Abstract
Buchnera aphidicola is an obligate, strictly vertically transmitted, bacterial symbiont of aphids. It supplies its host with essential amino acids, nutrients required by aphids but deficient in their diet of plant phloem sap. Several lineages of Buchnera show adaptation to their nutritional role in the form of plasmid-mediated amplification of key-genes involved in the biosynthesis of tryptophan (trpEG) and leucine (leuABCD). Phylogenetic analyses of these plasmid-encoded functions have thus far suggested the absence of horizontal plasmid exchange among lineages of Buchnera. Here, we describe three new Buchnera plasmids, obtained from species of the aphid host families Lachnidae and Pemphigidae. All three plasmids belong to the repA1 family of Buchnera plasmids, which is characterized by the presence of a repA1-replicon responsible for replication initiation. A comprehensive analysis of this family of plasmids unexpectedly revealed significantly incongruent phylogenies for different plasmid and chromosomally encoded loci. We infer from these incongruencies a case of horizontal plasmid transfer in Buchnera. This process may have been mediated by secondary endosymbionts, which occasionally undergo horizontal transmission in aphids.
Many insects maintain symbiotic associations with bacteria (1). These symbioses vary widely in properties like persistence, tissue distribution of the bacteria, mode of transmission, and level of integration between both partners. One end of the spectrum is represented by bacteriocyte symbioses, associations that are obligate, permanent, and mutualistic (2). They are estimated to occur in 10% of insect species (3). Bacteria in these associations are confined to a single, specialized cell type (bacteriocyte) and are transmitted maternally (2). In general, they play a nutritional role by complementing certain deficiencies in the diets of their hosts (4).
A prime example of bacteriocyte symbiosis is the association between aphids and Buchnera aphidicola (5). The former are plant-sap sucking insects of the order Hemiptera and Buchnera are γ-proteobacteria closely related to the Enterobacteriaceae (6). Their symbiosis resulted from a single bacterial infection of the common ancestor to all extant aphids, 100–250 million years ago (7). Host and symbiont lineages have evolved in parallel ever since. Besides Buchnera, aphids often harbor additional bacteria in their guts and in tissues surrounding the bacteriocytes (3, 8). The latter are commonly referred to as secondary endosymbionts. These too have been shown to be subject to vertical transmission (9), but their patchy distribution among aphid populations implies that they, unlike Buchnera, also undergo horizontal transmission (3).
Buchnera provisions its host with essential amino acids, nutrients in short supply in the aphids' diet of plant-phloem sap (4, 5). The genetic organization of amino acid biosynthetic pathways suggests that at least some lineages of Buchnera are adapted to overproduce these nutrients. Genes encoding key enzymes in the pathways leading to tryptophan and leucine (trpEG and leuABCD, respectively) have been found to be amplified through their translocation from the chromosome to plasmids (10, 11). In the case of the leucine genes, this translocation occurred to a resident plasmid that contained a repA1-replicon responsible for replication initiation (12). This replicon is evolutionarily related to the IncFII replicon, which occurs on a number of enterobacterial virulence and antibiotic resistance plasmids (13–15). In Buchnera, the replicon is defined by a replication initiation gene (repA1) and a distinct origin of replication located immediately downstream of this gene (12). Most plasmids contain an additional, shorter variant of the replication initiation gene (repA2), which is thought to have originated from a duplication of repA1 (16, 17), i.e., they are paralogous genes. Apart from its association with the leucine genes, the repA1-replicon also has been found on a small, cryptic plasmid in a Buchnera species in which leuABCD is still encoded on the chromosome (12). This small plasmid is thought to resemble the ancestral form that was used for the amplification of the leucine genes in different lineages of Buchnera.
Although the number of independent translocation events of leuABCD is disputed (12, 17), previous phylogenetic analyses have all suggested the absence of horizontal plasmid exchange in Buchnera. Here we present the finding and characterization of three plasmids containing the repA1-replicon. Based on phylogenetic analyses of all currently characterized repA1-replicons, we present evidence for gene conversion between directly oriented, duplicated repA genes and for horizontal or “postsymbiotic” acquisition of a plasmid carrying the replicon in one lineage of Buchnera. To our knowledge, this represents the first report of horizontal gene transfer in an insect–bacterial symbiosis.
Materials and Methods
General Methods.
Plant material carrying aphids was collected in the field and kept at 4°C until further manipulation of the animals. The aphids Pemphigus spyrothecae (collected from galls on Populus trees) and Geoica urticularia (collected from galls on Pistacea trees) were available in quantities sufficient to perform isolations of Buchnera cells and subsequent Buchnera plasmid DNA and genomic DNA isolations. Only a limited amount of aphids from Tuberolachnus salignus (collected from Salix trees) were available, and a whole-aphid DNA isolation method was used to maximize DNA yield. Methods used for Buchnera isolation, Buchnera plasmid DNA isolation, whole-aphid DNA isolation, inverse long PCR, routine cloning, and nucleotide sequencing have been described previously (12). Small plasmids isolated from Buchnera of the aphids P. spyrothecae and G. urticularia were linearized with EcoRI and XbaI, respectively, cloned, and completely sequenced. A Buchnera plasmid carrying leuA was amplified by inverse long PCR from whole aphid DNA (12) of the species T. salignus. The PCR product was 6.2 kb in size. It was digested with BamHI, cloned, and physically mapped, and portions were sequenced.
