Abstract
Linezolid, which targets the ribosome, is a new synthetic antibiotic that is used for treatment of infections caused by Gram-positive pathogens. Clinical resistance to linezolid, so far, has been developing only slowly and has involved exclusively target site mutations. We have discovered that linezolid resistance in a methicillin-resistant Staphylococcus aureus hospital strain from Colombia is determined by the presence of the cfr gene whose product, Cfr methyltransferase, modifies adenosine at position 2503 in 23S rRNA in the large ribosomal subunit. The molecular model of the linezolid-ribosome complex reveals localization of A2503 within the drug-binding site. The natural function of cfr likely involves protection against natural antibiotics whose site of action overlaps that of linezolid. In the chromosome of the clinical strain, cfr is linked to ermB, a gene responsible for dimethylation of A2058 in 23S rRNA. Co-expression of these two genes confers resistance to all the clinically-relevant antibiotics that target the large ribosomal subunit. The association of the ermB/cfr operon with transposon and plasmid genetic elements indicate its possible mobile nature. This is the first example of clinical resistance to the synthetic drug linezolid which involves a natural resistance gene with the capability of disseminating among Gram-positive pathogenic strains.
INTRODUCTION
An oxazolidinone drug linezolid is one of the newest clinically-important antibiotics. Approved for clinical use in 2000, it represented the first new class of antibiotics introduced into medical practice in more than a quarter of a century (Hutchinson, 2003). Linezolid is a synthetic inhibitor of protein synthesis that is active against many Gram-positive bacteria, including such pathogens as methicillin- and vancomycin-resistant staphylococci, vancomycin-resistant enterococci and penicillin-resistant pneumococci. It is often used for treatment of complicated infections when other therapies have failed (Bozdogan and Appelbaum, 2004).
Linezolid inhibits protein synthesis by binding to the large ribosomal subunit (Lin et al., 1997). Mutational and crosslinking studies revealed that the ribosomal peptidyl transferase center is the target of drug action (Kloss et al., 1999; Xiong et al., 2000; Prystowsky et al., 2001; Meka et al., 2004; Colca et al., 2003; Sander et al., 2002). The drug binding site is composed of rRNA residues that form the inner shell of the peptidyl transferase active site (Colca et al., 2003; Skripkin et al., 2005; Leach et al., 2007). Although the binding site of linezolid overlaps with those of several other peptidyl transferase inhibitors, its interactions with the ribosome are specific enough to allow for linezolid activity against bacterial pathogens that developed resistance to other antibiotics that target the ribosome (Shinabarger, 1999).
One of the critical advantages of linezolid over currently used antibiotics is its entirely synthetic nature. All other inhibitors of protein synthesis are derived from natural antibiotics of microbial origin whose producers serve as the natural reservoirs of resistance genes (Cundliffe, 1989; Poehlsgaard and Douthwaite, 2005). Acquisition of these genes through horizontal gene transfer is one of the most common ways through which clinical pathogens develop antibiotic resistance (Franceschi et al., 2004; Weisblum, 1995). Often associated with mobile genetic elements, natural resistance genes can rapidly spread among pathogenic strains and impede the clinical value of many important drugs (Bozdogan et al., 2004; Reyes et al., 2007; Roberts et al., 1999; Robicsek et al., 2006). Since linezolid is a synthetic compound which does not have natural prototypes it was expected that there is no natural pool of resistance genes which could facilitate the development of clinical resistance. Indeed, the only mechanism of resistance to linezolid in clinical isolates reported to date is due to mutations in the drug target site, primarily the rRNA of the large ribosomal subunit (Meka and Gold, 2004; Prystowsky et al., 2001). This type of resistance appears rarely, develops slowly because of the redundancy of rRNA genes in bacteria, and is not transferable between pathogenic species. In all the reported cases, the resistance was apparently generated de novo through spontaneous mutations rather than via genetic exchange.
We report here the first example of a linezolid resistance mechanism in a clinical MRSA strain that is based on acquisition of a natural and potentially transferable resistance gene. The product of this gene modifies a specific rRNA nucleotide located in the site of the drug action. In the chromosome of the clinical MRSA strain, this gene is apparently associated with mobile genetic elements which raises the possibility of its transmission to other pathogenic strains.
RESULTS
The linezolid-resistant MRSA isolate lacks known resistance mutations
A MRSA strain CM05 isolated in 2005 in Medellin, Colombia from a patient with a fatal ventilator-associated pneumonia exhibited resistance to linezolid (16 µg/ml) as well as to several other antibiotics (Table 1). This patient received only two doses of linezolid, and thus, the emergence of linezolid resistance was rather unexpected since this phenomenon has been previously associated with prolonged treatment with the antibiotic. This prompted us to investigate the mechanism of linezolid resistance in the CM05 MRSA strain. All known cases of linezolid resistance in clinical isolates of Gram-positive pathogens have been associated with mutations in the drug target site: domain V of 23S ribosomal RNA or protein L4 (Meka and Gold, 2004; Wolter et al., 2005). Nonetheless, sequencing of the PCR-amplified domain V of 23S rRNA of the CM05 isolate did not reveal any mutation (the sensitivity of the assay was sufficient to detect the presence of a mutation even in a single out of five or six rrn alleles present in staphylococci (Wada et al., 1993)). Similarly, no mutation was found in the L4 protein gene PCR-amplified from genomic DNA of the CM05 isolate. The lack of target site mutations in the linezolid-resistant MRSA strain indicated that a new molecular mechanism was responsible for resistance of the isolate to linezolid.
Table 1.
