Abstract
Background
Knowledge on the clinical features of infections caused by Escherichia coli producing plasmid-mediated AmpC β-lactamase is limited. Of the several groups of plasmid-mediated AmpC β-lactamase, CMY-type β-lactamase is the most common in the United States.
Methods
We prospectively identified E. coli producing CMY-type β-lactamase and collected clinical data over a seven-month period. A retrospective cohort study was performed to identify features associated with these cases, using cases due to extended-spectrum β-lactamase (ESBL)-producing E. coli as controls. Pulsed-field gel electrophoresis (PFGE), plasmid analysis and phylogenetic typing were performed.
Results
Twenty-two cases with CMY-producing E. coli and 25 cases with ESBL-producing E. coli were identified. The demographics of the patients were similar between the CMY and ESBL cohorts. CMY cases were significantly more likely to represent symptomatic infection compared with ESBL cases (P=0.028). The CMY-type β-lactamase was identified as CMY-2 or its variants. Ninety-four percent of the CMY-producing isolates belonged to E. coli phylogenetic groups B2 and D, which are associated with virulence. Many of them shared similar plasmid profiles, whereas the PFGE profiles were diverse. Co-resistance to non-β-lactam antimicrobials was common.
Conclusion
In Pittsburgh, CMY-producing E. coli is almost as common as ESBL-producing E. coli and causes symptomatic infection in the majority of cases.
Keywords: CMY-type β-lactamase, Escherichia coli, community-associated infection, virulence
TEXT
Plasmid-mediated AmpC-β-lactamases, since the initial description in 1989 [1], have spread in the family Enterobacteriaceae worldwide [2, 3]. While several groups of plasmid-mediated AmpC-β-lactamases have been identified, the most common of them has been the CMY-type produced by Escherichia coli and non-typhoidal Salmonella [2, 3]. These enzymes typically render the bacteria resistant to penicillins, penicillin/β-lactamase inhibitor combinations, cephalosporins including cephamycins and aztreonam. Unlike extended-spectrum β-lactamases (ESBLs), the method to detect CMY-type β-lactamase in the clinical laboratory has not been standardized [4]. Consequently, the prevalence of bacteria producing these enzymes is difficult to determine. In a population-based surveillance in Calgary, Canada, an increasing incidence of infections due to CMY-producing E. coli has been noted between 2000 and 2003 [5]. A recent study conducted in Nebraska also noted that CMY-producing E. coli was not uncommon and often originated in nursing homes [6]. However, information on clinical characteristics of infections caused by CMY-producing E. coli is scarce to date.
The objective of this study was to systematically identify clinical cases involving CMY-producing E. coli and describe their clinical features in comparison with cases involving ESBL-producing E. coli, which share in common resistance to third-generation cephalosporins.
PATIENTS, MATERIALS AND METHODS
Cohort study
The CMY cohort was constituted by all patients from whom CMY-producing E. coli was isolated from any clinical sample between September 1, 2006, and March 31, 2007. That cohort was compared with the ESBL cohort, which was constituted by all patients from whom ESBL-producing E. coli were isolated in the same period of time. The study was performed at the University of Pittsburgh Medical Center (Presbyterian-Shadyside Campuses), a 1,300-bed tertiary teaching hospital with affiliated outpatient clinics. The CMY cohort was further clinically and microbiologically characterized.