PCR and Sequencing of groE.
Primers for amplification of the groE (groES–groEL) cluster (≈1.9 kb) were designed on the basis of bacterial GroE sequences available at the European Molecular Biology Laboratory (EMBL) database: groExF1 5′-ATGAAWATTCGTCCRTTRCAYGATCG-3′ and groExR1 5′-TTACATCATKCCRCCCATRCCACCCA-3′. The groE cluster from Buchnera of the aphids P. spyrothecae, G. urticularia, and T. salignus was amplified from the same DNA preparations used in the previous analyses. In addition, the same fragment was amplified by using DNA from the aphids Pterocomma populeum, Tetraneura caerulescens, and Thelaxes suberi, which was extracted as in ref. 12. PCR products were cloned in pGEM-T Easy vector (Promega) and partially sequenced. The sequences obtained comprised the complete groES gene, the intergenic region, and 5′ partial (248 nt) sequence of groEL gene. For phylogenetic reconstruction, we used the coding sequences mentioned above, and groE from Buchnera of the aphid Acyrtosiphon pisum, and the Enterobacteria Yersinia enterocolitica, Escherichia coli, and Hemophilus influenzae, with EMBL accession nos. X61150, D14078, X07850, and U32736, respectively.
Analysis of DNA and Protein Sequences.
DNA sequence data were assembled and analyzed with the Genetics Computer Group (GCG) program package version 8.0 (18). blastp searches (19) were done at the network server of the National Center for Biotechnology Information, RepA sequences were aligned with dialign (20) and manually improved at regions of high divergence. Ibp sequences (see Results) were aligned as previously described (12), and groE sequences were aligned with clustalv (21). Phylogenetic reconstructions were obtained by using a maximum likelihood approach, with programs paml version 2.0 (22) and puzzle 4.0 (23). Additional analyses were performed by using the neighbor-joining algorithm (24) as implemented in the program neighbor from the phylip package (25). The amino acid substitution model was based on an update to Dayhoff's PAM250 matrix (26). The Kimura 2-parameter model was used to take superimposed nucleotide substitutions into account (27). All analyses used pairwise gap deletion options to deal with alignment gaps.
In the neighbor-joining method, the significance of reconstructions was tested by bootstrap resampling (1,000 replicates) to obtain estimates of the support for each node (28). The significance of the reconstructions in the maximum likelihood approach was tested by two methods. First, branches were tested for being longer than zero by using the corresponding standard errors derived from the information matrix (22). Branches were considered longer than zero when their lengths were larger than twice the standard error. Second, quartet puzzling was used to obtain the proportion of times for which each internal branch was recovered as significant in each possible quartet of sequences (23). This gives an estimate of the total support for each internal branch.
Tests of alternative topologies were carried out using the Resampling Estimate of Log-Likelihood (RELL) method (30, 31), which gives a bootstrap probability for candidate trees without performing a maximum likelihood estimation for each resampled data set. The package molphy 2.2 (32) was used to perform the RELL tests.
Relative rate tests of nucleotide substitutions were done using Wu and Li's (33) method complemented with Muse and Weir (34) as implemented in program k2wuli (35). This method considers tests among pairs of sequences and, consequently, multiple comparisons were dealt with using Bonferroni's sequential correction (36).
Results
Description of the Plasmids.
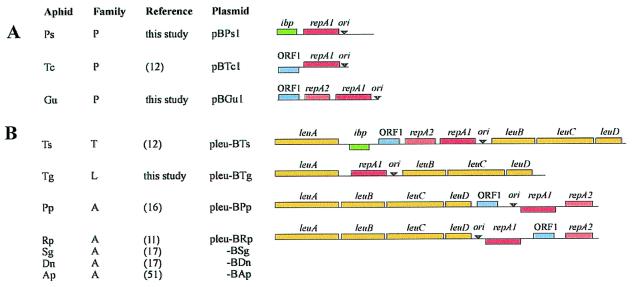
Similar to previously studied Buchnera of the aphid Tetraneura caerulescens (12), Buchnera of Pemphigus spyrothecae, and Geoica urticularia were each found to contain a small, cryptic plasmid. The two new plasmids, designated pBPs1 and pBGu1, are 2308 and 2387 bp in length and have average GC contents of 24.3 and 23.0%, respectively. pBPs1 contains two ORFs that are homologous to genes encoding plasmid replication initiation protein RepA1 and small heat-shock protein Ibp (37). Ibp also is carried by the leucine plasmid of Buchnera of Thelaxes suberi (12) but is chromosomally encoded in Buchnera from the aphid Schizaphis graminum (38). pBGu1 contains three ORFs, two of which are homologous to the paralogs repA1 and repA2 of the same replication initiation gene. The third ORF is homologous to ORF1, which encodes an inner membrane protein and which is commonly present on Buchnera leucine plasmids (11, 12, 16, 17). Physical maps of pBPs1 and PBGu1 are presented in Fig. 1A, together with the previously characterized plasmid from Buchnera of T. caerulescens (pBTc1) (12). The respective aphid hosts all belong to the family Pemphigidae (39, 40).