Antibiotic sensitivity profile of the MRSA strain CM05 and of the laboratory S. aureus strain transformed with the cfr-expressing plasmid pLXM1.
| MIC (µg/ml) /phenotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
 |
||||||||||||
| MRSA CM05 | 16/R | >64/R | 32/R | >64/R | 0.12/S | 64/R | 0.0075/S | 64/R | >32/R | 1/S | 0.5/S | 0.5/9.5 |
| ATCC a) 29213 | 2 | 0.12 | 0.06 | 4 | 0.5 | 0.25 | 0.0075 | 0.25 | 0.25 | 1 | 0.5 | 0.06/1.18 |
| RN4220 (pLXM1) | 4 | nd | nd | >10 | nd | nd | nd | nd | nd | nd | nd | nd |
| RN4220 a) (pLI50) | 1 | nd | nd | >10 | nd | nd | nd | nd | nd | nd | nd | nd |
A control S. aureus strain
Cells transformed with an empty vector
An unusual posttranscriptional modification at position 2503 causes linezolid resistance in the MRSA CM05
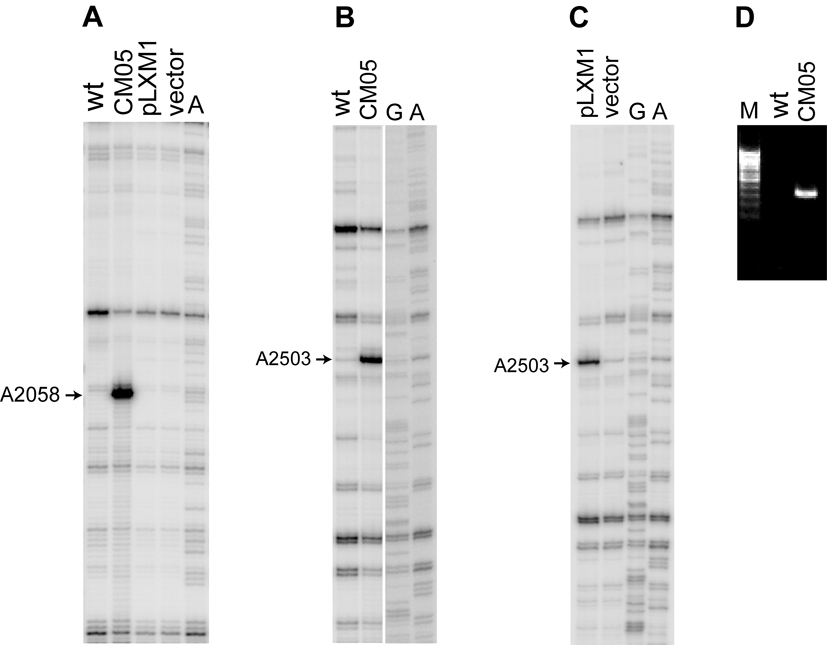
Resistance to ribosome-targeting antibiotics is often conferred by modification of specific nucleotide residues in rRNA (Kehrenberg et al., 2005; Mann et al., 2001; Treede et al., 2003; Weisblum, 1995). When rRNA from the CM05 isolate was examined by primer extension, we noticed a reverse transcriptase stop at A2058 in 23S rRNA (Figure 1A). Subsequent analysis showed that this effect is explained by the presence of ermB methyltransferase gene in the MRSA CM05 strain. The ErmB methyltransferase dimethylates A2058 conferring resistance to macrolides, lincosamides and streptogramins B (MLSB resistance) and may account, at least in part, for the observed resistance of the CM05 isolate to erythromycin and clindamycin (Table 1). The second strong reverse transcriptase stop indicated that another 23S rRNA nucleotide, A2503, carries an unusual posttranscriptional modification (Figure 1B). A2503 is a component of the linezolid binding site (Leach et al., 2007) and it is conceivable that modification of this rRNA residue may affect binding or action of the drug. A recent report by Kehrenberg et al. (2005) indicated that in staphylococci isolates from animals, A2503 can be a subject of posttranscriptional modification by a methyltransferase Cfr. The initial reports suggested that this modification could confer resistance to the peptidyl transferase targeting antibiotics florfenicol, chloramphenicol and clindamycin (Kehrenberg et al., 2005; Schwarz et al., 2000). Even though the cfr gene was never before found in clinical isolates, the finding of Kehrenberg et al. prompted us to test the presence of this gene in the clinical MRSA strain. PCR analysis carried out with cfr-specific primers indicated the presence of the cfr gene in the CM05 genome and thus revealed it as a putative cause of linezolid resistance of the clinical MRSA (Figure 1C).
Figure 1.
An unusual posttranscriptional modification of A2503 in 23S rRNA of the clinical MRSA isolate. Primer extension analysis of 23S rRNA isolated from a control wild type strain RN6390B (wt) and clinical linezolid-resistant S. aureus strain CM05 (CM05) or a laboratory strain RN4220 transformed with an empty vector pLI50 (vector) or cfr-expressing plasmid pLXM1 (pLXM1). Sequencing lanes are marked (G, A). The position of the reverse transcriptase stop corresponding to characteristic posttranscriptional modifications at A2058 and A2503 are indicated by arrows. A, ErmB-specific dimethylation of A2058. B and C, Cfr-specific modification of A2503. D. PCR amplification of a 330 bp portion of the cfr gene from the genome of CM05 cells. Similar amount of template DNA prepared from wild type S. aureus cells was used in the control (wt).
