Abstract
In oxidative environments, biomembranes contain oxidized lipids with short, polar acyl chains. Two stable lipid oxidation products are PoxnoPC and PazePC. PoxnoPC has a carbonyl group, and PazePC has an anionic carboxyl group pendant at the end of the short, oxidized acyl chain. We have used MD simulations to explore the possibility of complete chain reversal in OXPLs in POPC-OXPL mixtures. The polar AZ chain of PazePC undergoes chain reversal without compromising the lipid bilayer integrity at concentrations up to 25% OXPL, and the carboxyl group points into the aqueous phase. Counterintuitively, the perturbation of overall membrane structural and dynamic properties is stronger for PoxnoPC than for PazePC. This is because of the overall condensing and ordering effect of sodium ions bound strongly to the lipids in the PazePC simulations. The reorientation of AZ chain is similar for two different lipid force fields. This work provides the first molecular evidence of the “extended lipid conformation” in phospholipid membranes. The chain reversal of PazePC lipids decorates the membrane interface with reactive, negatively charged functional groups. Such chain reversal is likely to exert a profound influence on the structure and dynamics of biological membranes, and on membrane-associated biological processes.
Abbreviations: AZ, azelaoyl; DLPC, dilinoleoyl phosphatidylcholine; HazPC, 1-hexadecyl-2-azelaoyl-sn-glycero-3-phosphocholine; LDL, low-density lipoprotein; MD, molecular dynamics; OX, oxo-nonanoyl; OXPLs, oxidized phospholipids; PazePC, 1-palmitoyl-2-azelaoyl-ns-glycero-3-phosphocholine; PLPC, 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine; PME, particle mesh Ewald; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; PoxnoPC, 1-palmitoyl-2-(9′-oxo-nonanoyl)-sn-glycero-3-phosphocholine
Introduction
Mammalian phospholipids have mostly monounsaturated or polyunsaturated acyl chains (1). Unsaturated phospholipids are prone to oxidation at the location of the carbon-carbon double bonds. Phospholipid oxidation can be enzyme mediated or can result from reaction of lipids with reactive oxygen species including free radicals present in various tissues (2). Lipid oxidation products can bind receptors (3) and proteins, and influence signaling processes (2). Furthermore, owing to an atypical lipid structure (shorter and more polar acyl chains), OXPLs are expected to alter the physical properties of the lipid bilayer membrane, the effects of which can cascade down to several essential cellular processes. For example, lipid oxidation products can trigger mitochondrial damage (4) and have been implicated in mediating several diseased states, including cancer (5) and atherosclerosis (6).
PoxnoPC and PazePC are two stable lipid oxidation products. (Fig. 1). PoxnoPC and PazePC are OXPLs, which bear carbonyl and carboxyl groups at the end of their truncated sn-2 chains. In aqueous solution, the carbonyl and carboxyl groups are zwitterionic and anionic, respectively, at neutral pH (7). PoxnoPC is one of the key products of ozone-mediated oxidation of lung surfactant extract and promotes apoptosis and necrosis (8). PazePC has been detected in LDL and is a weak ligand for the peroxisome proliferator-activated receptor (3). HazPC, where the esterified acyl chains of PazePC is replaced by ether-linked alkyl residues, is also found in abundance in LDL (9) and has been implicated in the genesis of atherosclerosis. HazePC induces mitochondrial damage (4) and is a potent agonist to peroxisome proliferator-activated receptor γ, a protein that controls metabolic and cellular differentiation genes (10). The biochemical, biophysical, and physiological characterization of PoxnoPc and PazePC has been limited in comparison to HazPC and to other peroxidized lipid products (11).
Figure 1.
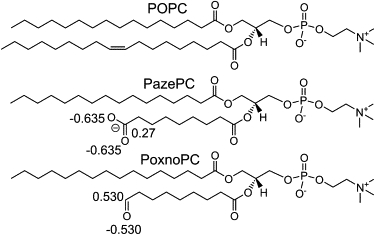
POPC and its two oxidation products investigated in this work. The partial charges for the carbonyl and carboxyl carbon, as well as oxygen atoms are shown.
The properties of PazePC were investigated using force-area (π-A) isotherms on a Langmuir balance in mixed monolayers with DPPC (12). It was proposed that the large free energy penalty of embedding a charged carboxylate moiety of the AZ in the hydrophobic core of a lipid bilayer might induce the reorientation of AZ into the aqueous phase, which would then adopt the so-called extended lipid conformation (13). Further evidence of AZ chain reversal came from measuring fluorescence resonance energy transfer as a result of the association of the water-soluble protein cytochrome c with PazePC micelles (14). The protein bound strongly to the micellar aggregates. The extent of binding reduced upon lowering pH (which would protonate the carboxylic acid on AZ) or upon addition of Ca2+ ions (which would bind competitively to the carboxylic acid group, thus inducing protein dissociation from the micelles). PoxnoPC and PazePC can be potential drug targets for antipsychotic compounds (15) and for antimicrobial peptides in cells under oxidative stress at inflammatory sites (16).
There is indirect evidence that chain reversal of the PazePC sn-2 chain might occur in PazePC monolayers (12) and in PC bilayers containing PazePC (14). Chain order parameters, density profiles, local curvature, and the electrostatic potential profiles across the bilayer are properties that might be directly affected by the presence of potentially chain-reversing charged lipids in a bilayer membrane. However, other than the preliminary evidence cited above, there have been no direct measurements of chain reversal in lipids and its impact on the properties of membranes. MD simulations have been previously used to characterize the impact of early peroxidation products of PLPC, including the carbonyl equivalent of PoxnoPC (11). Although complete chain reversal was not observed possibly because the oxidized chains were not hydrophilic enough, some of the oxidized sn-2 chains were found to be able to reach the lipid-water interface and make hydrogen bonds with water and lipid headgroups.
In this work, we employ fully atomistic MD simulations of POPC lipid bilayers doped with various amounts of PazePC and PoxnoPC to investigate the possibility of AZ-chain reversal and characterize the impact that such chain reversal might have on the overall physical properties of the lipid bilayer. MD simulations of lipid bilayer systems are now routinely used to study model lipid bilayer membranes to obtain a molecular perspective of the structural and dynamics properties of membranes, while providing detailed microscopic interpretations of experimental measurements (11,17–22).