Figure 1.
Linearized physical maps of Buchnera plasmids carrying the repA1-replicon. (A) Small, cryptic plasmids from Buchnera of members of the family Pemphigidae (P). (B) Leucine plasmids from Buchnera of members of the families Thelaxidae (T), Lachnidae (L), and Aphididae (A). Species abbreviations: Ps, Pemphigus spyrothecae; Tc, Tetraneura caerulescens; Gu, Geoica urticularia; Ts, Thelaxes suberi; Tg, Tuberolachnus salignus; Pp, Pterocomma populeum; Rp, Rhopalosiphum padi; Sg, Schizaphis graminum; Dn, Diuraphis noxia; and Ap, Acyrthosiphon pisum. Leucine plasmids from Buchnera of Sg, Dn, and Ap (17, 58) are highly similar to pleu-BRp. Genes above lines are transcribed in rightward direction and those below lines in leftward direction. Arrowheads indicate the approximate position of the conserved region in the origin of replication (ori) that contains three ATGC-repeats.
Inverse long PCR analysis revealed that Buchnera of Tuberolachnus salignus contains a plasmid that encodes genes involved in leucine biosynthesis. The plasmid is designated pleu-BTg and was found to carry only the repA1 gene in addition to leuABCD (Fig. 1B). The aphid host belongs to the family Lachnidae (39, 40), representing the third major lineage in which leucine plasmids have now been found.
The origin of replication of the Buchnera repA1-replicon is characterized by a conserved motif that contains three repeats of the tetranucleotide “ATGC” (5′-ATGC–N19–20–ATGC–N15–16–ATGC-3′) (12). All plasmids described here also contained this motif, suggesting that their replicons are homologous.
Alignment of RepA Sequences.
A comprehensive view of the evolution of the repA1-replicon in Buchnera can only be inferred through analysis of the repA genes because these are the only ones present on all plasmids. We therefore performed an elaborate phylogenetic analysis of these genes. Our dataset of RepA proteins comprised 10 RepA1 and seven RepA2 sequences (Fig. 1). The distantly related RepA sequences from Salmonella and Yersinia virulence plasmids and from E. coli plasmid R100 were included as outgroups. Individual sequence lengths and GC contents of repA genes are listed in Table 1. The length of the multiple alignment was 330 amino acids (complete alignment available upon request). RepA1 and RepA2 sequences differed from each other by two deletions (11 and 18 amino acids) in the C-terminal region of RepA2 (Fig. 2A).
Table 1.
Size and GC content of repA genes
| Species |
repA1
|
repA2
|
||
|---|---|---|---|---|
| S* | GC | S | GC | |
| Buchnera | ||||
| A. pisum | 283 | 29.23 | 250 | 24.97 |
| D. noxia | 280 | 28.23 | 251 | 25.13 |
| R. padi | 283 | 29.93 | 249 | 24.53 |
| S. graminum | 283 | 30.16 | 250 | 24.70 |
| P. populeum | 283 | 28.17 | 251 | 22.62 |
| T. salignus | 278 | 25.81 | ||
| T. suberi | 283 | 30.41 | 246 | 25.37 |
| T. caerulescens | 289 | 23.68 | ||
| G. utricularia | 305 | 25.71 | 204 | 20.49 |
| P. spyrothecae | 280 | 32.27 | ||
| Non-Buchnera | 285 | 57.23 | ||
| S. enteritidis | 288 | 56.90 | ||
| Y. enterocolitica | 289 | 54.56 | ||
*Size in amino acids.
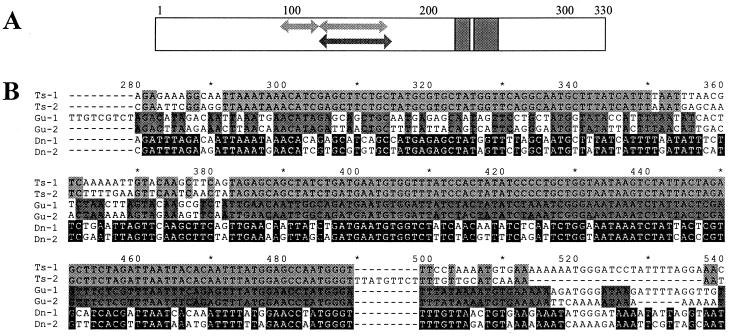
Figure 2.