In order to investigate whether the presence of cfr and the resulting modification of A2503 was indeed the cause of linezolid resistance of the CM05 isolate, the complete cfr ORF was PCR amplified, sequenced and cloned into the pLI50 vector under the control of Pspac promoter (Lee et al., 1991; Yansura and Henner, 1984). The 1050-nt long sequence of the cfr gene in the CM05 MRSA isolate was identical to that of cfr in the plasmid pSCFS1 reported by Schwarz and co-workers (Kehrenberg et al., 2004). Introduction of the resulting plasmid, pLXM1, into S. aureus RN4220 cells led to a 4-fold increase in MIC to linezolid (Table 1). Linezolid resistance of the transformants correlated with the appearance of the characteristic reverse transcriptase stop corresponding to the unusual modification of A2503 (Figure 1B). This result established a causative relationship between the presence of the cfr gene in the MRSA clinical strain, posttranscriptional modification of A2503, and linezolid resistance.
Location of cfr in the MRSA CM05 genome
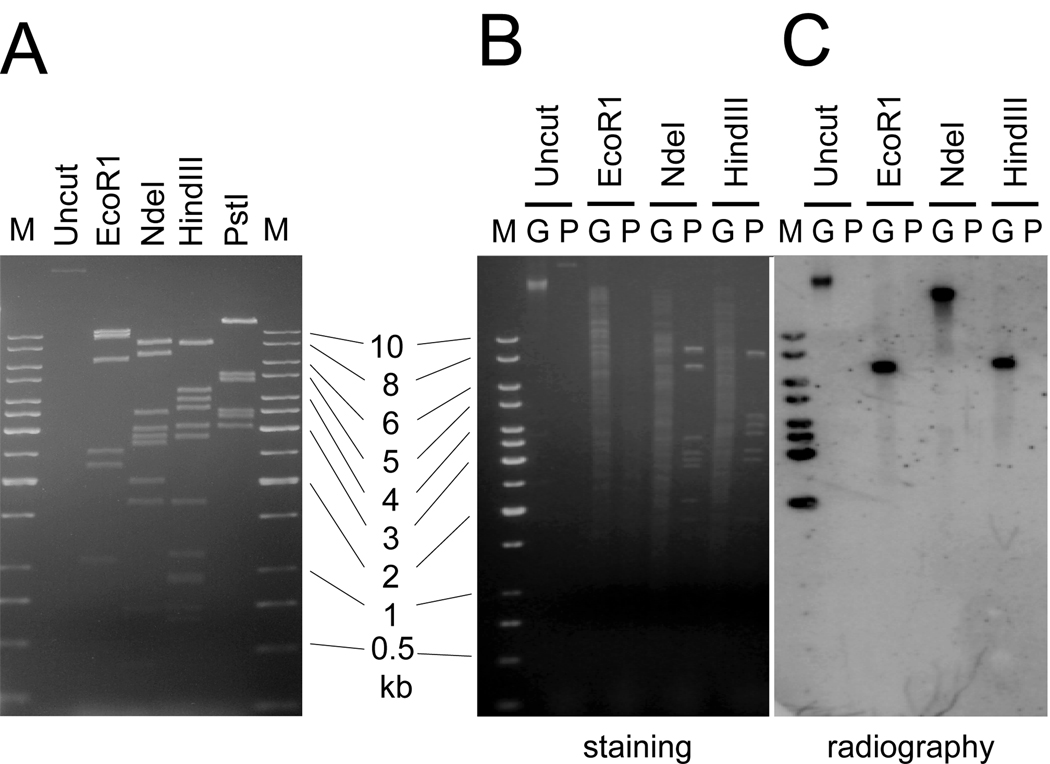
The cfr gene was previously found only in staphylococcal isolates from animal sources and almost exclusively on plasmids that ranged in size from 17 to 43 kb (Kehrenberg et al., 2004; Kehrenberg and Schwarz, 2006; Kehrenberg et al., 2007). Even though the CM05 MRSA carries a 32 kb plasmid (Figure 2A), Southern blotting analysis showed that in the CM05 genome, cfr is located on the chromosome (Figure 2 B and C). In agreement with this finding, the plasmid from the CM05 isolate failed to render recipient S. aureus cells resistant to linezolid (not shown).
Figure 2.
Localization of the cfr gene on the chromosome of the S. aureus CM05 strain. A), Restriction analysis of the 32-kb plasmid isolated from the CM05 strain. B), C), Southern blotting analysis of the total genomic (G) or plasmid (P) DNA prepared from the CM05 cells. The full-length cfr probe was used for hybridization. The sizes of DNA fragments in the DNA marker are indicated.
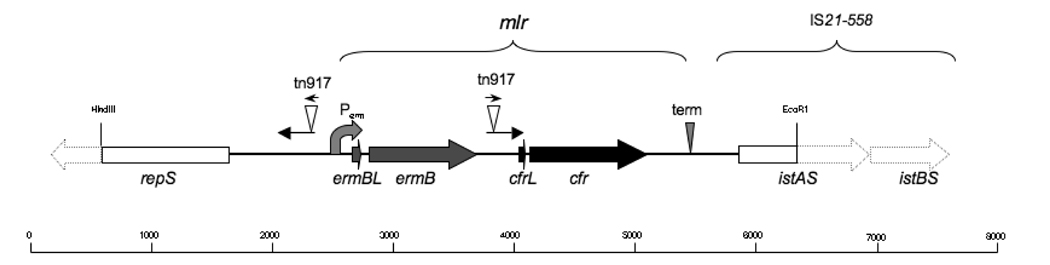
Sequencing of the cloned cfr-containing 5.8 kb HindIII/EcoRI chromosomal DNA fragment revealed that cfr was located downstream of the ermB gene which encodes a rRNA methyltransferase that dimethylates A2058 in the 23S rRNA and confers resistance to macrolides, lincosamides and streptogramins B (MLSB resistance) (Shaw and Clewell, 1985) (Figure 3). The ermB/cfr cluster is flanked on one side by the transposase/cointegrase gene istAS from the IS21-558 mobile genetic element. PCR analysis showed the presence of the complete IS21-558 (Smith and Mankin, unpublished). The IS21-558 element was shown to be implicated in the mobility of the cfr gene in animal isolates (Kehrenberg et al., 2007) and might contribute to its mobilization in the clinical strain.
