 10.1021/ja0535099. [DOI] [PubMed] [Google Scholar]
- 57.White S.H., King G.I., Cain J.E. Location of hexane in lipid bilayers determined by neutron-diffraction. Nature. 1981;290:161–163. [Google Scholar]
- 58.Deyoung L.R., Dill K.A. Partitioning of nonpolar solutes into bilayers and amorphous n-alkanes. J. Phys. Chem. 1990;94:801–809. [Google Scholar]
- 59.Lentz B.R. PEG as a tool to gain insight into membrane fusion. Eur. Biophys. J. Biophys. Lett. 2007;36:315–326. doi: 10.1007/s00249-006-0097-z. [DOI] [PubMed] [Google Scholar]
- 60.Tamm L.K., Han X., Li Y., Lai A.L. Structure and function of membrane fusion peptides. Biopolymers. 2002;66:249–260. doi: 10.1002/bip.10261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Figure S7
Figure S8
Data S1—DPD Simulation Method
Data S2—Stress Tensor Calculation
Data S3—Simulated Systems
Data S4—Geometry of Tense Membranes