Multiple alignment of Buchnera repA sequences. (A) Schematic representation of protein multiple alignment. Arrows in the region between amino acids 98 and 172 indicate location of segments that are identical among the repA1 and repA2 genes of plasmids pleu-BTs (upper arrows, positions 98–117 and 128–163) and pBGu1 (lower arrow, position 128–172). Gray blocks in the C-terminal domain indicate location and length of two deletions present in all repA2 genes. (B) Pairwise nucleotide sequence alignments of region of repA from plasmids pleu-BTs, pBGu1, and pleu-BDn, corresponding to amino acid positions 90–180, illustrating segments of complete identity in Ts-repAs (positions 292–350 and 383–489) and Gu-repAs (position 382–516).
Gene Conversion and Phylogenetic Analysis of RepA.
Pairwise alignments of repA1 and repA2 genes originating from a single plasmid revealed that the paralogs of Buchnera from T. suberi (Ts) and G. urticularia (Gu) contained long stretches between amino acid positions 98 and 172 that were completely identical at the nucleotide level between the two variants (Fig. 2A). In the Ts-repAs there were two such stretches, 59 and 107 nt long and in the Gu-repAs there was a single stretch of 126 nt. Although sequence similarity in this region was high across all species, other pairs of repA sequences from a single plasmid did not contain such long stretches of identity. This is illustrated in Fig. 2B, in which pairwise alignments are compared for the paralogs of Buchnera from T. suberi and G. urticularia and for those of Buchnera from Diuraphis noxia, a species in which divergence in this region was relatively high.
The fact that the identical segments in the Gu- and Ts-repA pairs did not even contain synonymous substitutions suggests that this identity did not result from selection on protein function. Neither is it likely that repA1 was only recently duplicated on these plasmids because divergences in other regions of the genes are comparable to those of other repA pairs. The sequence identity is most plausibly explained by assuming the action of a gene conversion mechanism (41). This finding is relevant in the present context because sequence conversion may obscure phylogenetic relationships for RepA. We therefore determined the direction of sequence conversion in the Ts- and Gu-repA pairs (data not shown) and subsequently compared phylogenetic reconstructions for three different multiple alignments. The first one was the original alignment. In the second, the deduced converted regions of Gu-RepA1 and Ts-RepA2 were removed, whereas in the third these two proteins were removed entirely.
The maximum likelihood tree estimated for the original dataset is presented in Fig. 3. Although most branches in this tree, as well as in those for the other two datasets, were significantly different from zero, support for internal nodes obtained in quartet puzzling and bootstrap resampling was very weak in most cases and local topologies differed considerably between trees. This was even more pronounced when different amino acid substitution models and methods of distance estimation and phylogenetic reconstruction were used for each alignment (data not shown). For the entire set of sequences, RepA1 and RepA2 never behaved as two monophyletic groups. Only sequences from the family Aphididae clustered into two groups of orthologous genes in most trees. Relationships within this family were also the only ones that conformed to established classifications of aphid hosts and previous molecular phylogenetic analyses (7, 16, 39, 40, 42).
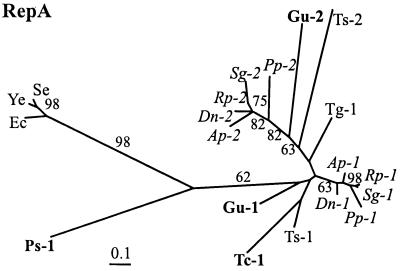
Figure 3.
Maximum likelihood tree for RepA proteins using the original multiple alignment. Figures near branches indicate support values larger than 50% obtained in quartet puzzling resampling. The number 1 designates RepA1 and 2 designates RepA2. Species abbreviations are given in the legend to Fig. 1. Sequences from Buchnera of the Pemphigidae are in bold face and those from the Aphididae are in italics. RepA outgroup sequences: Se, S. enteritidis virulence plasmid (U64797); Ye, Y. enterocolitica virulence plasmid (M55182); and Ec, E. coli plasmid R100 (P03066).
Aberrant Phylogenetic Position of Ps-RepA1.
One remarkable exception to the weakness of the phylogenetic signal in RepA was the placement of RepA1 from Buchnera of P. spyrothecae (Ps-RepA1) basal to the remaining Buchnera sequences (Fig. 3). This basal position was observed in each single analysis performed with the three multiple alignments. As a member of the Pemphigidae, sequences obtained from this species would be expected to show at least a tendency to cluster with other representatives of this family (Tc- and Gu-RepA1). Such results have indeed previously been obtained in studies of 16S rDNA and TrpEG sequences (7, 12, 43). However, RELL tests of alternative topologies, in which a monophyletic grouping of sequences derived from the Pemphigidae was enforced, showed that the placement of Ps-RepA1 was highly significant (relative support value of 97.18% for the topology shown in Fig. 3).
