Abstract
In this issue of Structure, Wiedenheft et al. describe the structure and activity of Cas1, the only protein associated with all CRISPR loci. Cas1 is a metal-dependent deoxyribonuclease, consistent with a role in the adaptation phase of CRISPR immunity against invading nucleic acids.
To maintain genetic identity, bacteria must prevent the acquisition of foreign genetic material often carried by phages and conjugative plasmids. Clustered, regularly interspaced, short palindromic repeats (CRISPR) systems have been recently characterized as a programmable barrier to phage infection (Barrangou, et al., 2007) and conjugation (Marraffini and Sontheimer, 2008). These clusters of repetitive elements have been found in about 40% of eubacterial and 90% of archaeal genomes and can contain hundreds of repeats (Sorek et al., 2008). Repeats are separated by short sequences called spacers that match sequences present in phages and plasmids and that specify the targets of CRISPR interference. CRISPR loci display a great deal of evolutionary plasticity; upon infection by lytic phages, occasional bacterial cells adapt and survive infection by incorporating additional repeat-spacer units containing sequences from the challenging phage (Barrangou, et al., 2007). Because the immunity is encoded in the genome of the adapted cell, its progeny are also resistant to the attacking phage.
CRISPR interference is assisted by a set of CRISPR associated (Cas) proteins that are encoded by the cas genes usually found immediately adjacent to the repeats. Cas proteins can be classified into 45 different types but their precise biochemical functions are largely unknown (Haft et al., 2005). Only one protein, Cas1, has orthologs in all CRISPR loci.
The molecular mechanisms of CRISPR interference are beginning to emerge. Accumulating evidence indicates that repeats and spacers are transcribed as a long precursor that is cleaved in each repeat into smaller CRISPR RNAs (crRNAs) containing a full spacer sequence (Brouns et al., 2008; Hale et al., 2008). In E. coli, this processing is carried out by the Cascade complex of Cas proteins (Brouns et al., 2008). The crRNAs thus generated target incoming DNA molecules (Brouns et al., 2008; Marraffini and Sontheimer, 2008). A cas3 mutant in E. coli is deficient in preventing phage infection but not in the generation of crRNAs (Brouns et al., 2008), suggesting that Cas3 mediates target destruction. In agreement with this hypothesis, Sulfobus solfataricus Cas3 has endonuclease activity against double-stranded substrates in vitro (Han and Krauss, 2009). In contrast to the interference phase of CRISPR defense, we know almost nothing about the adaptive component of the pathway in which new spacers are acquired in response to infection. In Streptococcus thermophilus, cas7 mutants apparently lose their ability to incorporate new repeat-spacer units, a result that suggests a role for Cas7 in the adaptation phase of CRISPR function (Barrangou et al., 2007).
Due to its universal presence in CRISPR loci, Cas1 is likely to have a fundamental role in CRISPR function. However, very little is known about this protein. Resistance to phage infection is not compromised in an E. coli strain lacking cas1, and Cas1 is not part of the Cascade complex (Brouns et al., 2008), indicating that it is dispensable for the functioning of existing repeats. The most plausible alternative explanation for its maintenance in all CRISPR loci is that it functions in the other critical phase of CRISPR defense, namely adaptation. In this issue of Structure, Wiedenheft et al. (2009) reveal Cas1 to be a metal-dependent deoxyribonuclease, consistent with a role for Cas1 in the acquisition of new CRISPR spacers.
The authors report the crystal structure of Pseudomonas aeruginosa Cas1, which reveals a unique three-dimensional fold consisting of a N-terminal β strand domain and a C-terminal α-helical domain. The α-helical domain contains a binding pocket for a divalent metal ion. Incubation of purified Cas1 with dsDNA, ssDNA, dsRNA, or ssRNA leads to specific degradation of dsDNA and ssDNA to ~80 base pair (bp) fragments, irrespective of substrate sequence and methylation state. Circular dsDNA is also subject to Cas1 degradation, indicating that the enzyme acts as an endonuclease. Multiple lines of evidence demonstrate that Mg2+ or Mn2+ binding is essential for Cas1 DNase activity.