Laboratory surveillance of CMY-producing E. coli
Prospective laboratory-based surveillance was conducted during the study period. Isolates that met the screening criteria for ESBL production (ceftriaxone or aztreonam zone size ≤ 27 mm on disk diffusion testing) but had negative phenotypic confirmatory test were collected [7]. Of these isolates, those showing non-susceptibility to ceftazidime on disk diffusion testing were checked for the presence of CMY-type β-lactamase gene by PCR analysis. Primers used in this study were CMY-F: 5’-CCG GAC ACC TTT TTG CTT TT-3’ and CMY-R: 5’-TAT CCT GGG CCT CAT CGT CAG TTA-3’. The amplification was conducted with an annealing temperature of 60°C for 30 cycles using a 9700 GeneAmp thermocycler (Applied Biosystems, Foster City, CA). The PCR products were sequenced on an ABI3100 instrument (Applied Biosystems). Susceptibility of each isolate against various β-lactam and non-β-lactam antimicrobials was tested using the disk diffusion method [7]. Phenotypic detection of plasmid-mediated AmpC β-lactamase using a boronic acid compound was also performed as described previously [8]. In brief, 300 µg of 3-aminophenyl boronic acid hydrochloride (3-APB) was applied to a ceftazidime disk placed on a Mueller-Hinton (MH) agar plate (BD, Sparks, MD) inoculated with the test isolate. An increase in zone diameter of ≥ 5 mm compared with a ceftazidime disk without 3-APB was interpreted as a positive test [8].
For the ESBL cohort, E. coli isolates which had positive phenotypic confirmatory test for ESBL production were also collected during the same period. PCR analyses for detection of TEM, SHV and CTX-M-type ESBL genes were conducted as described previously [9], followed by sequencing of the PCR products. For those negative for any of these ESBL genes, we conducted PCR analysis for CMY-type β-lactamase gene as described above.
Study sample and data collection
The study was approved by the Institutional Review Boards of the University of Pittsburgh. Medical records of patients from whom E. coli producing CMY-type β-lactamase or ESBL was identified were collected to document patient demographic characteristics, contact with healthcare system, underlying medical conditions, prior administration of antibiotics, role of the organism (symptomatic infection vs. colonization), type of infection, antibiotic treatment received, clinical and microbiologic outcome. The role of the organisms was determined by detailed chart review and in accordance with the CDC/NNIS guidelines whenever applicable [10]. These data were supplied “de-identified” to the investigators. The same patient was re-enrolled only when a positive culture was identified ≥30 days from the initial enrollment. The case definitions of hospital-acquired, healthcare-associated and community-associated infection or colonization were as follows:
Hospital-acquired
Positive culture obtained from a patient who had been hospitalized for ≥48 hours. If a patient was transferred from another hospital, the duration of inpatient stay was calculated from the date of the first hospital admission.
Healthcare-associated
Positive culture obtained from a patient at the time of hospital admission or within 48 hours of admission if the patient fulfilled any of the following criteria: (1) Received intravenous therapy at home; received wound care or specialized nursing care through a health care agency, family, or friends; or had self-administered intravenous medical therapy in the 30 days before the infection. Patients whose only home therapy was oxygen were excluded. (2) Attended a hospital or hemodialysis clinic or received intravenous chemotherapy in the 30 days before the infection. (3) Was hospitalized in an acute care hospital for 2 or more days in the 90 days before the infection. (4) Resided in a nursing home or long-term care facility.
Community-associated
Positive culture obtained at the time of hospital admission or <48 hours after admission who did not meet the criteria for a healthcare-associated infection.
Phylogenetic and molecular typing
The phylogenetic groups of the study isolates were determined by multiplex PCR analysis [11]. Among the four main phylogenetic groups (A, B1, B2, and D) in E. coli, virulent extra-intestinal strains belong mainly to group B2 and D [12, 13].
To determine the genetic relatedness of the study isolates, pulsed-field gel electrophoresis (PFGE) analysis was performed using XbaI as a restriction endonuclease and electrophoresing the genome in a CHEF DR III system (Bio-Rad, Hercules, CA) at 6 V with pulse times of 2.2 – 54.2 s and linear ramping at a temperature of 14°C for 22 h. Digitalized gel images were saved and subjected to analysis with BioNumerics software version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium). Cluster analysis was performed by using the unweighted pair-group method based on Dice coefficients to quantify the similarities.
Plasmid analysis
Plasmids encoding CMY-type β-lactamase genes were first transferred to E. coli DH10B by electroporation. The presence of the CMY genes in the transformants was confirmed by PCR. The plasmids in the transformants were extracted by the standard alkaline lysis method [14], digested with restriction enzyme PstI (New England Biolabs, Ipswich, MA) and subjected to electrophoresis in 0.8% agarose gel. The DNA ladders were then hybridized with a digoxigenin-labeled DNA probe specific for CMY-type β-lactamase gene using PCR DIG detection system (Roche Diagnostics, Indianapolis, IN). Cluster analysis for quantification of similarities was performed as in PFGE.