Methods
The starting coordinates of a POPC bilayer were obtained from a 100 ns preequilibrated system (23), the starting structure for which was derived from coordinates available at http://moose.bio.ucalgary.ca/. Force-field parameters for POPC lipids were taken from the united atom force field of Berger et al. (24), available at http://moose.bio.ucalgary.ca/. Force-field parameters of PazePC were derived from the POPC force field, by truncating the oleoyl chain at the double bond, replacing the last carbon by a carboxyl group, and adjusting the geometry appropriately. For the carboxyl group, partial charges of amino acids were used. For the carbonyl group of PoxnoPC, the parameters were taken from previous simulations (11). The hydrogen atom of the carbonyl group was treated implicitly.
Random lipid molecules in the pure POPC bilayer were replaced with OXPLs, keeping the number of OXPLs equal in both lipid monolayers. Six different concentrations were used for both PazePC and PoxnoPC, ranging from 1/128 mol fraction OXPL to 1/4 mol fraction OXPL (Table 1). The total number of lipids in each system was 128. Sodium counterions were used to keep the PazePC systems electrostatically neutral.
Table 1.
List of completed simulations
| No. PazePC or PoxnoPC∗ | Time Simulated (ns) PazePC/PoxnoPC |
|
|---|---|---|
| GROMACS | CHARMM† | |
| 0 | 120 | 43 |
| 1 | 102/127 | 78 |
| 2 | 118/118 | 74 |
| 4 | 102/127 | 70 |
| 8 | 118/118 | 67 |
| 16 | 102/127 | 64 |
| 32 | 102/126 | - |
No. OXPL + No. POPC = 128 for each system.
CHARMM simulations were carried out for PazePC lipids only.
MD simulations were carried out using the GROMACS 3.3.1 package in the isobaric-isothermal (NPT) ensemble at 1 bar and 320 K, using the Berendsen (25) thermostat (relaxation time 0.1 ps) and barostat (coupling constant 1.0) with semiisotropic pressure coupling. The Z axis was parallel to the bilayer normal. A time step of 2 fs was used, and coordinates were saved every 10 ps. The LINCS (26) algorithm was used to constrain bonds with hydrogen atoms. The PME (27) method was used to calculate long-range electrostatic interactions with a fourth-order spline and a grid spacing of 0.1. The relative error for the Ewald sum in the real and reciprocal space was set to 10−5. The short-range van der Waals and real space Coulomb interactions were cutoff at 10 Å. Periodic boundary conditions were applied in all three directions. The Simple Point Charge model (28) was water was used.
The area per lipid (AL) in the PazePC simulations equilibrated within ∼50 ns. The simulations were carried out for more than 100 ns in each system after initial energy minimization. For calculation of ensemble averages, the first 50 ns of each simulation were discarded.
To evaluate the influence of force field and simulation ensemble (NPT versus NPzAT), simulations were also carried out using the CHARMM param27 force field for PazePC. These simulations were carried out in the NPzAT ensemble, where the area was kept constant in the XY plane to 65.5 Å2 per lipid, and only the Z dimension (along the bilayer normal) of the simulation cell was allowed to fluctuate. As a result of an oversight in the configuration files, these simulations were carried out at 313 K, instead of 320 K, which was used in the GROMACS simulations. However, the slightly lower temperature should not affect the structure of the system significantly because 313 K is still well above the main-phase transition temperature of POPC. Simulations with the CHARMM27 parameter set for lipids (29) were performed with NAMD v2.6 (30). A procedure similar to that described above for GROMACS was used to develop the force field for PazePC. The SHAKE algorithm (31) was used to constrain bonds with hydrogen atoms. A time-step of 2 fs was used, but full electrostatic calculations were computed every 4 fs. The PME method (32) was used for computation of electrostatic forces. The grid spacing was kept below 1.0 Å, and a fourth-order spline was used for interpolation. Van der Waals interactions were smoothly switched off over a distance of 4.0 Å, between 10 Å and 14 Å. The Langevin piston method (33) with a damping coefficient of 5 ps−1 and a piston period of 100 fs was used to maintain constant pressure and temperature conditions. The ratio of the cell dimensions was kept constant in the XY plane.
In total, 6 PazePC + 6 PoxnoPC + 1 pure POPC = 13 simulations were implemented in GROMACS, and 5 PazePC + 1 pure POPC = 6 simulations were implemented in CHARMM (Table 1). The simulations were run on a Linux cluster with Dell PowerEdge 2950, 2 × 2, 66 Ghz Intel Woodcrest CPUs, connected with 2× Gigabit Ethernet. The performance was ∼18ns/day (4 cpus) and 3 ns/day (8 cpus) for the GROMACS and CHARMM simulations, respectively.
To resolve the effect of sodium counterions on the structure of the bilayer, an additional simulation with 25% PazePC without sodium ions was implemented. The sodium ions were replaced by a uniform neutralizing charge density, so that PME calculations could be performed.
Analysis of the GROMACS simulations were carried out in the GROMACS suite of programs, whereas the analysis of the NAMD trajectories was carried out in the CHARMM package (36). Visual Molecular Dynamics (VMD) was used for molecular graphics (37). Unless otherwise mentioned, all results discussed refer to the GROMACS simulations. Analysis of the NAMD simulations were used for force-field comparison purposes only.
Most of the following analysis and discussion focuses on the 25% OXPL systems, for which the changes in the properties of the membrane are most remarkable. OXPLs are present in various tissues at different concentrations. Accounting for the possibility of OXPL-induced phase separation (see later discussion), it is possible that local concentrations of OXPLs in membranes can be as high 25% physiologically.
Results
Chain reversal
The AZ chain of PazePC lipids reoriented such that the carboxyl group pointed out into the aqueous phase (Fig. 2). The effect was more pronounced at higher concentrations of PazePC. The tilt angle (Fig. 3) was defined as the angle between the bilayer normal and the vector from the last to the first carbon atom of the oxidized sn-2 acyl chain. Even at the lowest AZ concentration, the average tilt angle was at least 90°, indicating that the AZ chain was at least parallel to the interface. At 25% PazePC, the tilt angle of the chains approached a uniform distribution peaking at 160°, indicating nearly complete chain reversal. In PoxnoPC lipids, the OX chains carrying the terminal carbonyl group also reoriented, but not to the extent of the AZ chains of PazePC. The OX chains rarely extended out of the interface, but were able to reorient to reach the lipid headgroups. The distribution of tilt angles of the OX chains is shown in Fig. 3, b.
Figure 2.