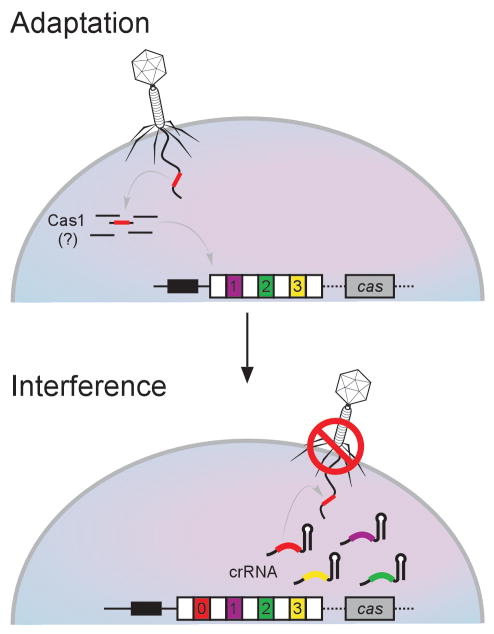
These results provide an initial glimpse into the role of Cas1 in CRISPR function. The “immunization” of bacterial strains by the incorporation of new spacers into the CRISPR locus would require DNase activity at multiple points, any or all of which could involve Cas1 (Figure 1). In the first of these mutually non-exclusive roles, Cas1 could use its DNase activity to cleave invasive DNA in preparation for incorporation into the CRISPR locus. Cas1 could also cleave the CRISPR locus itself to enable incorporation of invasive DNA sequences.
Figure 1. Possible Roles for Cas1 in CRISPR Immunity.
CRISPR loci include a set of repeats (white boxes) separated by spacers (colored boxes) with sequences that match phage and plasmid DNA that often invade bacteria. These clusters are preceded by a “leader” sequence and are usually flanked by cas genes. When a phage that does not carry a sequence identical to any CRISPR spacers infects bacteria, most cells succumb to the infection. At a low frequency, however, a fragment of the phage DNA can become incorporated as a new repeat-spacer unit at one end of the CRISPR locus (top panel). Cas1 is a likely candidate in the adaptation process, and Weidenheft et al. (2009) demonstrate DNase activity of Cas1 consistent with this possibility. The new spacer sequence is then included among the crRNAs generated by the CRISPR locus, leading to immunity against subsequent infections by the phage (bottom panel).
As it is often the case with exciting findings, the DNase activity of Cas1 opens many new questions. In vitro, Cas1 does not exhibit a detectable preference for any DNA sequence or methylation pattern. What then keeps Cas1 DNase activity in check in the absence of invaders? Similarly, what enables the occasional infected cell to recognize incoming DNA as foreign and adapt in time to survive infection, and does Cas1 function directly in this initial phase of defense? Notably, Cas1 showed in vitro activity toward both dsDNA and ssDNA, suggesting that ssDNA injected by some bacteriophages and conjugative plasmids could also be attacked by Cas1. Does the enzyme associate with other factors that provide substrate specificity? If Cas1 is directly involved in cleaving incoming phage or plasmid DNA, how do we reconcile the ~80 bp product size with the 32-bp length of CRISPR spacers in this strain of P. aeruginosa? Do the conserved sequences (“CRISPR motifs”) that flank target sites (Deveau et al., 2008) somehow constrain Cas1’s hypothesized role in spacer acquisition? Whatever the answers to these and related questions may be, the discovery and characterization of Cas1’s DNase activity represent a critical inroad into its role in this fascinating pathway of sequence-directed defense.
References
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. Science . 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Science . 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. RNA . 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Krauss G. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. Nat. Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structure. 2009;17(this issue):904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]