A profound divergence of Ps-RepA1 is illustrated by maximum likelihood distances to other Buchnera sequences, which on average were nearly twice as high as the average maximal distances within the latter group. This divergence cannot be explained by a possible recombination event, which might have produced a chimaeric gene, because amino acid differences among Ps-RepA1 and other Buchnera and enteric RepA1 proteins are distributed evenly over the entire sequence. Moreover, relative rate tests revealed that the evolutionary rate of Ps-repA1 was not aberrant compared to other Buchnera repA sequences: analysis of transversion substitutions among pairs of sequences showed a divergent evolutionary rate between repA from E. coli and those from Buchnera, including Ps-repA1, but there were no significant differences in evolutionary rate for this gene among Buchnera species, including the one isolated from P. spyrothecae (Table 2). This result is consistent with an acceleration of evolutionary rates in Buchnera compared to its free-living relatives (44). However, the apparent homogenous evolutionary rates among all Buchnera repA sequences suggests that the basal phylogenetic position of Ps-repA1 can unlikely be attributed to an artifact in phylogenetic reconstruction.
Table 2.
Results for comparisons of evolutionary rates (only transversions) among pairs of repA sequences
| Value in range | Taxon 1 | Taxon 2 | Taxon 3 | K13–K23 | z |
|---|---|---|---|---|---|
| Max | E. coli | Buchnera (Rp-1) | S. enteritidis | −0.464 ± 0.060 | −7.805* |
| Min | E. coli | Buchnera (Gu-2) | Y. enterocolitica | −0.865 ± 0.152 | −5.685* |
| E. coli | Buchnera (Ps) | S. enteritidis | −0.560 ± 0.073 | −7.618* | |
| Max | Buchnera (Tg-1) | Buchnera (Ps) | E. coli | 0.183 ± 0.121 | 1.515ns |
| Min | Buchnera (Sg-2) | Buchnera (Ps) | E. coli | −0.006 ± 0.108 | −0.060ns |
| Max | Buchnera (Rp-1) | Buchnera (Tg-1) | E. coli | −0.303 ± 0.101 | −2.993ns |
| Min | Buchnera (Sg-1) | Buchnera (Ts-1) | E. coli | 0.000 ± 0.062 | 0.000ns |
Four sets of comparisons are shown. The first two rows show the maximum (Max) and minimum (Min) deviations (z) from constancy of evolutionary rates between E. coli and Buchnera isolated from species other than P. spyrothecae. The next row shows the comparison between E. coli and Buchnera from P. spyrothecae. Rows 4 and 5 represent the extreme deviations in comparisons between Buchnera from P. spyrothecae and those from the remaining Buchnera species, and the last two rows show the extreme values among these. Significance of z values was obtained after application of Bonferroni's sequential correction procedure. Species names as in Fig. 1.
ns, not significant; *, P < 0.001.
Phylogenetic Analysis of GroESL and Ibp.
To provide an independent test for the incongruent phylogenetic relationships of the plasmid-encoded RepA from our presumed Buchnera isolate from P. spyrothecae relative to published phylogenetic schemes, we performed analyses of the chromosomally encoded groE cluster in relevant Buchnera species used in this study, as well as of available Ibp sequences. The groE cluster was amplified by PCR from genomic DNA samples that were obtained from the same bacterial preparations as the plasmid DNA isolations. Resulting PCR fragments were partially sequenced, yielding a GroESL multiple alignment with a total of 179 aa positions. A maximum likelihood tree estimated for this dataset is presented in Fig. 4A. In contrast to the RepA phylogenies, groupings in this tree were well-supported by bootstrap and quartet puzzling analyses. In accordance with of 16S rDNA and plasmid-encoded trpEG phylogenetic analyses (7, 12, 43), the sample obtained from P. spyrothecae clustered with the two other Buchnera sequences from members of the Pemphigidae.
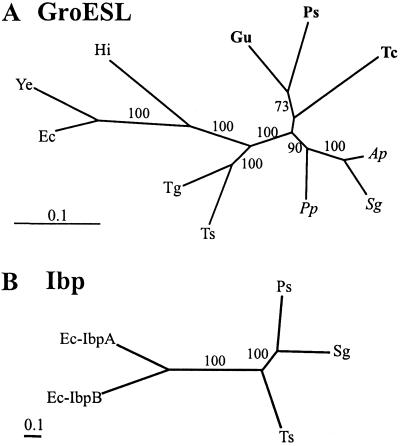
Figure 4.
Maximum likelihood trees for GroESL and Ibp proteins. (A) GroESL. Outgroup sequences: Hi, H. Influenzae; Ye, Y. enterocolitica; and Ec, E. coli. (B) Ibp. Outgroup sequences: Ec-IbpA and B, E. coli IbpA and IbpB. Figures near branches indicate support values greater than 50% obtained in quartet puzzling. Species abbreviations are given in the legend to Fig. 1.