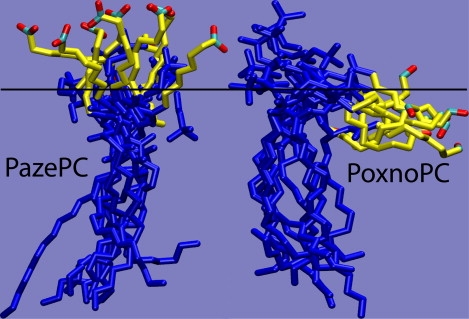
Simulation snapshots of a single PazePC and PoxnoPC molecule from the 25% OXPL simulations. The images were taken at uniform intervals during the last 50 ns and superimposed. The oxidized sn-2 chains are shown in yellow, except for the terminal functional group. The rest of the atoms are shown in blue. The horizontal black line represents the approximate average location of the phosphate groups in one bilayer leaflet.
Figure 3.
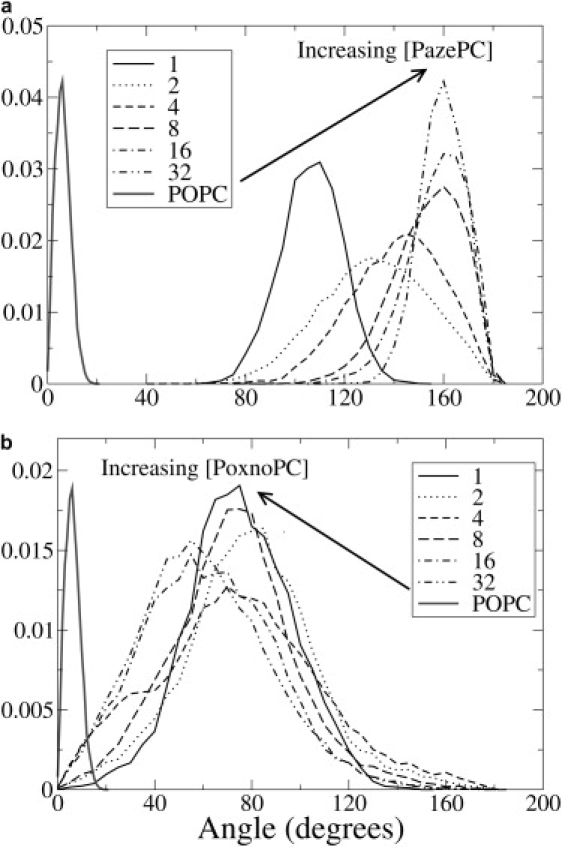
The concentration-dependent tilt angle distribution of the oxidized sn-2 chains of PazePC (a) and PoxnoPC (b). The numbers in the legend refer to the number of OXPLs present in each simulation. The tilt angle was defined as the angle between the bilayer normal and the vector from the last to the first carbon atom of the oxidized sn-2 acyl chain. A tilt angle of 0° indicates an sn-2 chain in the hydrocarbon interior of the bilayer and antiparallel to the bilayer normal. A tilt angle of 180° indicates an sn-2 chain pointing into the aqueous phase and parallel to the bilayer normal. The distribution for a POPC oleoyl chain (thick gray line) is from the pure POPC simulation.
Fig. 4 shows the density distribution of the carboxyl and carbonyl groups in the 25% PazePC and PoxnoPC simulations, relative to the density of phosphate for the pure POPC simulation. The distribution of the carboxyl group is nearly 30 Å wide in each leaflet and has two peaks: one in the aqueous phase, and the other just below the phosphate density. The carboxyl groups of some PazePC lipids were found to fluctuate between an interfacial position and a completely water-exposed conformation. However, there were some PazePC lipids for which the water-exposed conformation was not accessible, and other PazePC lipids for which the interfacial conformation was not accessible. Thus, the carboxyl groups of an individual PazePC lipid could adopt either an interfacial or aqueous conformation, or both. There was a vanishing density near the center of the bilayer. The carbonyl groups, being less hydrophilic, remained mostly confined below the phosphate interface. Radial distribution functions and hydrogen-bonding analysis indicated that the carbonyl groups of OX chains were closest to the ester region and made hydrogen bonds with both water and the esters (data not shown). The extent of reorientation of OX chains in this work is similar to that reported of OX chains in mixtures of OXPLs with PLPC bilayers (11).
Figure 4.
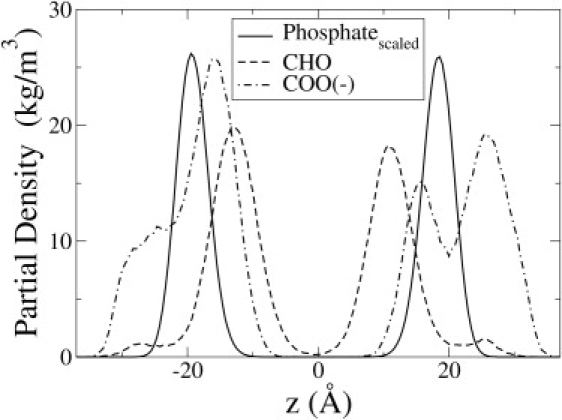
Density distribution of carboxyl (COO-) group of PazePC and carbonyl (CHO) group of PoxnoPC for the 25% OXPL simulations. The density of the phosphate is from the pure POPC simulation and was scaled by a factor of ∼0.067 to facilitate comparison.
Area, thickness, and density
The overall electron density of the bilayers with 25% OXPLs is shown in Fig. 5. The two main features of a higher density at the center of the bilayer, and a shift of the maxima toward the bilayer center, are in agreement with previous MD simulations of Wong-Ekkabut et al. (11) and x-ray scattering experiments on peroxidized DLPC lipid bilayers (38). In PazePC, there is also a slightly increased density in the aqueous region owing to the sn-2 chain reversal. Fig. 6 shows the average area per lipid (AL) and the thickness of the bilayers. Error estimates were calculated using a block averaging procedure (39). Interestingly, the perturbation of the electron density profile, and the overall AL and thickness is greater for PoxnoPC than for PazePC. In PoxnoPC, the sn-2 chains reorient such that the terminal carbonyl groups intercalate into the head-group region of the lipids, resulting in a higher AL and a significantly lower bilayer thickness. The lateral expansion of the bilayer with PoxnoPC is in qualitative agreement with monolayer expansion experiments of PoxnoPC and PazePC mixed with DPPC (12). The simulations thus support the model proposed by Sabatini et al. (12) that accommodation of polar moieties of the oxidized chain in the head-group region leads to expansion of the lipid monolayer. However, the simulations are in apparent conflict with the experimental observation that monolayer expansion was greater in the presence of PazePC (12). The conflict is resolved by taking into account the effect of sodium countercations in the PazePC simulation. The sodium cations were found to bind strongly to the interfacial glycerol region of the bilayer (data not shown). Interactions between the sodium cations and PC membranes result in membrane compression (18) using the Berger (24) force field for lipids.