Figure 3.
The gene cfr and its environment on the chromosome of the S. aureus strain CM05. The complete genes present within the sequenced regions are shown as solid arrows and incomplete genes are shown as solid rectangles. The unsequenced parts of the identified genes are shown as contoured arrows. The presence of the presumably complete istAS and istBS genes constituting the IS21–558 insertion sequence was verified by PCR (Smith and Mankin, unpublished). The location of the ermB and cfr leader ORFs (ermBL and cfrL, respectively) which might be involved in translation attenuation is shown. The ermB promoter (Perm) upstream of the ermB gene and a putative terminator (term) downstream from the cfr gene which might control transcription of the mlr operon are shown. The 290 bp-long inverted repeats flanking the ermB gene, which include the 28 bp-long transposon tn917 repeats, are shown by arrows. The scale is in bp.
Upstream from ermB, a 5’ segment of the gene repS is present. Its product, the RepS protein, is involved in initiation of plasmid replication (Hashiba et al., 1993). A close association of the cfr gene with a characteristic plasmid gene indicates that integration of a plasmid carrying the cfr gene in the chromosome of CM05 cells, was the likely route of acquisition of linezolid resistance by the MRSA human isolate.
DISCUSSION
We presented evidence that resistance of a clinical MRSA strain CM05 to a synthetic antibiotic linezolid is caused, at least in part, by acquisition of a gene whose product is responsible for a specific posttranscriptional modification of a 23S rRNA nucleotide located in the site of drug action.
A causative relationship between the presence of cfr and resistance of the clinical MRSA strain to linezolid is demonstrated by the fact that introduction of cfr into a linezolid-sensitive S. aureus strain notably increased its resistance to the drug and led to the appearance of an unusual posttranscriptional modification at A2503 in 23S rRNA but not at A2058. (Table 1 and Figure 1B). A similar effect was recently demonstrated by Long et al. (Long et al., 2006) who used a different construct to show that a cfr-containing DNA segment from animal isolates of staphylococci rendered recipient cells resistant to several peptidyl transferase-targeting antibiotics, including oxazolidinones. Since cfr-based resistance affects specifically the large ribosomal subunit, it is obvious that other determinants are present in the clinical MRSA CM05 strain that render it resistant to antibiotics affecting different targets (Table 1).
The exact nature of posttranscriptional modification introduced by the Cfr enzyme is unknown. In wild type E. coli, A2503 is mono-methylated at the adenine ring and is present in the ribosome in the form of m2A (Kowalak et al., 1995). Though the status of A2503 in wild type S. aureus is unknown, the presence of a weak reverse transcriptase stop at this position (Figure 1) suggests that, similar to E. coli, it might also carry a posttranscriptional modification. Closely homologous genes of methyltransferase enzymes, which are likely responsible for the ‘wild type modification’ of A2503, are present in the E. coli and S. aureus genomes (Toh and Mankin, in preparation). Expressed in E. coli, Cfr was shown to add an extra methyl group to m2A2503 (Kehrenberg et al., 2005). However, the exact location of the extra methyl group added by Cfr remained ambiguous. By extrapolation, we expect that S. aureus cells carrying the cfr gene also end up having two methyl groups at A2503.
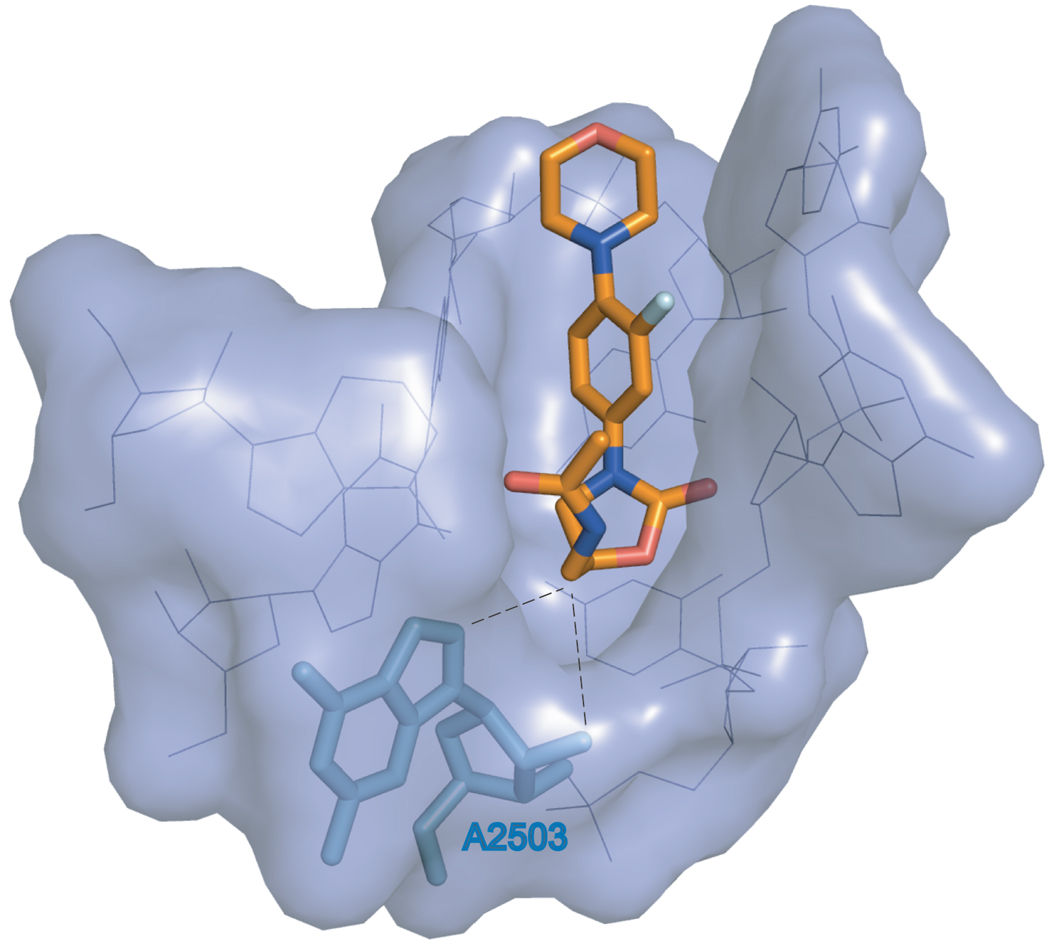