Statistical analysis
Categorical variables were compared using chi-squared test. Continuous variables were compared using the Mann-Whitney U test. P values < .05 were considered to be statistically significant.
RESULTS
E. coli clinical isolates producing CMY-type β-lactamase
During the study period, a total of 2583 E. coli isolates were identified in the microbiology laboratory. Of them, a total of 29 unique E. coli isolates were phenotypically confirmed as ESBL producers. Among the 18 unique ESBL screen-positive, confirmation-negative isolates and non-susceptible to ceftazidime that were identified, all isolates were found to possess CMY-type β-lactamase gene. E. coli isolates producing other types of plasmid-mediated AmpC-β-lactamase were found. The DNA sequences were consistent with CMY-2 for all of these isolates except one which was consistent with CMY-18, a variant of CMY-2. Out of 29 unique isolates phenotypically confirmed as ESBL-positive, 25 isolates possessed SHV, TEM or CTX-M-type ESBL genes. The remaining 4 isolates were found to possess CMY-type β-lactamase gene but not ESBL gene, indicating false-positive ESBL confirmatory tests. They included CMY-2 and its variants CMY-32 and 33. These four cases were added to the CMY cohort for the clinical analysis. Overall, 22 and 25 unique CMY and ESBL-producing E. coli isolates were identified and constituted the CMY cohort and ESBL cohort for this study, respectively.
All 22 CMY-producing isolates gave positive results with the phenotypic detection test of AmpC-β-lactamase production using 3-APB, whereas none of the 25 ESBL-producing isolates gave positive results, providing 100% sensitivity and specificity to this test among the study isolates.
Clinical features of CMY and ESBL-producing E. coli cases
Twenty-two cases in the CMY cohort and 25 cases in the ESBL cohort were included in the analysis (Table 1). The patients were predominantly female in both cohorts. The mean ages of the patients were 62.0 and 62.4 years, respectively. In the CMY cohort, 12 patients (55%) were admitted from home, whereas 11 patients (50%) had history of hospitalization within the previous 3 months.
Table 1.
Patient characteristics of CMY- and ESBL-producing E. coli cases. Number or proportion (%) of cases is shown unless specified otherwise.
| Characteristic | CMY cohort | ESBL cohort | P value |
|---|---|---|---|
| Mean age (SD) | 62.0 (21.0) | 62.4 (20.0) | 0.97 |
| Female | 18/22 (82) | 19/25 (76) | 0.63 |
| Caucasian | 20/21 (95) | 18/23 (78) | 0.10 |
| Transfer from another hospital | 3/22 (14) | 0/25 (0) | 0.056 |
| Previous hospitalization within 3 months | 11/21 (52) | 14/25 (56) | 0.81 |
| Previous location within 30 days | |||
| Home | 12/20 (60) | 13/24 (54) | 0.70 |
| Nursing home/LTCF | 5/20 (25) | 7/24 (29) | 0.76 |
| Hospital | 2/20 (10) | 4/24 (17) | 0.52 |
| Site of acquisition | |||
| Hospital-acquired | 11/21 (52) | 7/25 (28) | 0.091 |
| Healthcare-associated | 7/21 (33) | 14/25 (56) | 0.12 |
| Community-associated | 3/21 (14) | 3/25 (12) | 0.82 |
| Chronic underlying diseases | 19/21 (90) | 23/25 (92) | 0.86 |
| Immunosuppression | 13/21 (62) | 16/25 (64) | 0.88 |
| Previous antibiotic use within 30 days | 8/21 (38) | 9/25 (36) | 0.88 |
| Site of infection/colonization | |||
| Urinary tract | 17/22 (77) | 18/25 (72) | 0.68 |
| Symptomatic infection | 15/19 (79) | 10/22 (45) | 0.028 |
NOTE. LTCF; long-term care facility.