Figure 5.
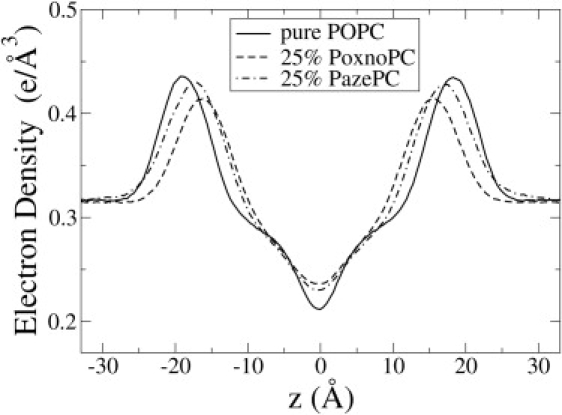
The overall electron density of the pure POPC bilayer, and for POPC bilayer with 25% OXPL.
Figure 6.
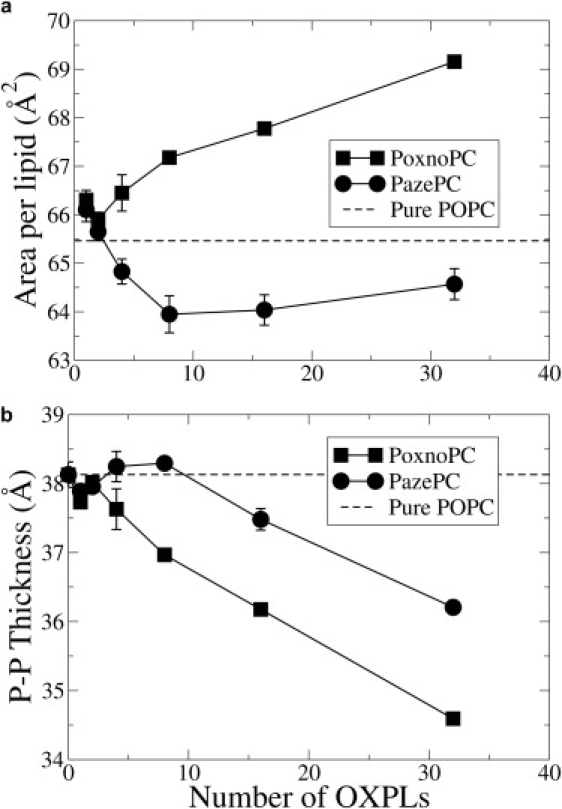
Average area per lipid (a) and thickness (b) of the lipid bilayers at various concentrations of OXPL. The errors were estimated using a block-averaging approach and cannot be seen in some cases because they are smaller than the size of the symbols. The highest block size was 14 ns. The thickness was calculated as the distance between the average positions of the phosphate groups in the two leaflets.
To better understand the effect of sodium counterions on the area and thickness of the PazePC simulations, the 25% PazePC simulation was carried out without sodium ions. The system now had a net charge of −32. To permit use of PME, the system was kept electrostatically neutral by adding a uniform neutralizing charge density in the simulation box. In the absence of sodium ions, the area per lipid of the 25% PazePC simulation increased to 72 ± 0.52 Å2, and the bilayer thickness decreased to 32.53 ± 0.19 Å. The corresponding numbers for area and thickness for the 25% PazePC simulations in presence of sodium ions (Fig. 6) were 64.57 ± 0.32 Å2 and 36.2 ± 0.097 Å, respectively. The area and thickness for the 25% PoxnoPC simulations (Fig. 6) were 69.11 ± 0.11 Å2 and 34.60 ± 0.06 Å, respectively. Thus, in the absence of sodium ions, the PazePC-containing lipid bilayer expands as a result of chain reversal, whereas in the presence of sodium ions, the expanding effect induced by chain reversal is offset by the condensing effect of the strong binding of sodium ions to the lipid-water interface. The expansion of the bilayer was more for PazePC than for PoxnoPC, in agreement with the monolayer experiments (12). The tilt angle profiles for the AZ chain in the 25% PazePC simulations with and without sodium counterions are compared in Fig. 7. The tilt angles were unaffected by the presence of sodium, indicating that the PazePC chain reversed completely in the simulations without sodium as well.
Figure 7.
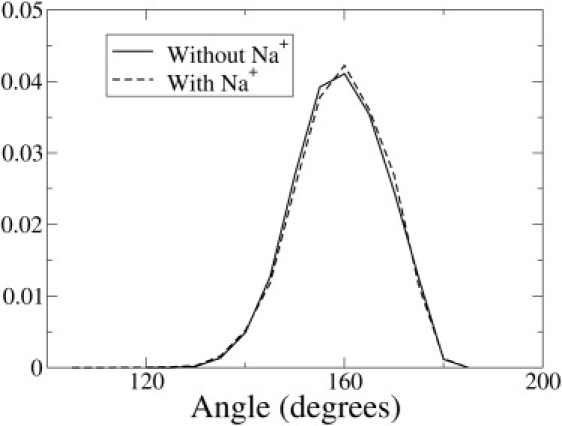
Tilt angle distribution for the AZ chain the 25% PazePC simulations, with and without salt. The definition of tilt angle is the same as in Fig. 3.
In Fig. 8, the density of the carboxyl and the phosphate groups are compared for 12.5% PazePC for the two force fields: CHARMM (param27 force field for lipids) and GROMACS (Berger force field for lipids). The comparison is not straightforward because the simulations in GROMACS were carried out in the NPT ensemble, whereas those with the CHARMM force field were carried out in the NPzAT ensemble, where the dimensions of the cell were fixed in the bilayer plane. The NPzAT ensemble was used with CHARMM, because the CHARMM force field leads to unreasonably low equilibrium values of area-per-lipid and gel-like behavior (40,41) in NPT. Nevertheless, the CHARMM simulations corroborated the two-peak chain reversal of the AZ side chain. In CHARMM, the interfacial density peaks of COO− are higher than those in GROMACS. In GROMACS, the COO− peaks are further out in the aqueous phase, and the carboxyl groups spend more time there relative to the CHARMM simulation. The complete chain-reversing effect is less pronounced in CHARMM because the area is fixed in the XY plane, and the lipid headgroups cannot freely accommodate the reverse orientation of the sn-2 AZ chain. However, there is qualitative agreement between the two force fields, and the AZ chain undergoes complete chain reversal in both cases.