The length of the multiple alignment of Ibp sequences was 172 positions. A maximum likelihood tree estimated for this dataset is presented in Fig. 4B. The three Buchnera Ibp sequences formed a strongly supported group, but relationships among them were inconsistent with previous phylogenetic schemes. These would predict a clustering of Ts-Ibp with either Ps- (12) or Sg-Ibp (7). The RELL test relative support value for the former topology was 12.64%, 2.5% for the second, whereas the one shown in Fig. 4B had a value of 84.82%. However, ibp is a small and variable gene (37) and phylogenetic reconstructions with a taxonomic sample as small as the present one should be interpreted with utmost caution. Nevertheless, in view of the observations made for RepA and GroESL, the significance of the present result lies in the fact that Ps-Ibp, similar to Ps-GroESL but contrary to Ps-RepA1, was highly similar to the two other Buchnera sequences.
Discussion
To date, Buchnera and all of its genetic information were thought to be transmitted strictly vertically. This finding was based on the demonstration of maternal transmission of symbionts in aphids (1, 45, 46) and supported by the strict congruence between phylogenies derived for aphids and Buchnera chromosomal and plasmid-borne genes (5, 7, 10, 16, 42). However, with the exception of 16S rDNA sequences, taxonomic sampling of Buchnera genes has been both limited and biased toward representatives of the Aphididae. Inferences made from such biased sampling on strictly vertical transmission of plasmids may hold for this particular lineage but are likely to miss processes that occurred in the early evolution of Buchnera or in other, more diverged lineages. Using a broader taxonomic sampling, we unexpectedly obtained a significant conflict among Buchnera gene phylogenies. Phylogenetic incongruence can arise merely from methodological errors or it may point to unexpected biological phenomena. We interpret the above conflicting gene phylogenies as evidence of horizontal transfer of plasmids in Buchnera. Our inferences hinge on the proposed monophyly of aphids and Buchnera from the family Pemphigidae (7, 12, 39, 40).
The repA1 gene of a plasmid (pBPs1) isolated from Buchnera of P. spyrothecae was found to be highly divergent relative to all other Buchnera repA sequences and was placed basal to these in RepA trees (Fig. 3). Previous phylogenies based on Buchnera 16S rDNA and TrpEG (7, 12, 43) and aphid morphology (7, 39) would predict a clustering with other members of the Pemphigidae. Such a clustering was further confirmed by our present analyses of GroE. Given the statistical significance of the present phylogenetic result and the finding that the evolutionary rate of Ps-RepA1 was not aberrant, which suggests that a systematic error accounting for its phylogenetic placement can be excluded, there remain two possible explanations for the incongruence: i) either an aphid-associated bacterium other than Buchnera was, contrary to our previous assumption, the source of plasmid pBPs1 in preparations made from P. spyrothecae or ii) although presently carried by Buchnera, Ps-repA1 was acquired horizontally by Buchnera from an unknown source in the P. spyrothecae-lineage after its divergence from the other Pemphigidae.
Several lines of evidence favor the second scenario of “postsymbiotic” plasmid acquisition. First, the low GC contents of Ps-repA1 (Table 1) and of pBPs1 as a whole (24.3%) are consistent with previous values reported for Buchnera DNA (5) but not with those of, e.g., aphid secondary endosymbiont DNA (3, 8, 47). Secondly, plasmid pBPs1 was isolated in relatively high amounts from P. spyrothecae. Both observations point to Buchnera as the most likely source of pBPs1. Thirdly, Ps-Ibp from the same plasmid, and Ps-GroESL from the same bacterial preparation, phylogenetically clustered strongly within the Buchnera lineage. Because repA1, unlike ibp, is part of a replicon, the segment of a plasmid that bears indispensable functions for replication, it is the most likely element to be transferred and to persist in different hosts. The phylogenetic position of Ps-Ibp therefore provides support for the assumption that pBPs1 is presently carried by Buchnera, but it also implies that this plasmid is an evolutionary chimaera of a horizontally acquired repA1-replicon and an ibp gene of Buchnera chromosomal origin. Fourthly, Buchnera of P. spyrothecae has recently been found to contain a tryptophan plasmid that also differs in a number of properties from those of other Buchnera, most notably with respect to the resident replicon (43). Like ibp, however, the trpEG genes from this plasmid were clearly of Buchnera origin. In the present context, these parallel cases of chimaeric plasmids contained within a single Buchnera lineage make a scenario of horizontal gene transfer as part of their evolutionary history most plausible.
Plasmid-mediated gene transfer is an important process in bacterial evolution (48–50) and is well-documented in plant-associated bacteria (51). A few recent studies have reported in insecta genetic exchange among plant-associated bacteria as well as among strains of the insect-pathogenic bacterium Bacillus thuringiensis (52, 53). Apart from a sometimes rich microbial gut-flora (8), aphids often harbor secondary endosymbionts in tissues bordering the bacteriocytes that contain Buchnera (1, 5, 47). Although these bacteria may be inherited maternally (9), their incidence and distribution suggest that they also undergo horizontal transmission among host lineages (3). It is conceivable that in the putative case of horizontal plasmid acquisition in Buchnera reported here, the donor may have been such a secondary endosymbiont.