The molecular model of linezolid bound to the bacterial ribosome shows close proximity of the C5 side chain of the drug to A2503 (Leach et al., 2007) (Figure 4). A methyl group at 2’ oxygen of the A2503 ribose or C8 carbon of the A2503 base will come into close proximity to the C5 side chain of linezolid and could affect its placement in the ribosome. Alternatively, an additional methyl group at the adenine base can change its conformation from syn, as modeled in the E. coli wild type ribosome, to anti which would result in the direct clash of the nitrogen base with the antibiotic (Schuwirth et al., 2005; Selmer et al., 2006). Since A2503 is also involved in binding of other peptidyl transferase targeting antibiotics, it is not surprising that its modification by the Cfr enzyme renders cells resistant to a large array of peptidyl transferase inhibitors (Hansen et al., 2003; Kehrenberg et al., 2005; Long et al., 2006; Schlunzen et al., 2001).
Figure 4.
The proximity of A2503, which is the target of Cfr-modification, to linezolid bound in the A-site of the bacterial ribosome. The orientation and placement of linezolid within its binding site in the translating bacterial ribosome was taken from the reference (Leach et al., 2007). RNA nucleotides constituting the linezolid binding site are shown as lines (blue) and are also rendered as a surface representation. Linezolid and A2503 are shown as sticks. A 2-methyl group has been computationally added to A2503. In linezolid, carbon atoms are colored orange, nitrogens - blue, oxygens - red and fluorine is light-blue. The proximity of 2’O and C8 of A2503 with the linezolid C5 side chain is indicated by dashed lines.
The genetic organization of the cfr gene in the chromosome of the strain CM05 is remarkable. The putative cfr promoter sequence identified in the pSCFS1, pSCFS3 and pSCFS6 plasmids in staphylococci from animal sources (Kehrenberg and Schwarz, 2006) has been eliminated in MRSA CM05 due to the insertion of the ermB gene (The latter was likely delivered by the tn917 transposon as indicated by the presence of the tn917 inverted repeats surrounding the ermB gene). No other obvious promoter sequence is found in the intergenic spacer between ermB and cfr. RT-PCR analysis showed the presence of transcripts that span both ermB and cfr genes indicating that cfr is likely co-transcribed with ermB (Smith and Mankin, unpublished). Both Cfr and ErmB are rRNA methyltransferases. ErmB dimethylates N6 of A2058 in 23S rRNA conferring the MLSB type of resistance. Methylation of A2503 by Cfr renders cells resistant to oxazolidinones, phenicols, lincosamides, streptogramins A and pleuromutilins (Long et al., 2006). Thus, expression of ermB and cfr as a part of one operon (which we designate mlr for modification of large ribosomal subunit) should result in modification of two 23S rRNA nucleotides, A2058 and A2503, and render cells resistant to all the clinical antibiotics acting upon the large ribosomal subunits. To our knowledge, the mlr operon represents the first example of a single transcription unit affording resistance to such a diverse spectrum of ribosomal antibiotics. Simultaneous modification of two rRNA nucleotides within a drug binding site may synergistically increase antibiotic resistance (Treede et al., 2003; Liu and Douthwaite, 2002). While A2058 is not a part of the linezolid binding site in the ribosome and thus its modification is not expected to contribute to linezolid resistance, simultaneous modification of A2503 by Cfr and of A2058 by ErmB may have a strong effect on cell susceptibility to streptogramins because in their ribosome-bound form, two streptogramin components, A and B, are in a close contact with A2503 and A2058, respectively (Harms et al., 2004; Tu et al., 2005; Long et al., 2006). Testing the effects of A2503 and A2058 methylation on susceptibility to streptogramins is currently in progress (Smith and Mankin, unpublished).