Figure 8.
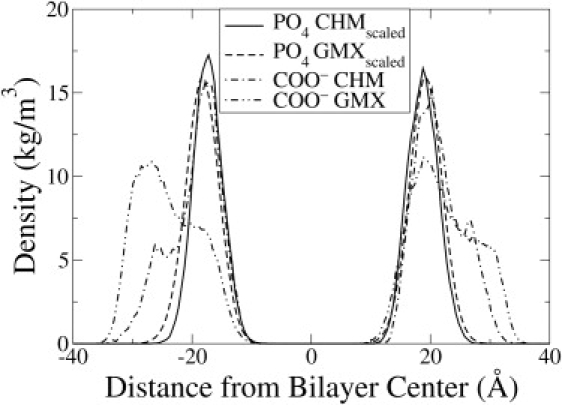
Partial density of the carboxyl (COO−) and phosphate (PO4) groups in the 12.5% OXPL simulations with the CHARMM (CHM) and the GROMACS (GMX) force fields. The density of phosphate was scaled by a factor of 0.05 to facilitate comparison.
Order parameters
The lipid chain order parameters were used to quantify the extent of chain disorder induced by the OXPLs. Order parameters were calculated as described in Wong-Ekkabut et al. (11). The order parameters for the palmitoyl chains of POPC lipids for varying concentrations of PoxnoPC and PazePC are shown in Fig. 9. Interestingly, the uncharged OX side chain induced more disorder in the bilayer compared to anionic AZ side chain. It could be envisioned that this is because the OX chains were oriented nearly perpendicular to the bilayer normal, which disturbed bilayer packing. In PazePC, on the other hand, the complete chain reversal of the AZ chains reduced the extent of disorder in the lipid bilayer. However, simulations of the sodium-free PazePC simulations suggest that the reduced disorder in PazePC-POPC compared to PoxnoPC-POPC bilayers was the result of the compression effect of tightly bound sodium counterions, which were present only in the PazePC simulations. In the 25% PazePC simulation without sodium ions, the acyl chain order of POPC lipids was lower than for 25% PazePC with sodium, and also lower than that in 25% PoxnoPC (see Fig. S1 in the Supporting Material). The overall trends were similar for the order parameters of the oleoyl chain of POPC, the results for which have not been shown.
Figure 9.
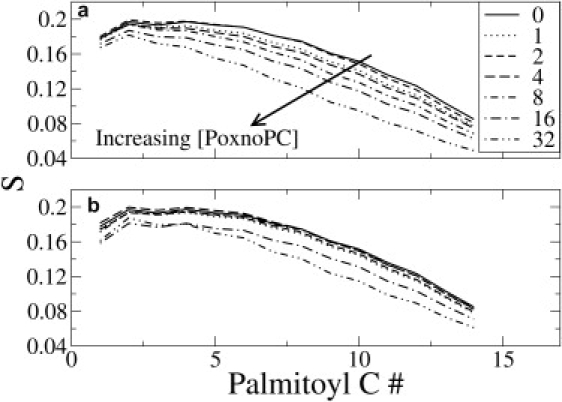
The order parameters for the palmitoyl chain of POPC lipids as a function of the amount of PoxnoPC (a) and PazePC (b) present. The numbers in the legend refer to the number of OXPL present in each simulation.
Electrostatic potential
The electrostatic potential profiles for the 25% PazePC and PoxnoPC are shown in Fig. 10, alongside that of a pure POPC bilayer. The electrostatic potential was calculated from the Poisson equation by a double integration of the charge density profile across the bilayer obtained from the MD simulation. Although both OXPLs were found to induce a lower dipole potential barrier near the interface compared to POPC, the effect was more pronounced for PoxnoPC than for PazePC. The distribution of the head-group orientation (P-N) vector (drawn from the phosphorus atom to the nitrogen atom of choline) with respect to the bilayer normal did not change much for POPC lipids, even at the highest concentration of OXPLs (data not shown). PazePC lipids had a P-N distribution that peaked at 25°. However, PoxnoPC lipids had a wider P-N distribution, which peaked at 55°.
Figure 10.
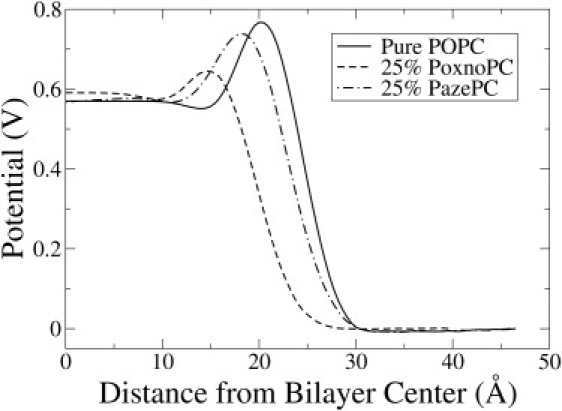
The electrostatic potential profile across a single monolayer for the pure POPC and the 25% OXPL simulations. The potential was calculated from double integration of Poisson's equation along the bilayer normal.
Discussion
The simulations reported in this work provide direct evidence that upon sufficient oxidation of a sufficiently large number of unsaturated lipids, the orientation of oxidized acyl chains of phospholipids in lipid bilayers can reverse completely. To our knowledge, this is the first direct molecular evidence of spontaneous chain reversal of free-standing phospholipid bilayers. Chain splaying has only been observed in previous simulations in interacting bilayers (42). The enthalpic penalty of embedding a charged group in the hydrophobic core of the membrane is thus sufficiently large to offset the entropic penalty of exposing approximately seven hydrophobic methylene groups of the AZ acyl chain to the aqueous phase and the interfacial region of the bilayer.