The source of CMY-producing E. coli was the urinary tract in 17 patients (77%). Three cases (14%) were community-associated, all of which were of urinary tract origin. The first case had cystitis and was treated with piperacillin-tazobactam. The second case had pyelonephritis and was treated with imipenem. Both had clinical cure. The third case represented colonization. Seven and 11 cases (32 and 50%) were healthcare-associated and hospital-acquired, respectively. The site of acquisition could not be determined in 1 case. Thirteen patients (59%) had at least one immunosuppressive condition, including diabetes, immunosuppressive therapy, malignancy and transplant. Whether the patient had symptomatic infection or colonization could be determined in 19 of 22 cases in the CMY cohort. Fifteen patients (86%) had symptomatic infection, significantly more frequent than those in the ESBL cohort (10 patients; 45%) (P = 0.028). Three patients in the CMY cohort, including 1 from the community, had pyelonephritis, whereas none in the ESBL cohort did.
CMY-producing E. coli clinical isolates
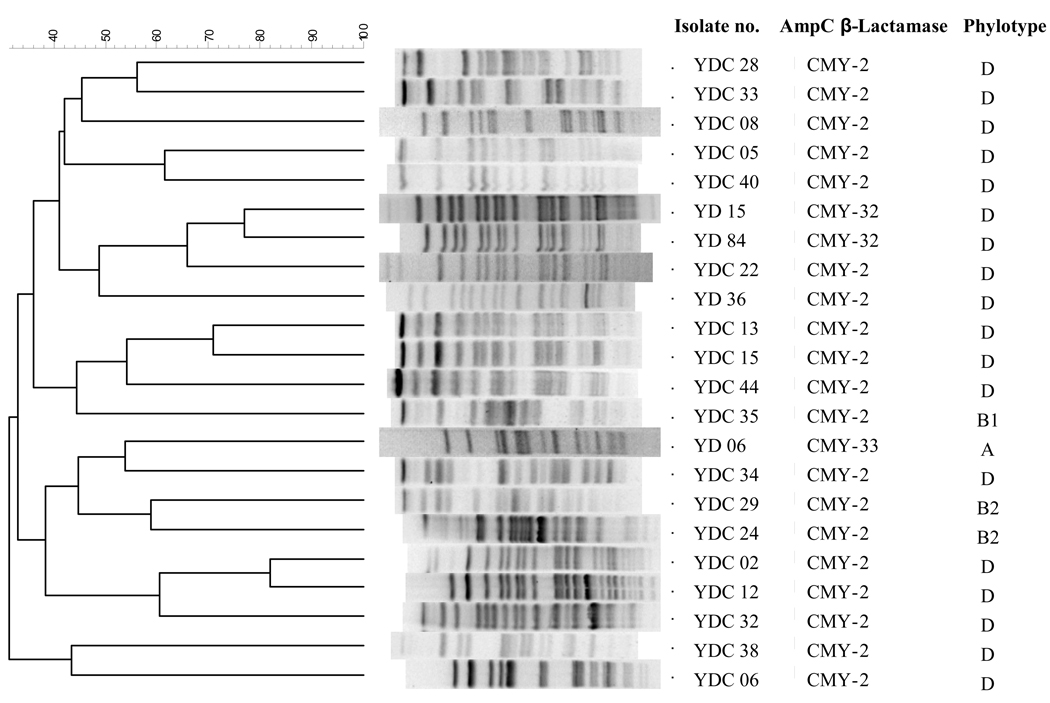
All CMY-producing clinical isolates were resistant to cefoxitin and cefpodoxime, intermediate or resistant to ceftazidime and variably resistant to cefotaxime. Co-resistance to non-β-lactam antimicrobials used for treatment of E. coli infection was common. Only 7 of 22 isolates (32%) in the CMY cohort and 5 of 25 isolates (20%) in the ESBL cohort were susceptible to ciprofloxacin. Fourteen (64%) and 8 (32%) were susceptible to gentamicin, and 11 (50%) and 8 (32%) were susceptible to sulfamethoxazole/trimethoprim in the CMY and ESBL cohorts, respectively. On PFGE, they constituted a relatively diverse population with 13 distinct PFGE types, as shown in Figure 1. In the CMY cohort, 20 of 22 isolates (91%) belonged to the two phylogenetic groups associated with virulence (2 isolates to B2 and 18 isolates to D), whereas 21 of 25 (88%) belonged to these two phylogenetic groups in the ESBL cohort (12 isolates to B2 and 9 isolates to D) (P = NS).
