Abstract
Purpose
Aberrant activation of the Notch signaling pathway is commonly observed in human pancreatic cancer, although the mechanisms for this activation have not been elucidated.
Experimental Design
A panel of 20 human pancreatic cancer cell lines was profiled for the expression of Notch pathway related ligands, receptors and target genes. Disruption of intracellular Notch signaling – either genetically by RNA interference targeting NOTCH1 or pharmacologically by means of the gamma secretase inhibitor GSI-18, was used for assessing requirement of Notch signaling in pancreatic cancer initiation and maintenance.
Results
Striking overexpression of Notch ligand transcripts was detectable in the vast majority of pancreatic cancer cell lines, most prominently, JAGGED2 (18/20 cases; 90%) and DLL4 (10/20 cases; 50%). In two cell lines, genomic amplification of the DLL3 locus was observed, mirrored by overexpression of DLL3 transcripts. In contrast, coding region mutations of NOTCH1 or NOTCH2 were not observed. Genetic and pharmacological inhibition of Notch signaling mitigated anchorage independent growth in pancreatic cancer cells, confirming that sustained Notch activation is a requirement for pancreatic cancer maintenance. Further, transient pre-treatment of pancreatic cancer cells with GSI-18 resulted in depletion in the proportion of tumor-initiating aldehyde dehydrogenase (ALDH)-expressing subpopulation, and was associated with inhibition of colony formation in vitro and xenograft engraftment in vivo, underscoring a requirement for the Notch-dependent ALDH-expressing cells in pancreatic cancer initiation.
Conclusions
Our studies confirm that Notch activation is almost always ligand-dependent in pancreatic cancer, and inhibition of Notch signaling is a promising therapeutic strategy in this malignancy.
Keywords: Pancreatic cancer, Notch, NOTCH-1, gamma secretase inhibitor, aldehyde dehydrogenase, tumor initiating cells
Statement of Translational Relevance
Potent therapeutic strategies are urgently needed for pancreatic cancer, a disease of near uniform lethality. Activation of the Notch signaling pathway is commonly observed in pancreatic cancer suggesting that pathway blockade with small molecule inhibitors might be a feasible therapeutic strategy. In this study, we first systematically document the mechanisms of Notch activation in pancreatic cancer, and demonstrate this to be ligand-driven rather than mutationally activated. Second, we demonstrate the requirement of sustained Notch signaling for pancreatic cancer maintenance using genetic and pharmacological approaches towards inhibition of the pathway. Finally, we confirm the presence of a highly Notch-dependent tumor-initiating population in pancreatic cancer that has been implicated as the putative source for disease recurrence and systemic metastases. Our studies underscore the emerging paradigm that cancers are heterogeneous populations comprised of tumor-initiating “stem cells” and the “bulk” tumor population, and therapeutic success will be engendered by dual targeting of both compartments.
Introduction
Pancreatic cancer is an almost uniformly lethal disease with an overall five-year survival of approximately 5%, and this dire prognosis has not markedly improved over the last few decades (1). In the United States, approximately 34,000 individuals succumb to this malignancy each year. To date, the only potentially curative therapeutic option is complete surgical resection, but unfortunately, the majority of patients are diagnosed at a locally advanced or distant metastatic stage, thus precluding surgical cure (2). Currently available treatment options for advanced pancreatic cancer, such as gemcitabine, have had minimal impact in ameliorating survival. Identification of aberrant signalling pathways that can also form the substrate for targeted therapies has thus become an area of foremost priority.
The re-activation of embryonic signal transduction pathways such as Notch and Hedgehog have been reported in a variety of human cancers (3, 4); further, the availability of potent small molecule inhibitors has meant that these pathways can be targeted in these cancers, as we and others have recently shown (5–7). The Notch signaling pathway is an evolutionarily conserved pathway that plays a major role in cell fate decisions in various tissues during the development of multicellular organisms (8). In adult tissues, Notch signaling prevents cells from undergoing terminal differentiation, thus maintaining pools of undifferentiated stem/progenitor cells (9, 10). Activation of the Notch signaling pathway has previously been described in several human malignancies, including pancreatic cancer (4, 11, 12). For example, our group has shown that expression of Notch gene targets is observed not only in invasive pancreatic cancers, but also in the non-invasive precursor lesions of this malignancy (13). In a series of elegant studies, Sarkar and colleagues have demonstrated a requirement for active Notch signalling for tumor maintenance in pancreatic cancer, with downregulation of Notch-1 contributing to growth inhibition and apoptosis of cancer cells through inhibition of key survival pathways like nuclear factor kappa B (NFκB) (14–17). However, the underlying mechanisms causing aberrant Notch signaling in pancreatic cancer are poorly understood.