Chain reversal of oxidized unsaturated chains presents potent reactive functional groups to the aqueous phase, in this case, an anionic carboxylic acid (COO−) moiety, the presence of which on the membrane interface can have myriad effects on the properties of integral and peripheral membrane proteins (14); on peptides; as well as on the binding of ions, drugs (15), hormones, and small antimicrobial peptides (16). The negative charge of most anionic phospholipids resides on the phosphate group. The impact of the reversed AZ chain is expected to be different from that due to the presence of conventional anionic lipids, because the COO− group of PazePC can extend into the aqueous phase up to 10 Å beyond the average plane of the phosphate groups (Fig. 4). Furthermore, the COO− group is unshielded, and being pendant on a flexible acyl chain, it can adopt a variety of conformations, depending upon the specific interaction with the ligand it binds. Depending upon the reaction enthalpy available, COO− can also participate in covalent reactions, including formation of esters (with alcohols), anhydrides (with other carboxylic acid chains), or amides (with amines). Oxidized LDL participates in the genesis of atherosclerosis (43). Free fatty acids like oleic acid are known to react with cholesterol in oxidized LDL to form cholesteryl esters, the accumulation of which has been also associated with atherosclerosis (44). PazePC is known to be present in LDL (3). It is tempting to speculate that ester formation between the AZ chain of PazePC and cholesterol might be one possible modulation route of OXPL-mediated heart disease, and of cholesterol trafficking in general. However, such PazePC-cholesterol esters have not been detected in LDL, and neither have enzymes that can catalyze the reaction been isolated or characterized.
Langmuir balance and fluorescence spectroscopy experiments have shown that the intercalation of positively charged antimicrobial peptides into lipid monolayers and liposomes was increased by addition of anionic lipids or PoxnoPC, but was not enhanced by addition of PazePC (16). In contrast, the simulations presented here suggest that presence of PazePC should also enhance association of cationic antimicrobial peptides by presenting negatively charged groups on the membrane interface. The disagreement can be partly reconciled by inspecting the electrostatic potential profiles for PazePC and PoxnoPC in Fig. 10. A positively charged particle will experience a lower dipolar potential barrier near a PoxnoPC membrane than near a PazePC membrane. The observation is counterintuitive considering the charged nature of PazePC. The deep binding of sodium ions (see next paragraph) is partly responsible for the attenuation of the electrostatic force that a peptide might experience as is approaches the PazePC interface.
The comparison of the impact of PoxnoPC and PazePC on lipid bilayers must be interpreted with one caveat: there were no sodium counterions present in the PoxnoPC simulations. The reversal of AZ chains into the aqueous phase was very fast, on the time scale of 10 s of picoseconds. The area per lipid relaxed to equilibrium values quickly (∼5 ns) in PoxnoPC-containing bilayers. However, in the PazePC simulations, the area per lipid relaxed slowly, in ∼40–50 ns, because of the slow binding of cations to the interface, similar to that observed in past simulations (18,45–49). In all these cases, cations bound preferentially to the ester region of the phospholipids and led to overall compression of the lipid bilayer accompanied by an increase in order parameters of the acyl chain carbon atoms close to the headgroups (18,45–53). Thus, in simulations with the Berger force field (24) for lipids, cations have an overall condensing effect on the lipid bilayer. Developing force fields for ions have proven to be an unexpectedly difficult proposition (54–57), and it is not entirely resolved whether or not the condensing effect of salt is exaggerated in the Berger force field, because similar detailed studies on long time scales have not been performed with other force fields. The structural properties derived from the PazePC simulations are thus controlled by two competing forces: the condensing effect of sodium cations and the expansion of the bilayer induced by reorientation of the carboxyl chains.
When the sodium ions in the 25% PazePC simulation were replaced by a uniform charge neutralizing plasma, the membrane thinned along the bilayer normal, and the area per lipid increased significantly. Without sodium counterions, the expansion of the lipid bilayer was higher in the PazePC-containing bilayer than in the PoxnoPC-containing bilayer. This is in agreement with Langmuir monolayer observations, where PazePC expanded the monolayer more than PoxnoPC (12). Moreover, the order parameters dropped significantly in 25% PazePC when the sodium ions were replaced by the uniform charge density. The results thus establish that the condensation effect seen in lipid bilayers is a result of the strong binding of sodium to the lipid headgroups. The agreement of the sodium-free simulations with the monolayer experiments suggest that the intercalation of sodium ions in the lipid headgroups, and the consequent condensing effect might be a force-field artifact.
In this work, the carboxylic acid group of the AZ chain of PazePC was kept deprotonated. Is it possible that the carboxylic group can pick up a proton in the lipid environment? Free energy profiles of amino acids across lipid bilayers showed that the carboxylic acid group remains protonated in the hydrophobic core of the bilayer (58). However, the protonated carboxylic acid group is more polar than the carbonyl group, and therefore even the protonated AZ chain is expected to reorient to the interfacial region of the lipid bilayer. The apparent pKa of carboxylic acid group of phosphatidylserine lipids is 5.5 (59), so it is reasonable to assume that the carboxylic acid group of PazePC will be mostly deprotonated near the lipid-water interface.
When the orientation of the AZ chain reverses, the headgroup of PazePC can be envisioned to become extraordinarily large, because it now comprises the conventional lipid headgroup plus the reversed AZ chain residing in the aqueous phase. The shape of PazePC then resembles the wedge shape of lysolipids, which only have a single hydrophobic acyl chain pendant on the headgroup. Lysolipids are nonlamellar-forming lipids and, owing to their conical shape, have tendency to promote hexagonal HI structures (60). Lysolipids can thus alter the curvature stress profile in mixed bilayers (61,62). The presence of PazePC in membranes could alter the curvature stress field of the bilayer, which could eventually lead to domain separation, depending on the local lipid composition and other environmental factors such as temperature and ionic strength. Given the larger size of the PazePC headgroup, the effect might be greater than for lysolipids. Domain separation in DLPC bilayers could be induced at much lower concentrations of cholesterol when the lipids were subject to peroxidation (63). Interestingly, the small headgroup of cholesterol gives it an inverted conical shape and promotes the formation of inverted hexagonal HII structures (60). We are pursuing efforts to investigate the phase behavior of cholesterol-PazePC-DLPC mixtures using coarse-grained MD methods (35,64,65). Coarse-grained MD methods will also allow for measurement of the mechanical properties of bilayers, specifically the area compressibility modulus and the bending rigidity. The bending rigidity of giant unilamellar vesicles of POPC was decreased in the presence of lysolipids in micropipette aspiration experiments (John H. Ipsen, personal communication, 2008), and it is possible that PazePC and PoxnoPC also have significant impact on the mechanical strength of membranes.
Conclusion
This work provides the first molecular-scale evidence that oxidized lipid acyl chains can undergo complete chain reversal and adopt the long-debated “extended lipid conformation” without the requirement of interaction with external hosts. Although chain reversal has a disordering effect on the bilayer, the overall integrity of the bilayer was preserved up to 25% mole fraction OXPL.