Figure 1.
Pulsed-field gel electrophoresis (PFGE) profiles of E. coli clinical isolates producing CMY-type β-lactamase.
Plasmid analysis
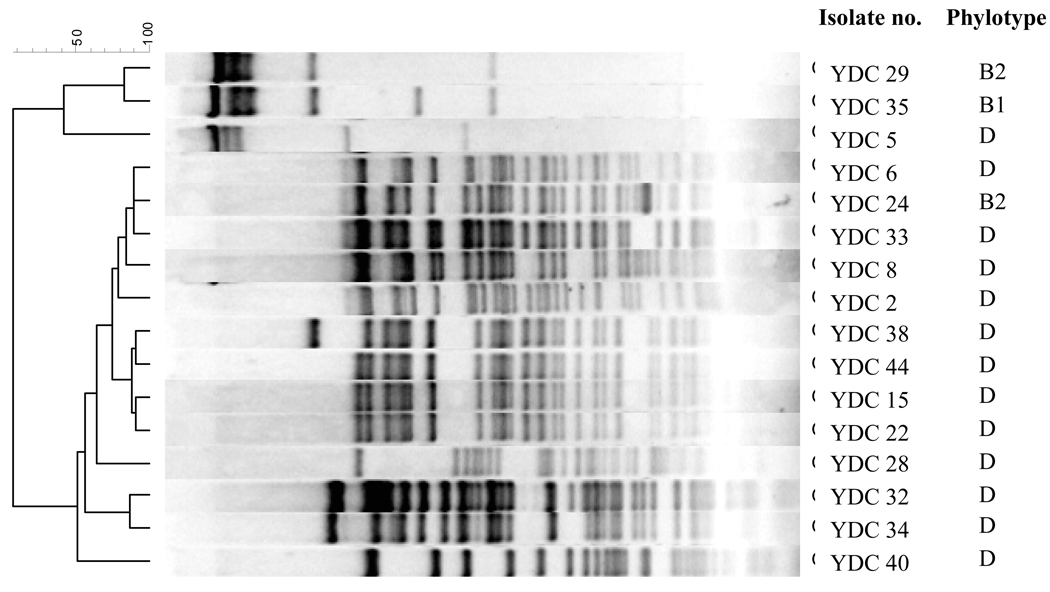
The results of restriction analysis of the isolates in the CMY cohort are shown in Figure 2. Plasmids from the isolates belonging to phylogenetic group D, the most commonly observed group in the CMY cohort, shared restriction patterns with greater than 70% similarity, suggestive of a common origin. The plasmid from one of the two isolates in phylogenetic group B2 also belonged to this cluster.
Figure 2.
Restriction profiles of plasmids encoding the CMY genes.
DISCUSSION
Despite the broad spectrum β-lactam resistance they confer, there have been many unanswered questions about plasmid-mediated AmpC β-lactamase in E. coli. First is that of its prevalence. One surveillance study conducted in the United States reported detection of plasmid-mediated AmpC β-lactamase and ESBL in 4 and 40% of E. coli isolates with reduced susceptibility to broad-spectrum cephalosporins collected between 1992 and 2000, respectively [15]. In the present study, a total of 2583 E. coli isolates were identified. Twenty-two unique cases due to CMY-producing E. coli thus translate into approximately 0.9% of all E. coli, though the actual rate is likely slightly higher given the exclusion of repeat isolates for the cases. Meanwhile, at least 1.0% of E. coli isolates were found to produce ESBL during the same period. Though the number of cases is much smaller in the present study, our data suggest that the incidence of cases due to CMY-producing E. coli may be almost as high as that of ESBL-producing E. coli in certain epidemiologic contexts, results that are consistent with recent findings in Nebraska [6]. Of note, the majority of isolates in the CMY cohort (18 of 22; 82%) belonged to phylogenetic group D and shared common restriction profiles of the plasmids encoding the CMY genes. This was in contrast to findings in a small study from France, where all CMY-producing E. coli isolates belonged to phylogenetic group B1 [16]. Our findings suggest that certain groups of extraintestinal E. coli strains may have affinity with CMY-bearing plasmids and warrants further investigation.