The putative mobile nature of the mlr operon is worrisome, since it can compromise treatment of various Gram-positive infections in humans. Previously, the cfr gene was found primarily on plasmids which could be easily exchanged between staphylococcal strains in animals(Kehrenberg and Schwarz, 2006; Kehrenberg et al., 2007). In the clinical MRSA strain, the cfr gene is present on the chromosome but is positioned next to the repS gene whose product is involved in the control of plasmid copy number (Hashiba et al., 1993). This proximity indicates that in the CM05 chromosome cfr resides on an integrated plasmid which could retain its ability for excision and transmission. The repS gene in the CM05 chromosome shows high homology to repS from streptococcal and enterococcal plasmids (Ceglowski and Alonso, 1994; Paulsen et al., 2003). This opens the general possibility of transmission of the mlr operon to a variety of pathogenic strains. Interestingly, the ermB gene, which in the MRSA CM05 strain is found next to cfr, is prevalent in enterococcal strains while only ermA has been found so far in Colombian MRSA isolates (Reyes et al., 2007). This finding may indicate that mlr operon was actually formed in an enterococcal strain and subsequently was brought on a plasmid into the genome of MRSA CM05. Pulse-field gel electrophoresis analysis of DNA of the CM05 strain revealed its close relation to the so called Chilean MRSA clone that emerged within the last five years and is currently accounting for the majority of hospital S. aureus infections in Colombia (Arias et al., 2006). Neither of the other tested Colombian S. aureus clones, including those which show a DNA fragmentation pattern similar to CM05, showed resistance to linezolid and thus are unlikely to carry cfr (Arias et al., 2006). Thus, it appears that acquisition of cfr by the Chilean clone is a recent event. In CM05 chromosome, the mlr operon is adjacent to the IS21-558 mobile genetic element which was previously shown to be implicated in cfr transposition in pSCFS3 and pSCFS6 plasmids in animal isolates (Kehrenberg and Schwarz, 2006; Kehrenberg et al., 2007). This proximity opens another route – through genetic transposition – for dissemination of the mlr operon to other pathogenic strains.
One of the perceived advantages of linezolid was its entirely synthetic nature which was expected to help evade natural resistance mechanisms. However, since linezolid binds to the same ribosomal site that is used by several classes of peptidyl transferase inhibitors (Leach et al., 2007; Moazed and Noller, 1987; Porse and Garrett, 1999; Poulsen et al., 2001), it is feasible that a natural resistance mechanism that targets the peptidyl transferase center renders cells resistant to this synthetic antibiotic. Even though the origin of cfr is unknown, the gene has likely originated in a microbial producer of one of the natural peptidyl transferase inhibitors. The acquisition, maintenance and spread of cfr among staphylococci that infect farm animals could be promoted by the use of phenicols and lincosamides in veterinary practice as well as the use of these antibiotics as growth promoters (Aarestrup et al., 2000; Schwarz et al., 2004). Our data re-emphasize the fact that a reservoir of resistance genes in the animal industry could potentially jeopardize the usefulness of clinical antibiotics, even those that do not have natural prototypes.
EXPERIMENTAL PROCEDURES
The linezolid-resistant clinical isolate of S. aureus and its microbiological testing
Linezolid- and methicillin-resistant S. aureus was isolated in 2005 in Medellin, Colombia from the sputum of a patient with a fatal ventilator-associated pneumonia. The isolate was designated CM05. The patient had received only two doses of linezolid, 600 mg each, one day prior to isolation of the culture. Typing of the isolate as methicillin-resistant S. aureus was based on the BactiStaph latex agglutination assay (Remel, Lenexa, KS) and growth on Mueller-Hinton plates containing 6 µg/ml oxacillin, in accordance with CLSI guidelines (Standards, 2003). Antibiotic sensitivity testing was done using a CLSI-approved broth microdilution protocol or E-test (for clindamycin). S. aureus strain ATCC 29213 was included as control.
Isolation of genomic DNA, plasmid DNA and total cellular RNA
Genomic DNA was isolated from S. aureus strains using a cetyltrimethylammonium bromide (CTAB) method (Ausebel et al., 1987). The only difference with the published protocol was the addition of lysostaphin to 40 µg/ml and incubation for 30 min at 37°C prior to addition of RNase.
Plasmid DNA was extracted from the CM05 strain following the maxi-prep alkaline lysis procedure (Sambrook et al., 1989) with the addition of a lysostaphin treatment step. Supercoiled circular DNA was further purified by CsCl centrifugation. The size of the plasmid was calculated as the sum of the fragment sizes obtained after digestion of the plasmid with restriction enzymes.
For isolation of total RNA, S. aureus cells (the clinical isolate CM05 or wild type RN6390B strain (Balaban and Novick, 1995) were grown in tryptic soy broth to an optical density of A550 = 1.0. RNA was isolated using the UltraClean Microbial RNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer’s protocol.
Testing for the presence of known linezolid resistance mutations
Domain V of 23S rRNA gene was PCR amplified from total DNA of the CM05 isolate and a linezolid-sensitive control, S. aureus strain (ATCC 29213) using primers, GTGAAATCATAGTACCTGTGAAG and TTGATTAAGTCTTCGATCGATTAG, corresponding to positions 2010–2032 and 2880–2904 of 23S rRNA. The resulting 887 bp DNA fragment was sequenced using the same primers. In addition, the PCR fragment was treated with restriction enzyme NheI and analyzed by agarose gel electrophoresis to confirm the absence of the most common linezolid resistance mutation (G2576T) which generates a new NheI site. The gene of ribosomal protein L4 was PCR-amplified from DNA of the CM05 isolate using primers CAGTGATTACGGGGCGCTTAAG and GTAATAAATAATAAGAAGTGAAAGG and sequenced.