OXPLs are known to participate in the genesis of a variety of serious ailments including cancer, apoptosis, and heart disease. The reorientation of polar acyl chains presents reactive functional groups near the membrane, which are capable of noncovalent and covalent, specific and nonspecific interactions with a large variety of membrane hosts, ranging from single ions to large proteins. Our work provides a clear structural model that will help comprehend and visualize such interactions better, which will lead to a better understanding of how OXPLs participate in the genesis of diseased states that are especially prevalent in tissues with oxidative environments, such as inflammation sites in cancer and in microbial infections.
Acknowledgments
The Danish Center for Scientific Computing at the University of Southern Denmark, Odense is acknowledged for computing resources. H.K. is supported by MEMBAQ, a Specific Targeted Research Project supported by the European Commission under the Sixth Framework Programme (Contract NMP4-CT-2006-033234). MEMPHYS - Center for Biomembrane Physics is supported by the Danish National Research Foundation.
Supporting Material
References
- 1.Hulbert A.J., Rana T., Couture P. The acyl composition of mammalian phospholipids: an allometric analysis. Comp. Biochem. Physiol. B. 2002;132:515–527. doi: 10.1016/s1096-4959(02)00066-0. [DOI] [PubMed] [Google Scholar]
- 2.Fruhwirth G.O., Loidl A., Hermetter A. Oxidized phospholipids: from molecular properties to disease. Biochim. Biophys. Acta. 2007;1772:718–736. doi: 10.1016/j.bbadis.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Davies S.S., Pontsler A.V., Marathe G.K., Harrison K.A., Murphy R.C. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. J. Biol. Chem. 2001;276:16015–16023. doi: 10.1074/jbc.M100878200. [DOI] [PubMed] [Google Scholar]
- 4.Chen R., Yang L., McIntyre T.M. Cytotoxic phospholipid oxidation products. Cell death from mitochondrial damage and the intrinsic caspase cascade. J. Biol. Chem. 2007;282:24842–24850. doi: 10.1074/jbc.M702865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cejas P., Casado E., Belda-Iniesta C., De Castro J., Espinosa E. Implications of oxidative stress and cell membrane lipid peroxidation in human cancer (Spain) Cancer Causes Control. 2004;15:707–719. doi: 10.1023/B:CACO.0000036189.61607.52. [DOI] [PubMed] [Google Scholar]
- 6.Tsimikas S., Witztum J.L. The role of oxidized phospholipids in mediating lipoprotein(a) atherogenicity. Curr. Opin. Lipidol. 2008;19:369–377. doi: 10.1097/MOL.0b013e328308b622. [DOI] [PubMed] [Google Scholar]
- 7.Itabe H., Kobayashi T., Inoue K. Generation of toxic phospholipid(s) during oxyhemoglobin-induced peroxidation of phosphatidylcholines. Biochim. Biophys. Acta. 1988;961:13–21. [PubMed] [Google Scholar]
- 8.Uhlson C., Harrison K., Allen C.B., Ahmad S., White C.W. Oxidized phospholipids derived from ozone-treated lung surfactant extract reduce macrophage and epithelial cell viability. Chem. Res. Toxicol. 2002;15:896–906. doi: 10.1021/tx010183i. [DOI] [PubMed] [Google Scholar]
- 9.Tokumura A., Toujima M., Yoshioka Y., Fukuzawa K. Lipid peroxidation in low density lipoproteins from human plasma and egg yolk promotes accumulation of 1-acyl analogues of platelet-activating factor-like lipids. Lipids. 1996;31:1251–1258. doi: 10.1007/BF02587909. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted in proof.
- 11.Wong-Ekkabut J., Xu Z., Triampo W., Tang I.M., Tieleman D.P. Effect of lipid peroxidation on the properties of lipid bilayers: a molecular dynamics study. Biophys. J. 2007;93:4225–4236. doi: 10.1529/biophysj.107.112565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabatini K., Mattila J.-P., Megli F.M., Kinnunen P.K.J. Characterization of two oxidatively modified phospholipids in mixed monolayers with DPPC. Biophys. J. 2006;90:4488–4499. doi: 10.1529/biophysj.105.080176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinnunen P.K.J. Fusion of lipid bilayers - a model involving mechanistic connection to Hii-phase forming lipids. Chem. Phys. Lipids. 1992;63:251–258. doi: 10.1016/0009-3084(92)90041-m. [DOI] [PubMed] [Google Scholar]
- 14.Mattila J.-P., Sabatini K., Kinnunen P.K.J. Interaction of cytochrome c with 1-palmitoyl-2-azelaoyl-sn-glycero-3-phosphocholine: evidence for acyl chain reversal. Langmuir. 2008;24:4157–4160. doi: 10.1021/la703940s. [DOI] [PubMed] [Google Scholar]
- 15.Mattila J.-P., Sabatini K., Kinnunen P.K.J. Oxidized phospholipids as potential novel drug targets. Biophys. J. 2007;93:3105–3112. doi: 10.1529/biophysj.107.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattila J.P., Sabatini K., Kinnunen P.K. Oxidized phospholipids as potential molecular targets for antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:2041–2050. doi: 10.1016/j.bbamem.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Pandit S.A., Chiu S.W., Jakobsson E., Grama A., Scott H.L. Cholesterol surrogates: a comparison of cholesterol and 16:0 ceramide in POPC bilayers. Biophys. J. 2007;92:920–927. doi: 10.1529/biophysj.106.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurtovenko A.A., Vattulainen I. Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. J. Phys. Chem. B. 2008;112:1953–1962. doi: 10.1021/jp0750708. [DOI] [PubMed] [Google Scholar]
- 19.Feller S.E. Acyl chain conformations in phospholipid bilayers: a comparative study of docosahexaenoic acid and saturated fatty acids. Chem. Phys. Lipids. 2008;153:76–80. doi: 10.1016/j.chemphyslip.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Klauda J.B., Roberts M.F., Redfield A.G., Brooks B.R., Pastor R.W. Rotation of lipids in membranes: molecular dynamics simulation, 31P spin-lattice relaxation, and rigid-body dynamics. Biophys. J. 2008;94:3074–3083. doi: 10.1529/biophysj.107.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hub J.S., Salditt T., Rheinstadter M.C., de Groot B.L. Short-range order and collective dynamics of DMPC bilayers: a comparison between molecular dynamics simulations, x-ray, and neutron scattering experiments. Biophys. J. 2007;93:3156–3168. doi: 10.1529/biophysj.107.104885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knecht V., Marrink S.J. Molecular dynamics simulations of lipid vesicle fusion in atomic detail. Biophys. J. 2007;92:4254–4261. doi: 10.1529/biophysj.106.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parry M.J., Alakoskela J.M., Khandelia H., Kumar S.A., Jaattela M., Mahalka A.K., Kinnunen P.K. High-affinity small molecule-phospholipid complex formation: binding of siramesine to phosphatidic acid. J Am. Chem. Soc. 2008;130:12953–12960. doi: 10.1021/ja800516w. [DOI] [PubMed] [Google Scholar]
- 24.Berger O., Edholm O., Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 26.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 27.Reference deleted in proof.