Second, unlike ESBLs, the detection method of plasmid-mediated AmpC β-lactamase including CMY-type β-lactamase has not been standardized by the CLSI or any other authorities, which is a major barrier in defining its epidemiology. As of now, the isolates producing this group of β-lactamases are typically labeled as ESBL-negative and would not be tested further. Recently, however, there has been a growing interest in using boronic acid compounds as specific AmpC inhibitors for detection of plasmid-mediated AmpC β-lactamase production in E. coli and Klebsiella spp. [8, 17, 18]. In the present study, the use of disk-based method adding 3-APB to a ceftazidime disk gave a sensitivity and specificity of 100% in detecting CMY-type β-lactamase production. Routine use of the boronic acid-based method on isolates which are positive for the initial screen test for ESBL production (i.e. reduced susceptibility to broad-spectrum cephalosporins) would greatly enhance detection of E. coli producing CMY or other types of plasmid-mediated AmpC-type β-lactamases. Four isolates producing CMY-type β-lactamase (1 CMY-2, 2 CMY-32 and 1 CMY-33) were misidentified as ESBL producers by the phenotypic confirmatory test in the clinical microbiology laboratory, which was reproducible in the research laboratory as well. These isolates all gave positive results with the boronic acid-based method as well but were definitely CMY producers and negative for ESBLs by genotypic tests. For the 2 isolates producing CMY-32, enhanced susceptibility of CMY-32 to clavulanic acid may account for this phenomenon [19]. We are currently investigating the mechanism underlying false positive ESBL testing for the other 2 isolates.
Third, largely as the consequence of the lack of a standardized detection method, the clinical significance of CMY-producing E. coli has not been known. In the present study, about half of the cases were acquired outside hospital, with urine being the most common site of infection or colonization. Of note, CMY-producing E. coli was significantly more likely to cause true infection (as opposed to colonization) than ESBL-producing E. coli. This was a surprising finding, which needs to be confirmed in a larger number of cases. What is the basis for the virulence of CMY-producing E. coli? There was no difference in membership in a virulence-associated phylogenetic group (group B2 or D) between the two groups. Therefore, it will be worth investigating the specific virulence gene contents of these isolates. In particular, it has been suggested that some of the plasmids carrying the CMY-type β-lactamase genes also carry a cluster of genes encoding the type IV pili, which contributes to adhesion and invasion [20]. The clinical implications of the potentially enhanced virulence of CMY-producing E. coli, if confirmed, are paramount. We also noted high rates of co-resistance of these isolates to non-β-lactam antimicrobials commonly used to treat E. coli infections, much higher than in a previous report [5], likely due to the difference in patient populations. Although our study was too small to systematically assess clinical outcome, the multidrug-resistant nature of CMY-producing E. coli is a concern in terms of empiric management of these infections.
Our study has several limitations. The study was performed at a single center in the United States, and we only had relatively small number of cases in both cohorts. Because of the retrospective nature of the clinical study, some of the clinical data were not available, leaving room for potential bias, though the number of these cases was minimal.
In conclusion, we have shown that clinical cases due to E. coli producing CMY-type broad-spectrum β-lactamase are almost as frequent as those due to E. coli producing ESBLs in Pittsburgh, Pennsylvania. Routine screening for CMY-producing E. coli using a simple phenotypic method will help identify these clinical cases in the clinical microbiology laboratory. A larger, multi-center clinical study is warranted to further assess the clinical significance of these E. coli isolates.