Buchnera genome evolution is governed by three major processes: (i) a mutational bias that has led to an increase in A + T content (5, 54); (ii) the accumulation of mildly deleterious mutations in protein-coding genes, which has been attributed to mutational bias, small population size and the absence of recombination (55); and (iii) reductive genome evolution (56). Moran (55) has suggested that, collectively, these processes may determine an evolutionary fate for Buchnera, and possibly for other endosymbiotic bacteria, of “long-term deterioration.” As noted by the author, however, the fact that the aphid/Buchnera symbiosis has persisted for more than 100 million years implies that this process is either extremely slow or that compensatory processes counteract or retard the evolutionary outcome. In addition to high level expression of protein chaperonines (54) and recombination that might occur in the polyploid Buchnera genome (57), maintenance of a capacity for genetic exchange with free-living bacteria or insect secondary symbionts over a long period of evolutionary time, for which the present study has provided evidence, might also represent one such compensatory process. A broader implication is that if even a tightly integrated symbiont like Buchnera has long retained a capacity for genetic exchange, it is likely that bacteria more loosely associated with insects (secondary endosymbionts) still have unrestrained access to the wider microbial gene pool.
Previous studies have suggested that the duplication of repA1 was ancestral to the lineage leading to Buchnera of the families Thelaxidae and Aphididae (17). The current taxonomic sampling of repA sequences encompasses a wider range of evolutionary divergences but our phylogenetic analyses failed to reproduce a perfect clustering of repA1 and repA2 genes. We suspect that this is due primarily to the high variability of these genes and that indeed, the duplication of repA1 occurred only once. Rather than by basic similarity at the amino acid level, this is perhaps most strongly supported by structural similarities in the C-terminal region among RepA1 and RepA2 proteins, respectively. The length of this region is highly conserved in the former, whereas the RepA2 sequences show great length variation and seem to share at least two major deletions (Fig. 2A). The repA2 gene is not known from the distantly related, enterobacterial IncFII replicon and seems to be a unique but dispensable addition to the Buchnera repA1-replicon. However, the ancestral lineage in which the duplication of repA1, followed by major deletions in the C-terminal region of the resulting paralog, must have occurred, cannot still be inferred with certainty from the present dataset. To elucidate the complete evolutionary history of the repA1-replicon, including the origin of the leucine plasmids, the disputed positioning of the root within the Buchnera phylogeny must first be resolved (12, 17).
Acknowledgments
We thank J. M. Michelena for identification of aphid species and Colin Dale for comments on an earlier version of the manuscript. This work was supported by an European Union Human Capital Mobility research grant (CHRX-CT94–0660 to R.V.H.) and Grant PB96–0793 C0401 from Dirección General de Ensen̄anza Superior (to A.M.).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the EMBL database [accession nos. AJ404864 (pBPs1), AJ404863 (pBGu1), AJ404865 (BTg-repA1), AJ401307 (BPs-groE), AJ401305 (BGu-groE), AJ401309 (BTg-groE), AJ401306 (BPp-groE), AJ401308 (BTc-groE), and AJ401310 (BTs-groE)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180310197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180310197
References
- 1.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: Wiley Interscience; 1965. [Google Scholar]
- 2.Douglas A E. Biol Rev. 1989;69:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 3.Moran N A, Telang A. BioScience. 1998;48:295–304. [Google Scholar]
- 4.Douglas A E. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran N A, Clark M A. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 6.Munson M A, Baumann P, Kinsey M G. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 7.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171. [Google Scholar]
- 8.Harada H, Oyaizu H, Ishikawa H. J Gen Appl Microbiol. 1996;42:17–26. doi: 10.2323/jgam.43.349. [DOI] [PubMed] [Google Scholar]
- 9.Chen D Q, Purcell A H. Curr Microbiol. 1997;34:220–225. doi: 10.1007/s002849900172. [DOI] [PubMed] [Google Scholar]
- 10.Lai C-Y, Baumann L, Baumann P. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracho A M, Martinez-Torres D, Moya A, Latorre A. J Mol Evol. 1995;41:67–73. doi: 10.1007/BF00174042. [DOI] [PubMed] [Google Scholar]
- 12.van Ham R C H J, Moya A, Latorre A. J Bacteriol. 1997;179:4768–4777. doi: 10.1128/jb.179.15.4768-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Pena J M, Buisan M, Ibañez M, Rotger R. Gene. 1997;25:53–61. doi: 10.1016/s0378-1119(96)00776-7. [DOI] [PubMed] [Google Scholar]