- 28.Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces (Jerusalem Symposia) Springer; 1981. pp. 331–342. [Google Scholar]
- 29.MacKerell A.D., Jr., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 30.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 32.Reference deleted in proof.
- 33.Feller S.E., Zhang Y., Pastor R.W., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 34.Reference deleted in proof.
- 35.Monticelli L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 36.Brooks B.R., Bruccoleri R.E., Olfson B.D., States D.J., Swaminathan S. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 37.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 14. 1996;33–38:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 38.Mason R.P., Walter M.F., Mason P.E. Effect of oxidative stress on membrane structure: Small-angle x-ray diffraction analysis. Free Radic. Biol. Med. 1997;23:419–425. doi: 10.1016/s0891-5849(97)00101-9. [DOI] [PubMed] [Google Scholar]
- 39.Hess B. Determining the shear viscosity of model liquids from molecular dynamics simulations. J. Chem. Phys. 2002;116:209–217. [Google Scholar]
- 40.Jensen M.O., Mouritsen O.G., Peters G.H. Simulations of a membrane-anchored peptide: structure, dynamics, and influence on bilayer properties. Biophys. J. 2004;86:3556–3575. doi: 10.1529/biophysj.103.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siu S.W.I., Vacha R., Jungwirth P., Bockmann R.A. Biomolecular simulations of membranes: Physical properties from different force fields. J. Chem. Phys. 2008;128:125103. doi: 10.1063/1.2897760. [DOI] [PubMed] [Google Scholar]
- 42.Stevens M.J., Hoh J.H., Woolf T.B. Insights into the molecular mechanism of membrane fusion from simulation: evidence for the association of splayed tails. Phys. Rev. Lett. 2003;91:188102. doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 43.Steinberg D., Parthasarathy S., Carew T.E., Khoo J.C., Witztum J.L. Beyond cholesterol - modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 1989;320:915–924. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- 44.Leitinger N. Cholesteryl ester oxidation products in atherosclerosis. Mol. Aspects Med. 2003;24:239–250. doi: 10.1016/s0098-2997(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 45.Pandit S.A., Berkowitz M.L. Molecular dynamics simulation of dipalmitoylphosphatidylserine bilayer with Na+ counterions. Biophys. J. 2002;82:1818–1827. doi: 10.1016/S0006-3495(02)75532-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bockmann R.A., Hac A., Heimburg T., Grubmuller H. Effect of sodium chloride on a lipid bilayer. Biophys. J. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay P., Monticelli L., Tieleman D.P. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys. J. 2004;86:1601–1609. doi: 10.1016/S0006-3495(04)74227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurtovenko A.A. Asymmetry of lipid bilayers induced by monovalent salt: atomistic molecular-dynamics study. J. Chem. Phys. 2005;122:244902. doi: 10.1063/1.1942489. [DOI] [PubMed] [Google Scholar]
- 49.Zhao W., Rog T., Gurtovenko A.A., Vattulainen I., Karttunen M. Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidylglycerol lipid bilayers with Na+ counterions. Biophys. J. 2007;92:1114–1124. doi: 10.1529/biophysj.106.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S.J., Song Y., Baker N.A. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophys. J. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elmore D.E. Molecular dynamics simulation of a phosphatidylglycerol membrane. FEBS Lett. 2006;580:144–148. doi: 10.1016/j.febslet.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 52.Cascales J.J.L., delaTorre J.G., Marrink S.J., Berendsen H.J.C. Molecular dynamics simulation of a charged biological membrane. J. Chem. Phys. 1996;104:2713–2720. [Google Scholar]
- 53.Sachs J.N., Nanda H., Petrache H.I., Woolf T.B. Changes in phosphatidylcholine headgroup tilt and water order induced by monovalent salts: molecular dynamics simulations. Biophys. J. 2004;86:3772–3782. doi: 10.1529/biophysj.103.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossfield A., Ren P., Ponder J.W. Ion solvation thermodynamics from simulation with a polarizable force field. J. Am. Chem. Soc. 2003;125:15671–15682. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 55.Patra M., Karttunen M. Systematic comparison of force fields for microscopic simulations of NaCl in aqueous solutions: Diffusion, free energy of hydration, and structural properties. J. Comput. Chem. 2004;25:678–689. doi: 10.1002/jcc.10417. [DOI] [PubMed] [Google Scholar]
- 56.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamoureux G., Roux B. Absolute hydration free energy scale for alkali and halide ions established from simulations with a polarizable force field. J. Phys. Chem. B. 2006;110:3308–3322. doi: 10.1021/jp056043p. [DOI] [PubMed] [Google Scholar]
- 58.MacCallum J.L., Bennett W.F., Tieleman D.P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cevc G., Watts A., Marsh D. Titration of the phase-transition of phosphatidylserine bilayer-membranes - effects of pH, surface electrostatics, ion binding, and headgroup hydration. Biochemistry. 1981;20:4955–4965. doi: 10.1021/bi00520a023. [DOI] [PubMed] [Google Scholar]
- 60.Mouritsen O.G. Springer-Verlag; New York: 2005. Life-As a Matter of Fat: The Emerging Science of Lipidomics. [Google Scholar]
- 61.Kooijman E.E., Chupin V., de Kruijff B., Burger K.N.J. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 62.May E.R., Kopelevich D.I., Narang A. Coarse-grained molecular dynamics simulations of phase transitions in mixed lipid systems containing LPA, DOPA, and DOPE lipids. Biophys. J. 2008;94:878–890. doi: 10.1529/biophysj.107.113951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacob R.F., Mason R.P. Lipid peroxidation induces cholesterol domain formation in model membranes. J. Biol. Chem. 2005;280:39380–39387. doi: 10.1074/jbc.M507587200. [DOI] [PubMed] [Google Scholar]
- 64.Marrink S.J., de Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 65.Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