In the present study we examine the mechanisms of Notch pathway activation in the setting of pancreatic cancer. We find that endogenous overexpression of Notch ligands, specifically JAGGED2 and DLL4, appears to be the most common mechanism; uncommonly, genomic amplification of the DLL3 locus on chromosome 19q13 contributes to Notch activation in this malignancy. In contrast to hematological malignancies like T-cell leukemia (18), mutational activation of Notch is rare to absent in pancreatic cancer. Our studies also demonstrate that sustained Notch signalling is required for the viability of a subpopulation of pancreatic cancer cells with tumor initiation properties (i.e., “cancer stem cells”), further supporting the utility of targeting this pathway as a therapeutic strategy in this malignancy.
Materials and methods
Cell lines and culture conditions
Twenty pancreatic cancer cell lines (PANC-1, CAPAN-1, Colo-357, CFPAC, MIAPaCa-2, BxPC-3, AsPc-1, L3.6PL, PL-4, PL-5, PL-8, PL-9, PL-12, PL-13, XPA-1, XPA-3, XPA-4, Panc-8.13, Panc-3.27, and Panc-4.30) were grown as previously described (19). Immortalized non-malignant human pancreatic epithelial cells (hTERT-HPNE) were cultured as described elsewhere (20). The hTERT-HPNE cells were used for normalization of expression levels for Notch pathway components amongst the 20 cancer cell lines.
RNAi-mediated transcript knockdown
For knockdown of NOTCH1 transcripts, PANC-1 and CAPAN-1 cells were transiently transfected with gene specific or scrambled siRNA using Oligofectamine (Invitrogen) following the standard procedure recommended by the manufacturer. Efficacy of knockdown was confirmed by qRT-PCR, as described below. The sequences for the synthetic siRNAs against NOTCH1 (Dharmacon, Lafayette, CO, USA) have been previously described (21). Similarly, RNAi against DLL3 was performed in PANC-1 and SU86.86 cell lines using SMARTPool™ siRNA (Dharmacon), followed by qRT-PCR to confirm efficacy of DLL3 knockdown.
Stable overexpression of NICD in PANC-1 cells
Generation of PANC-1 cells stably overexpressing the Notch-1 intracytoplasmic domain (N1ICD) was accomplished as previously described (21). Empty vector was used for mock transfection.
Notch pathway inhibitor GSI-18
Synthesis of the gamma-secretase inhibitor [11-endo]-N-(5,6,7,8,9,10-hexahydro-6,9-methanobenzo[a][8]annulen-11-yl)-thiophene-2-sulfonamide (a.k.a. GSI-18) and its ability to block Notch pathway activity in cancer cells have been previously described (21–23).
Notch reporter assays
Assessment of Notch activity following GSI-18 administration was performed using a CBF-1 binding site luciferase reporter (8X-Luc), as previously described, in PANC-1 cells (13). Renilla luciferase was used as transfection control.
Cell viability assay
Growth inhibition was measured using the CellTiter 96® Aqueous Cell Proliferation Assay (Promega, Madison, WI, USA), which relies on the conversion of a tetrazolium compound (MTS) to a colored formazan product by the activity of living cells. Briefly, 2000 cells/well were plated in 96 well plates, and were treated with 2, 5 and 10 µM concentrations of GSI-18, for 96 hours, at which point the assay was terminated, and relative growth inhibition compared to vehicle-treated cells measured using the CellTiter 96® reagent, as described in the manufacturer's protocol. A panel of six human pancreatic cancer cell lines were examined (PANC-1, CAPAN-1, BxPC-3, MIAPaca-2, PANC-8.13, PANC-3.27) in the MTS assays. Cell viability assays were also performed for PANC-1 and SU86.86 cells following RNAi against DLL3, using scrambled siRNA as control. All experiments were set up in triplicates to determine means and standard deviations.
Anchorage independent growth
Anchorage independent growth was assessed in PANC-1 and CAPAN-1 cells following either genetic inhibition (NOTCH1 siRNA) or with pharmacological inhibition of Notch signaling with GSI-18 (5 µM). Soft agar assays were set up in 6-well plates, each well containing a bottom layer of 1% agarose (Invitrogen), a middle layer of 0.6% agarose including 10,000 cells, and a top layer of medium only. For the pharmacological inhibition experiments, mixtures in each well were supplemented with GSI-18 at the respective concentration or solvent only, and the plates were incubated for three weeks. An independent series of colony assays was performed in PANC-1 and SU86.86 cells, following genetic knockdown of DLL3 using siRNA. To assess colony formation, the medium was removed, and 1.5 ml of 0.5% Wright’s staining solution was added to each well. After incubation at 4 °C for 12 h, removal of the staining solution and washing twice with PBS, colonies were visualized by trans-UV illumination and counted using the analysis software Quantity One (BioRad, Hercules, California, USA).