Acknowledgments
The authors thank research coordinator Diana L. Pakstis; data analyst Lloyd G. Clarke; and the clinical microbiology laboratory staff at the University of Pittsburgh Medical Center.
Financial support. This study was supported by a joint fellowship grant from the National Foundation of Infectious Diseases and the Infectious Diseases Society of America and in part by Ministerio de Sanidad y Consumo (PI070190 and Spanish Network for the Research in Infectious Diseases, REIPI RD06/0008), Junta de Andalucía (PI0048/2008) and Training Grant (T32AI007333) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Potential conflicts of interest. No conflicts.
REFERENCES
- 1.Bauernfeind A, Chong Y, Schweighart S. Extended broad spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection. 1989;17:316–321. doi: 10.1007/BF01650718. [DOI] [PubMed] [Google Scholar]
- 2.Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walther-Rasmussen J, Hoiby N. Plasmid-borne AmpC β-lactamases. Can J Microbiol. 2002;48:479–493. doi: 10.1139/w02-039. [DOI] [PubMed] [Google Scholar]
- 4.Doi Y, Paterson DL. Detection of plasmid-mediated class C beta-lactamases. Int J Infect Dis. 2007;11:191–197. doi: 10.1016/j.ijid.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Pitout JD, Gregson DB, Church DL, Laupland KB. Population-based laboratory surveillance for AmpC β-lactamase-producing Escherichia coli, Calgary. Emerg Infect Dis. 2007;13:443–448. doi: 10.3201/eid1303.060447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanson ND, Moland ES, Hong SG, Propst K, Novak DJ, Cavalieri SJ. Surveillance of community-based reservoirs reveals the presence of CTX-M, imported AmpC, and OXA-30 β-lactamases in urine isolates of Klebsiella pneumoniae and Escherichia coli in a U.S. community. Antimicrob Agents Chemother. 2008;52:3814–3816. doi: 10.1128/AAC.00877-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement M100-S17. 2007. [Google Scholar]
- 8.Yagi T, Wachino J, Kurokawa H, et al. Practical methods using boronic acid compounds for identification of class C β-lactamase-producing Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 2005;43:2551–2558. doi: 10.1128/JCM.43.6.2551-2558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Dde O, Doi Y, Szabo D, et al. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum β-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob Agents Chemother. 2008;52:1790–1793. doi: 10.1128/AAC.01440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horan TC, Gaynes RP. Surveillance of nosocomial infections. In: CG M, editor. Hospital epidemiology and infection control. 3rd ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2004. [Google Scholar]
- 11.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 13.Picard B, Garcia JS, Gouriou S, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Russell DW. Molecular cloning. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 15.Alvarez M, Tran JH, Chow N, Jacoby GA. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–537. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri H, Eb F, Berkani A, Nordmann P. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered in a French hospital. J Antimicrob Chemother. 2008;61:498–503. doi: 10.1093/jac/dkm538. [DOI] [PubMed] [Google Scholar]
- 17.Jacoby GA, Walsh KE, Walker VJ. Identification of extended-spectrum, AmpC, and carbapenem-hydrolyzing β-lactamases in Escherichia coli and Klebsiella pneumoniae by disk tests. J Clin Microbiol. 2006;44:1971–1976. doi: 10.1128/JCM.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenwald NP, Jevons G, Andrews J, Ang L, Fraise AP. Disc methods for detecting AmpC β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother. 2005;56:600–601. doi: 10.1093/jac/dki278. [DOI] [PubMed] [Google Scholar]
- 19.Endimiani A, Doi Y, Bethel CR, et al. CMY-32, a novel β-lactamase of the CMY-2 family with enhanced susceptibility to sulfone inhibitors [abstract C1-888]; Program and abstracts of the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy; Washington, DC: American Society for Microbiology; 2008. [Google Scholar]
- 20.Garcia-Fernandez A, Chiaretto G, Bertini A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008;61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]