- 14.Vanooteghem J-C, Cornelis G R. J Bacteriol. 1990;172:3600–3608. doi: 10.1128/jb.172.7.3600-3608.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Womble D D, Rownd R H. Microbiol Rev. 1988;52:433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva F J, van Ham R C H J, Sabater B, Latorre A. FEMS Microbiol Lett. 1998;168:43–49. doi: 10.1111/j.1574-6968.1998.tb13253.x. [DOI] [PubMed] [Google Scholar]
- 17.Baumann L, Baumann P, Moran N A, Sandstrom J, Thao M L. J Mol Evol. 1999;48:77–85. doi: 10.1007/pl00006447. [DOI] [PubMed] [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern B. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 21.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z H. palm, Phylogenetic Analysis by Maximum Likelihood. London: University College; 1999. , Version 2.0. [Google Scholar]
- 23.Strimmer K, von Haeseler A P. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 24.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. phylip, Phylogenetic Inference Package. Seattle: University of Washington; 1993. , Version 3.5c. [Google Scholar]
- 26.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 27.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Evolution (Lawrence, KS) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. Syst Biol. 1994;43:329–342. [Google Scholar]
- 30.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 31.Kishino H, Miyata M, Hasegawa M. J Mol Evol. 1990;31:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 32.Adachi J, Hasegawa M. molphy, A Program Package for Molecular Phylogenetic. The Graduate University for Advanced Study, Tokyo: Department of Statistical Science; 1994. , Version 2.2. [Google Scholar]
- 33.Wu C-I, Li W-H. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muse S V, Weir B S. Genetics. 1992;132:269–276. doi: 10.1093/genetics/132.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jermiin L S. K2WuLi. Australian National University, Canberra, Australia: John Curtin School of Medical Research; 1996. , Version 1.0. [Google Scholar]
- 36.Rice W R. Evolution (Lawrence, KS) 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 37.Caspers G-J, Leunissen J A M, de Jong W W. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- 38.Clark M A, Baumann P, Baumann L. Curr Microbiol. 1999;38:309–311. [Google Scholar]
- 39.Heie O E. In: Aphids: Their Biology, Natural Enemies and Control, World Crop Pests. Minks A K, Harrewijn P, editors. 2a. Amsterdam: Elsevier; 1987. pp. 367–391. [Google Scholar]
- 40.Blackman R L, Eastop V F. Aphids on the World's Crops. Chichester, U.K.: Wiley; 1984. [Google Scholar]
- 41.Kobayashi I. Adv Biophys. 1992;28:81–133. doi: 10.1016/0065-227x(92)90023-k. [DOI] [PubMed] [Google Scholar]
- 42.Rouhbakhsh D, Lai C-Y, von Dohlen C D, Clark M A, Baumann L, Baumann P, Moran N A, Voegtlin D J. J Mol Evol. 1996;42:414–421. doi: 10.1007/BF02498635. [DOI] [PubMed] [Google Scholar]
- 43.van Ham R C H J, Martinez-Torres D, Moya A, Latorre A. Appl Environ Microbiol. 1999;65:117–125. doi: 10.1128/aem.65.1.117-125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brynnel E U, Kurland C G, Moran N A, Andersson S V G. Mol Biol Evol. 1998;15:574–582. doi: 10.1093/oxfordjournals.molbev.a025958. [DOI] [PubMed] [Google Scholar]
- 45.Hinde R. J Insect Physiol. 1971;17:1791–1800. doi: 10.1016/0022-1910(71)90147-8. [DOI] [PubMed] [Google Scholar]
- 46.Brough C N, Dixon A F G. Tissue Cell. 1990;22:51–63. doi: 10.1016/0040-8166(90)90089-r. [DOI] [PubMed] [Google Scholar]
- 47.Unterman B M, Baumann P, McLean D L. J Bacteriol. 1989;171:2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto K, Takahashi N, Fujitani Y, Yoshikura H, Kobayashi I. Genetics. 1996;143:27–36. doi: 10.1093/genetics/143.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazodier P, Davies J. Annu Rev Genet. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 50.Cohan F M. Trends Ecol Evol. 1994;9:175–180. doi: 10.1016/0169-5347(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 51.Jain R, Rivera M C, Lake J A. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farrand K S. In: Gene Transfer in the Environment. Levy S A, Miller R V, editors. New York: McGraw–Hill; 1989. pp. 261–285. [Google Scholar]
- 53.Watanabe K, Hara W, Sato M. J Invertebr Pathol. 1998;72:104–111. doi: 10.1006/jipa.1998.4764. [DOI] [PubMed] [Google Scholar]
- 54.Jarrett P, Stephenson M. Appl Environ Microbiol. 1990;56:1608–1614. doi: 10.1128/aem.56.6.1608-1614.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charles H, Ishikawa H. J Mol Evol. 1999;48:142–50. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- 57.Komaki K, Ishikawa H. J Mol Evol. 1999;48:717–722. doi: 10.1007/pl00006516. [DOI] [PubMed] [Google Scholar]
- 58.Soler T, Latorre A, Sabater B, Silva F J. Curr Microbiol. 2000;40:264–268. doi: 10.1007/s002849910052. [DOI] [PubMed] [Google Scholar]