Evaluation of aldehyde dehydrogenase (ALDH) activity
ALDH expression was determined at baseline and after pharmacological Notch inhibition in two pancreatic cancer cell lines, E3LZ10.7 and CAPAN-1, where we have previously demonstrated that inhibition of Hedgehog signaling selectively depletes the ALDH “bright” subpopulation (5, 6). After incubation with either vehicle or GSI-18 (5µM) for 24 hours, E3LZ10.7 and CAPAN-1 cells were stained for ALDH expression using the Aldefluor reagent (StemCell Technologies, Vancouver, Canada) according to the manufacturer’s instructions and analyzed on a FACSCalibur flow cytometer (Becton Dickinson). ALDH positive cells were quantified by calculating the percentage of total cells that displayed greater fluorescence compared to a control staining reaction containing the ALDH inhibitor diethylamino-benzaldehyde (DEAB).
Pre-treatment with GSI-18
We have previously shown that transient ex vivo pre-treatment with Hedgehog antagonists inhibits both anchorage independent growth and in vivo tumorigenicity of pancreatic cancer and glioblastoma cell lines (6, 24). In order to determine the effects of Notch antagonism on tumor initiation, pancreatic cancer cell lines E3LZ10.7 and CAPAN-1 cells were pre-treated with either vehicle or GSI-18 for 24 hours (2 and 5 µM), and allowed to recover in full serum for 24 hours. Thereafter, equal numbers of viable cells from each condition, quantified using trypan-blue dye exclusion assay, were plated in soft agar for colony assays, as described above. Pre-treated and serum-recovered E3LZ10.7 and PANC-1 cells were also injected in athymic (nude) mice for tumor engraftment studies, as described below.
Colony assays with LY294002, an Akt/PI-3-kinase pathway inhibito
In order to confirm the specificity of Notch inhibition against the tumor initiating component and exclude the potential for artefact, we performed a series of experiments using CAPAN-1 cells treated with LY294002, a small molecule inhibitor of the oncogenic Akt/PI-3-kinase pathway. Two parallel sets of anchorage independent assays were performed: first, a “pre-treatment” experiment mirroring the GSI-18 study, with two doses of LY294002 (5 and 10 µM). In this experiment, CAPAN-1 cells were exposed to the drug for 24 hours, followed by full serum recovery and plating in soft agar. The second set of experiments, with the same dosages, utilized a “continuous” (conventional) approach, where the cells were incubated in soft agar with continuous exposure to LY294002 for two weeks. Colony counts were performed as described above.
Generation of murine subcutaneous xenografts
All animal experiments conformed to the guidelines of the Animal Care and Use Committee of Johns Hopkins University and animals were maintained in accordance to guidelines of the American Association of Laboratory Animal Care. A total of 5×106 E3LZ10.7 or PANC-1 cells in a volume of 200 µl of 1/1 (v/v) PBS/matrigel, pre-treated with either vehicle or with GSI-18 at 5µM, and allowed to recover in full serum for 24 hours, were injected subcutaneously into male CD1 nu/nu athymic mice (Charles River). Tumor volumes (V) were determined after measuring the larger (a) and smaller (b) diameters as , as previously described (5, 6).
Statistical analysis
Kruskal-Wallis analysis was performed using SPSS version 15.0.1 for Microsoft Windows, two-tailed t-test, one way ANOVA and linear regression analysis (Pearson’s test) were performed using GraphPad Prism for Windows version 5. P<0.05 was regarded as statistically significant. Results in bar diagrams are plotted as means and standard deviations if not otherwise indicated.