Primer-extension analysis of rRNA
5 pmol of one of the 5’[32P]-labeled primers, GTCCATCCCGGTCCTCTCGTAC, GCCGACATCGAGGTGCC or TAGTATCCCACCAGCGTCTC, was annealed to 2 µg of total S. aureus RNA and extended using AMV reverse transcriptase (Seikagaku America) using the described protocol (Merryman and Noller, 1998). The cDNA products of the primer extension reactions were separated on 6% polyacrylamide sequencing gels.
Modeling the interaction of linezolid with A2503 in the ribosome
The modeling of linezolid placement in its ribosomal binding site based on the in vivo crosslinking data and crystallographic structure of the E. coli ribosome will be described elsewhere (Leach et al., 2007). In order to generate Figure 4, the C2-methyl group was added to the adenine 2503 base computationally.
Detection, cloning and sequencing of the cfr gene
The presence of the cfr gene in the CM05 isolate (Schwarz et al., 2000) was verified by PCR using a pair of primers ATGAATTTTAATAATAAAACAAAG and TACACCCAAAATTACATCCG. The complete cfr gene was PCR amplified using the primers ATGAATTTTAATAATAAAACAAAG and CTATTGGCTATTTTGATAATTACC and cloned into the shuttle vector pLI50 (Lee et al., 1991) under the control of a B. subtilis promoter Pspac (Yansura and Henner, 1984) to produce plasmid pLXM1. In order to investigate the physical arrangement of cfr on the chromosome of the CM05 strain, a 5.8 kb EcoRI/ HindIII fragment of CM05 genomic DNA, containing the cfr gene, was cloned into the corresponding sites of the pBR322 vector to produce plasmid pMS1. The cloned segment was sequenced by primer walking.
A 4.9 kb HindIII-XbaI fragment of the CM05 DNA insert was excised from pMS1 and cloned into the corresponding sites of pLI50. The resulting plasmid, pMS2, as well as pLXM1 and the empty pLI50 vectors were introduced in S. aureus RN4220 cells by electroporation (Schenk and Laddaga, 1992). The transformants were selected on tryptic soy agar plates supplemented with 10 µg/ml of chloramphenicol.
For Southern blotting analysis, total DNA or purified plasmid DNA from S. aureus CM05 was digested with restriction enzymes EcoR1, HindIII, PstI or their combinations. DNA was fractionated in a 0.8% agarose gel, transferred to Hybond-N+ membranes (Amersham Biosciences) and hybridized with the PCR-amplified and radiolabeled full-length cfr gene.
Nucleotide sequence accession numbers
The sequence of a 5,822-bp segment of S. aureus CM/05 including the ermB and cfr region and its flanking areas has been deposited to the NCBI database under accession number EF450709.
ACKNOWLEDGEMENTS
We thank Dr. Martha Vallejo (Hospital General de Medellin) and The Colombian Nosocomial Resistance Study Group for isolating and sending the linezolid-resistant MRSA strain, Ms. Reyes (Universidad El Bosque) for susceptibility testing, Dr. Jayaswal (Illinois State University) for providing the pLI50 plasmid, Drs. Lavie and Grum-Tokar (University of Illinois at Chicago) for help in preparing Figure 4 and L. Smith for proofreading. This work was supported in part by NIH grant U19 AI056575, project 1 (to A.S.M.).
REFERENCES
- Aarestrup FM, Kruse H, Tast E, Hammerum AM, Jensen LB. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb Drug Resist. 2000;6:63–70. doi: 10.1089/mdr.2000.6.63. [DOI] [PubMed] [Google Scholar]
- Arias CA, Villegas MV, Reyes J, Moreno J, Xiong L, Vallejo M, Lolans K, Mankin AS, Castañeda E, Quinn JP. Linezolid resistance in the absence of 23S rRNA mutations in methicillin-resistant Staphylococcus aureus. Washington, D.C. Abstracts of the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy; 2006. American Society for Microbiology, Abstract C2–1147. [Google Scholar]
- Ausebel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley/Greene Publishing; 1987. [Google Scholar]
- Balaban N, Novick RP. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdogan B, Appelbaum PC. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int J Antimicrob Agents. 2004;23:113–119. doi: 10.1016/j.ijantimicag.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Bozdogan B, Bogdanovich T, Kosowska K, Jacobs MR, Appelbaum PC. Macrolide resistance in Streptococcus pneumoniae: clonality and mechanisms of resistance in 24 countries. Curr Drug Targets Infect Disord. 2004;4:169–176. doi: 10.2174/1568005043340821. [DOI] [PubMed] [Google Scholar]
- Ceglowski P, Alonso JC. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the ORF eta-copS region. Gene. 1994;145:33–39. doi: 10.1016/0378-1119(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Thomasco LM, Gadwood RC, Lund ET, Cavey GS, Mathews WR, Adams LD, Cecil ET, Pearson JD, Bock JH, Mott JE, Shinabarger DL, Xiong L, Mankin AS. Crosslinking in the living cell locates the site of action of oxazolidinone antibiotics. J Biol Chem. 2003;278:21972–21979. doi: 10.1074/jbc.M302109200. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- Franceschi F, Kanyo Z, Sherer EC, Sutcliffe J. Macrolide resistance from the ribosome perspective. Curr Drug Targets Infect Disord. 2004;4:177–191. doi: 10.2174/1568005043340740. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Harms JM, Schlunzen F, Fucini P, Bartels H, Yonath A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004;2:4. doi: 10.1186/1741-7007-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiba H, Takiguchi R, Joho K, Aoyama K, Hirota T. Identification of the replication region of Streptococcus thermophilus No. 29 plasmid pST1. Biosci Biotechnol Biochem. 1993;57:1646–1649. doi: 10.1271/bbb.57.1646. [DOI] [PubMed] [Google Scholar]