Results
Endogenous overexpression of Notch ligands in pancreatic cancer
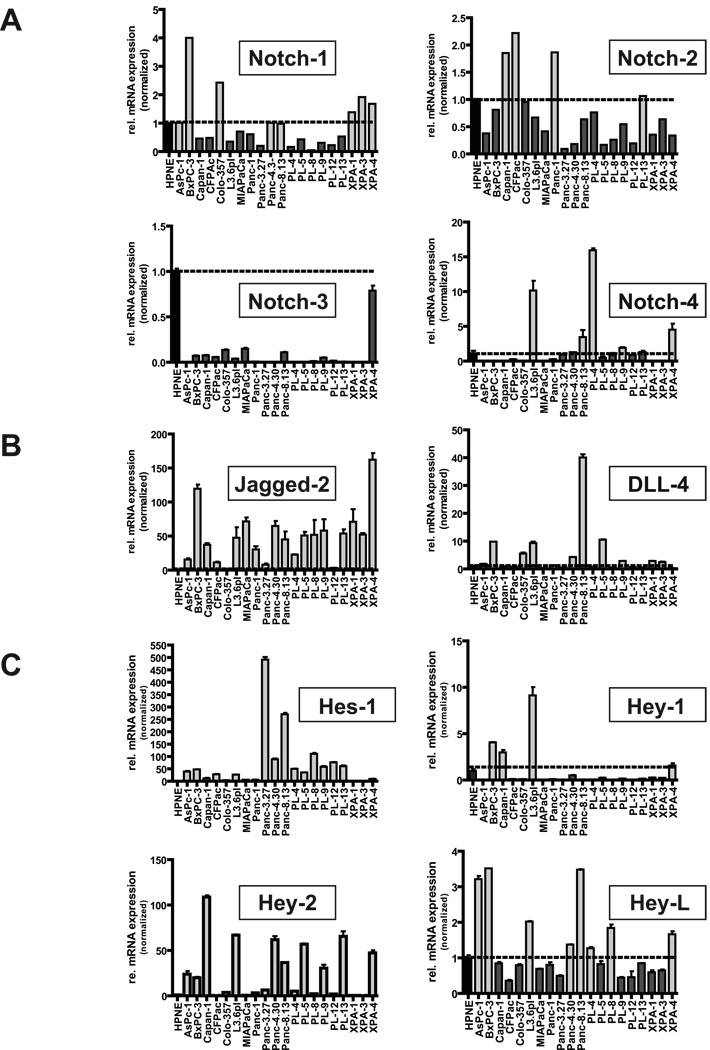
Quantitative real-time qRT-PCR analysis of 20 human pancreatic cancer cell lines compared with hTERT-HPNE cells confirmed variable expression of NOTCH1 through 4 transcripts, with most cell lines not demonstrating any evidence of receptor mRNA overexpression. Thus, compared to hTERT-HPNE cells, only 8 of 20 (40%) pancreatic cancer lines had equal or greater expression of NOTCH1, 5 of 20 (25%) had equal or greater expression of NOTCH4, and 4 of 20 (20%) had equal or greater expression of NOTCH2 transcripts, respectively (Figure 1A). Curiously, NOTCH3 mRNA expression was lower than hTERT-HPNE cells in all 20 cancer cell lines. In contrast to receptor mRNA levels, marked upregulation of two of four Notch pathway ligand transcripts (specifically, JAGGED2 and DLL4) were observed in the majority of pancreatic cancer cell lines. This was particularly striking for JAGGED2 where 18 of 20 (90%) of cell lines had higher transcript levels than observed in hTERT-HPNE (with the majority of cases at >50-fold elevation), and to a lesser extent with DLL4, with 10 of 20 lines (50%) demonstrating mRNA overexpression compared to hTERT-HPNE cells (Figure 1B). JAGGED1 and DLL1 transcripts were expressed at more attenuated levels (no higher than 10-fold relative overexpression compared to hTERT-HPNE cells), and were upregulated in fewer cell lines within the panel (Supplementary Figure 1). Consistent with Notch pathway activation, striking overexpression of the Notch target genes HES1 and HEY2 (HERP1) was seen in 16 of 20 (80%) and 13 of 20 (65%) of PC cell lines, respectively (Figure 1C). In contrast, overexpression of the remaining Notch gene targets, HEY1 (HERP2) and HEYL was observed in only a minority of cancer cell lines when compared with corresponding transcript levels in hTERT-HPNE cells. Comparable expression results were obtained when GUSB was used as housekeeping control instead of PGK1 (data not shown).Upon correlating Notch ligand levels with that of target genes, JAGGED2 mRNA expression was most closely and significantly correlated with that of HES1 transcripts (P = 0.045, Pearson correlation), further underscoring the importance of this basic helix-loop-helix (bHLH) transcription factor in the context of pancreatic neoplasia (13, 25). On the contrary, there was highly significant correlation between the patterns of expression of DLL4 and the Notch target gene HEYL in pancreatic cancer cell lines (P = 0.003, Pearson correlation), reiterating previous observations that despite the commonalities within the pathway, individual ligands have disparate effects on target genes (21).
Figure 1. Profiling the Notch pathway in pancreatic cancer cell lines.
RNA from a panel of 20 pancreatic cancer cell lines was assessed for expression of Notch receptors NOTCH1,NOTCH2, NOTCH3 and NOTCH4 (panel A), Notch ligands JAGGED2 and DLL4 (panel B) and Notch gene targets HES1, HEY1, HEY2 and HEYL (panel C), and relative fold levels compared to immortalized hTERT-HPNE cells. Horizontal line indicates normalized ratio of 1 in hTERT-HPNE cells. X-axis corresponds to individual cell line samples, and Y-axis to relative fold level of expression. Light grey bars indicate cancer cell lines with overexpression of corresponding mRNA compared to hTERT-HPNE cells, while dark grey bars indicate cell lines with lesser expression. All assays were performed in triplicate, using PGK1 as housekeeping control, and an independent set of assays was performed using GUSB as housekeeping control (data not shown).
Amplification of DLL3 is an uncommon “driver” for Notch signaling in pancreatic cancer
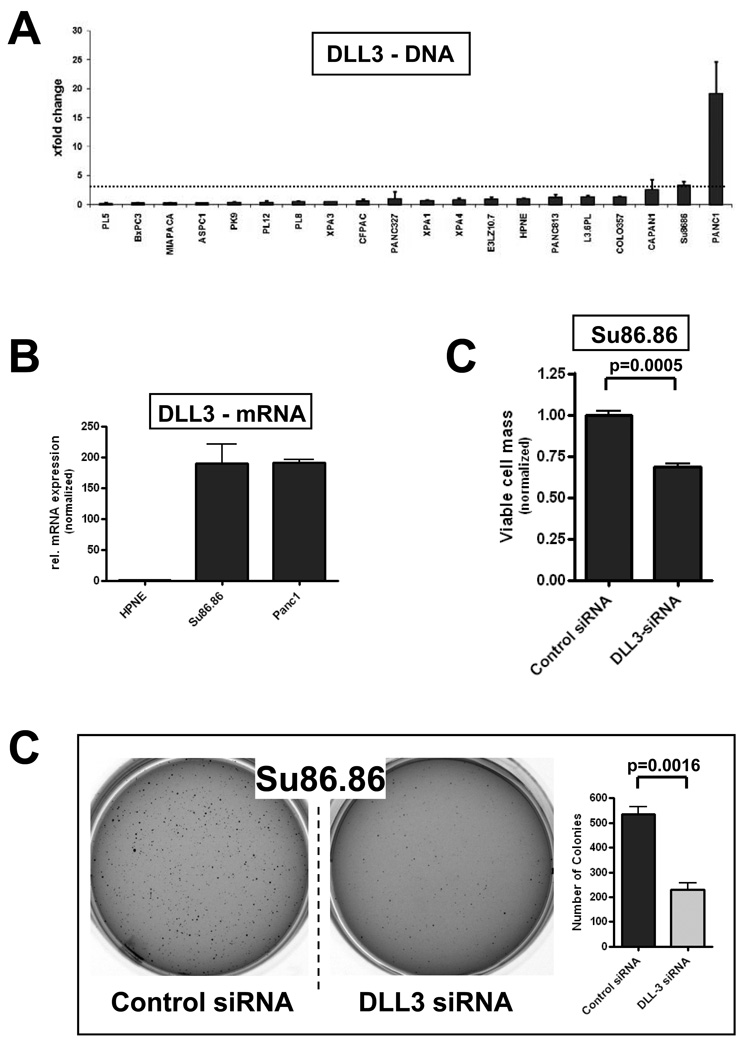
Previously published genomic copy number analyses of pancreatic cancer cell lines and xenografts by our group and others have shown that the DLL3 locus on chromosome 19q13 is included in a recurrent amplicon in this malignancy (19, 26, 27). Therefore, we assessed DLL3 gene dosage in a panel of 20 cell lines, and found two lines – PANC-1 and SU86.86 that demonstrated 3-fold or greater copy number by genomic quantitative PCR (qPCR), compared to hTERT-HPNE cells (Figure 2A). Transcript profiling confirmed that PANC-1 andSU86.86 had strikingly high expression of DLL3 mRNA, ~200-fold that of hTERT-HPNE cells (Figure 2B). DLL3 was downregulated by transient RNA interference (RNAi) in both cells lines, and effects on in vitro growth and anchorage independence were determined following validation of gene-specific knockdown. No significant effects were observed on either phenotype in PANC-1 cells with DLL3 RNAi (data not shown), suggesting redundant mechanisms for Notch pathway activation in this cell line. In contrast, knockdown of DLL3 in SU86.86 resulted in significant growth inhibition by MTS assay (Figure 2C, P=0.0005), as well as significant inhibition of anchorage independent growth in soft agar (Figure 2D, P=0.0016). Thus, in a minor subset of pancreatic cancers, Notch pathway activation is likely to be driven by increased DLL3 copy number and resulting endogenous overexpression of the ligand protein.
Figure 2. Copy number alteration of DLL3 in a subset of pancreatic cancer cell lines.
(A) Genomic qPCR for DLL3 copy number demonstrates two cell lines (PANC-1 and SU86.86) with an average gene dosage ratio of greater than three. The chromosome 19q13 gene KCL3 was used as a reference control. Genomic qPCR was performed in triplicate, and average ± SD calculated, with the latter expressed as error bars.