- Hutchinson DK. Oxazolidinone antibacterial agents: a critical review. Curr Top Med Chem. 2003;3:1021–1042. doi: 10.2174/1568026033452195. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Ojo KK, Schwarz S. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J Antimicrob Chemother. 2004;54:936–939. doi: 10.1093/jac/dkh457. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother. 2006;50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Aarestrup FM, Schwarz S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother. 2007;51:483–487. doi: 10.1128/AAC.01340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss P, Xiong L, Shinabarger DL, Mankin AS. Resistance mutations in 23S rRNA identify the site of action of protein synthesis inhibitor, linezolid, in the ribosomal peptidyl transferase center. J Molec Biol. 1999;294:93–101. doi: 10.1006/jmbi.1999.3247. [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J Biol Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- Leach KL, Swaney SM, Colca JR, McDonald WG, Blinn JR, Thomasco LM, Gadwood RC, Shinabarger D, Xiong L, Mankin AS. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell. 2007;25 doi: 10.1016/j.molcel.2007.04.005. XXX-XXX. [DOI] [PubMed] [Google Scholar]
- Lee CY, Buranen SL, Ye ZH. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- Lin AH, Murray RW, Vidmar TJ, Marotti KR. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob Agents Chemother. 1997;41:2127–2131. doi: 10.1128/aac.41.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Douthwaite S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc Natl Acad Sci USA. 2002;99:14658–14663. doi: 10.1073/pnas.232580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann PA, Xiong L, Mankin AS, Chau AS, Mendrick CA, Najarian DJ, Cramer CA, Loebenberg D, Coates E, Murgolo NJ, Aarestrup FM, Goering RV, Black TA, Hare RS, McNicholas PM. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol Microbiol. 2001;41:1349–1356. doi: 10.1046/j.1365-2958.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- Meka VG, Gold HS. Antimicrobial resistance to linezolid. Clin Infect Dis. 2004;39:1010–1015. doi: 10.1086/423841. [DOI] [PubMed] [Google Scholar]
- Meka VG, Pillai SK, Sakoulas G, Wennersten C, Venkataraman L, DeGirolami PC, Eliopoulos GM, Moellering RC, Jr, Gold HS. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J Infect Dis. 2004;190:311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- Merryman C, Noller HF. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith CWJ, editor. RNA:Protein Interactions, A Practical Approach. Oxford: Oxford University Press; 1998. pp. 237–253. [Google Scholar]
- Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- Porse BT, Garrett RA. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23S rRNA and the synergism of their inhibitory mechanisms. J Molec Biol. 1999;286:375–387. doi: 10.1006/jmbi.1998.2509. [DOI] [PubMed] [Google Scholar]
- Poulsen SM, Karlsson M, Johansson LB, Vester B. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol Microbiol. 2001;41:1091–1099. doi: 10.1046/j.1365-2958.2001.02595.x. [DOI] [PubMed] [Google Scholar]
- Prystowsky J, Siddiqui F, Chosay J, Shinabarger DL, Millichap J, Peterson LR, Noskin GA. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob Agents Chemother. 2001;45:2154–2156. doi: 10.1128/AAC.45.7.2154-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J, Hidalgo M, Diaz L, Rincon S, Moreno J, Venegas N, Castañeda E, Arias CA. Characterization of macrolide resistance in Gram-positive cocci from Colombian hospitals: a countrywide surveillance. Int J Infect Dis. 2007 doi: 10.1016/j.ijid.2006.09.005. in press. [DOI] [PubMed] [Google Scholar]
- Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43 doi: 10.1128/aac.43.12.2823. 2823–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning; A Laboratory Manual. Cold Spring Harbor , NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sander P, Belova L, Kidan YG, Pfister P, Mankin AS, Bottger EC. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol Microbiol. 2002;46:1295–1304. doi: 10.1046/j.1365-2958.2002.03242.x. [DOI] [PubMed] [Google Scholar]
- Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. 2000;44:2530–2533. doi: 10.1128/aac.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shaw JH, Clewell DB. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinabarger D. Mechanism of action of the oxazolidinone antibacterial agents. Expert Opin Investig Drugs. 1999;8:1195–1202. doi: 10.1517/13543784.8.8.1195. [DOI] [PubMed] [Google Scholar]
- Skripkin E, McConnell TS, King B, Devito J, Franceschi F, Sutcliffe J. Designer oxazolidinones bind to the 50S peptidyl-transferase region and can overcome ribosome-based linezolid resistance. Abstracts of the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy; Washington, D.C. 2005. American Society for Microbiology, Abstract F-1255. [Google Scholar]
- Standards NCFCL. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6. Wayne, P.A: 2003. [Google Scholar]
- Treede I, Jakobsen L, Kirpekar F, Vester B, Weitnauer G, Bechthold A, Douthwaite S. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol Microbiol. 2003;49:309–318. doi: 10.1046/j.1365-2958.2003.03558.x. [DOI] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Wada A, Ohta H, Kulthanan K, Hiramatsu K. Molecular cloning and mapping of 16S-23S rRNA gene complexes of Staphylococcus aureus. J Bacteriol. 1993;175:7483–7487. doi: 10.1128/jb.175.22.7483-7487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob Agents Chemother. 2005;49:3554–3557. doi: 10.1128/AAC.49.8.3554-3557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Kloss P, Douthwaite S, Andersen NM, Swaney S, Shinabarger DL, Mankin AS. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J Bacteriol. 2000;182:5325–5331. doi: 10.1128/jb.182.19.5325-5331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura DG, Henner DJ. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]