(B) Quantitative reverse transcription PCR (qRT-PCR) for DLL3 mRNA in PANC-1 and SU86.86 cells demonstrates striking overexpression (~200-fold), relative to levels in hTERT-HPNE cells.
(C) Knockdown of DLL3 by synthetic small interfering RNA (siRNA) significantly inhibits in vitro growth of SU86.86 cells, as measured by an MTS cell viability assay at 96 hours, compared to control (scrambled siRNA transfected) cells; (P =0.0005).
(D) Knockdown of DLL3 by siRNA significantly inhibits anchorage independent growth in SU86.86 cells, as assessed by colony formation in soft agar, compared to control (scrambled siRNA transfected) cells; (P =0.0016). Colony assays were performed in triplicate, and average ± SD calculated, with the latter expressed as error bars.
NOTCH1 or NOTCH2 mutations are rare to absent in pancreatic cancer
Activating mutations of the NOTCH receptors have been suggested to be the underlying driving force of Notch pathway activation in several malignancies, particularly in T-cell leukemias, wherein activating NOTCH1 mutations are found in as many as 50% of cases (18). To determine whether such coding sequence mutations of NOTCH1 or NOTCH2 exist in the setting of pancreatic cancer, mutational analysis of 20 pancreatic cancer cell lines, as well as 22 patient-derived pancreatic cancer xenografts, was performed by direct Sanger sequencing of the coding regions. All sequence variations from RefSeq (http://www.ncbl.nlm.nih.gov/RefSeq) were first confirmed by replicate PCR, and subsequently cross-matched against the single nucleotide polymorphism database dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP). A previously undescribed heterozygous L2458V alteration was identified in the C-terminal PEST domain of NOTCH1 in the MIAPACA-2 pancreatic cancer cell line (data not shown). However, gauged by the low expression levels of Notch pathway target genes in this line (see Figure 1C), the functional significance of this alteration was uncertain. We failed to find any evidence of activating mutations in any of the 42 cancer samples within the NOTCH1 and NOTCH2 coding regions.
Sustained Notch signaling is required for pancreatic cancer maintenance
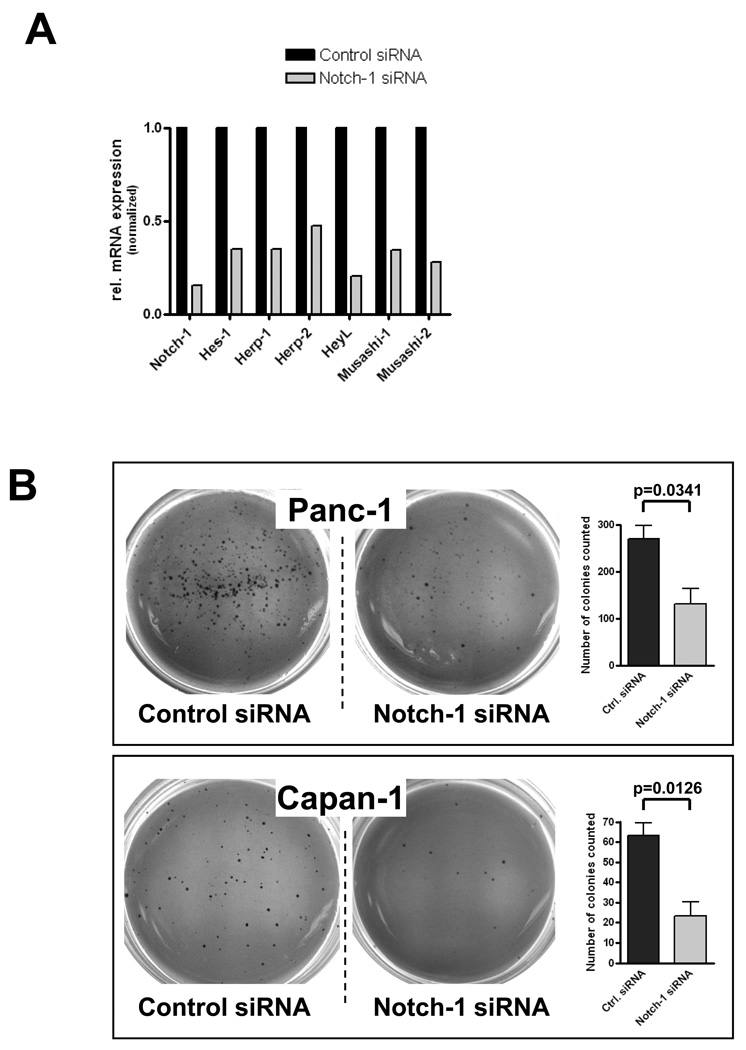
In light of the evidence suggesting ligand-dependent Notch activation in the majority of human pancreatic cancer cell lines, we then evaluated whether sustained Notch signaling is required for the maintenance of pancreatic cancer, and in particular, for anchorage independent growth, a property of transformed cells. We first used RNAi to downregulate NOTCH1 transcript levels in PANC-1 and CAPAN-1 cancer cell lines; efficacy of RNAi was confirmed by real-time PCR demonstrating downregulation of NOTCH1 transcripts, as well as multiple Notch target genes (Figure 3A). Both cell lines transfected with NOTCH1 siRNA demonstrated a significant reduction in the number of colonies formed in soft agar compared to scrambled siRNA transfected controls, confirming a requirement of active Notch signaling for anchorage independent growth (P=0.0341 and P=0.0126, respectively; Figure 3B).
Figure 3. Genetic knockdown of NOTCH1 function in pancreatic cancer cells inhibits anchorage independent growth.
(A) NOTCH1 RNAi in CAPAN-1 cells leads to >80% downregulation of gene specific transcript levels compared to scrambled siRNA transfected control cells. In addition, efficacy of functional knockdown is confirmed by reduced transcript levels for Notch gene targets, including HES1, HEY1 [HERP2], HEY2 [HERP1], HEYL, MUSASHI1 and MUSASHI2, compared to scrambled siRNA transfected control cells.
(B) NOTCH1 siRNA significantly inhibits anchorage independent growth in CAPAN-1 and PANC-1 cells, as assessed by colony formation in soft agar, compared to control (scrambled siRNA transfected) cells; (P <0.05). Colony assays were performed in triplicate, and average ± SD calculated, with the latter expressed as error bars.
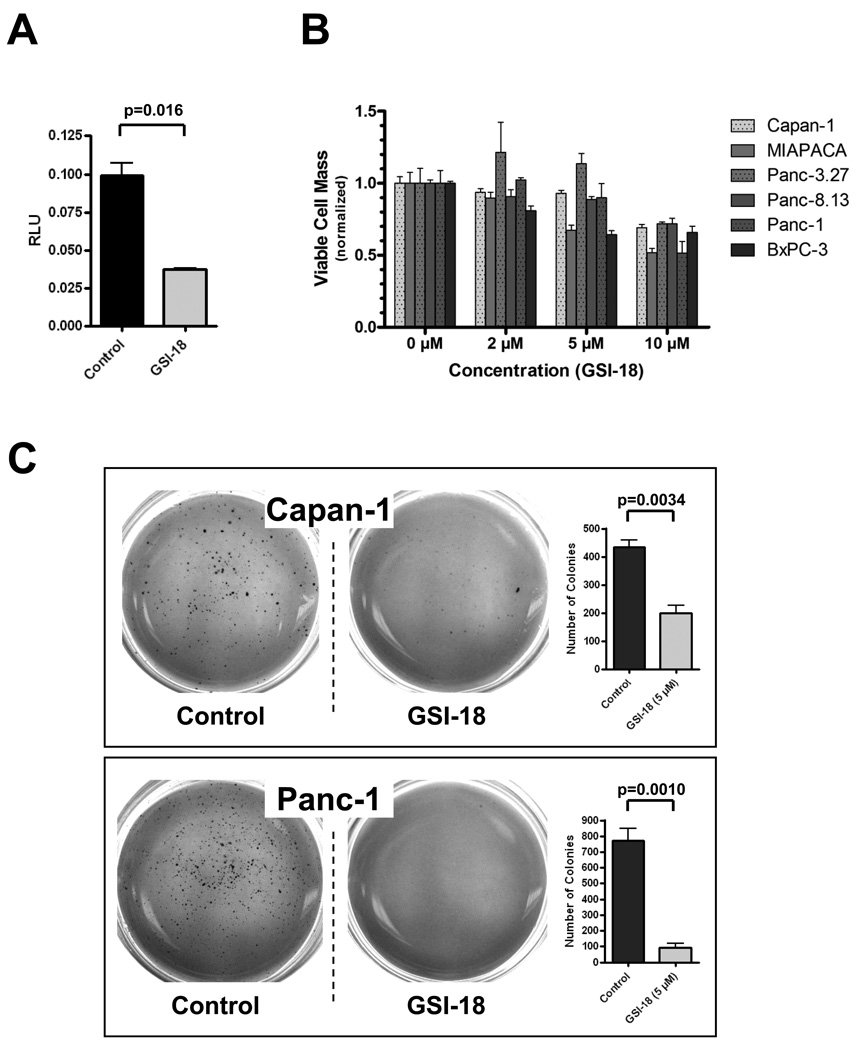
To complement the RNAi findings, we also studied the effects of pharmacological blockade of Notch signaling in pancreatic cancer cells on in vitro growth in monolayer and anchorage independent growth in soft agar. GSI-18 is a previously described gamma secretase inhibitor with potent inhibitory effects on Notch signaling (21–23). We first established that exposure of PANC-1 cells to GSI-18 leads to significant down-regulation of Notch activity, as observed using CBF1-binding site luciferase reporter assays (Figure 4A). A panel of six pancreatic cancer cell lines were used for in vitro growth (MTS) assays. As shown in Figure 4B, only modest growth inhibition was observed with GSI-18 at the highest dose (10 µM), and cell viability was largely unaffected at 2 and 5µM doses. In contrast, significant reduction in colony formation in soft agar was observed in both CAPAN-1 and PANC-1 cell lines when exposed to 5µM GSI-18 (P=0.0034 and P=0.0010, respectively; Figure 4C). Thus, based on the combined results of NOTCH1 RNAi and GSI-18 treatment, we conclude that continuous blockade of Notch signaling is deleterious for the anchorage independent growth of pancreatic cancer cells.
Figure 4. Pharmacological knockdown of Notch function in pancreatic cancer cells inhibits anchorage independent growth.
(A) Gamma secretase inhibitor GSI-18 (2µM) significantly downregulates CBF-1 binding site luciferase reporter activity (P=0.016) in PANC-1 cells, consistent with inhibition of intracellular Notch function. Control cells are treated with DMSO vehicle. Y-axis depicts relative luciferase activity (RLU).
(B) Modest dose dependent inhibition of in vitro cell growth (assessed by MTS cell viability assay at 96 hours) is observed in a panel of six pancreatic cancer cell lines (CAPAN-1, PANC-1, MIAPACA-2, BxPC-3, PANCA-8.13, and PANC-3.27) upon GSI-18 treatment. Three independent doses (2, 5, and 10µM) are used, and cell viability is normalized to DMSO vehicle treated cells (0µM column). All MTS assays are performed in triplicate, and average ± SD calculated, with the latter expressed as error bars.
(C) GSI-18 significantly inhibits anchorage independent growth in CAPAN-1 and PANC-1 cells, as assessed by colony formation in soft agar, compared to control (DMSO-treated) cells; (P <0.005). Colony assays were performed in triplicate, and average ± SD calculated, with the latter expressed as error bars.
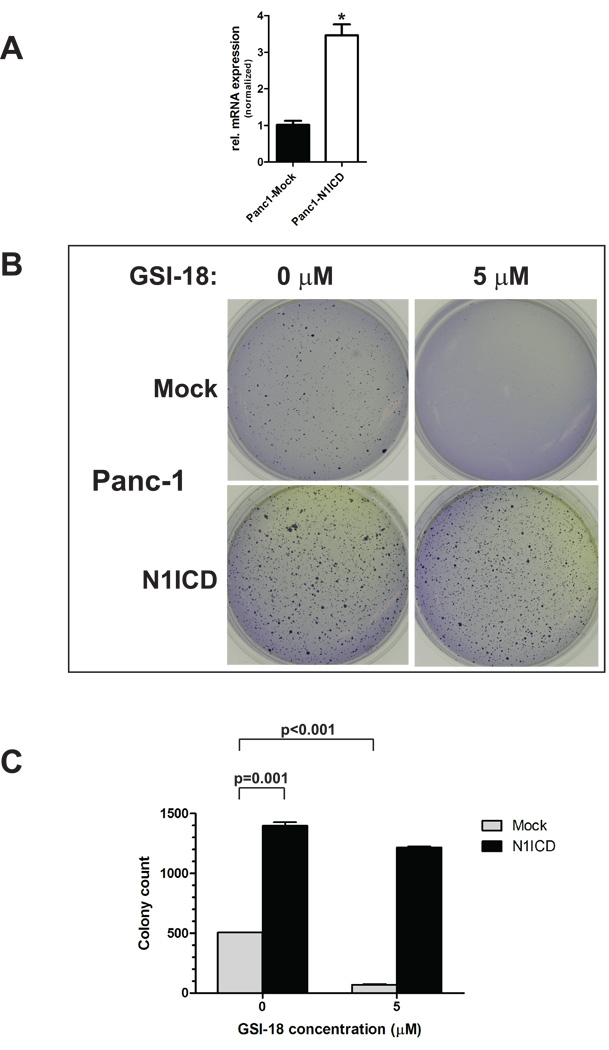
Overexpression of Notch 1 Intracellular Domain (N1ICD) rescues GSI-18-mediated inhibition of anchorage independent growth in PANC-1 cells
A mammalian expression vector encoding N1ICD was stably transfected in PANC-1 cells (‘PANC-1-N1ICD’), and overexpression of N1ICD as compared to empty vector transfected cells was confirmed by qRT-PCR (Figure 5A). Of note, enforced N1ICD expression per se markedly enhanced anchorage independent growth of PANC-1 cells in soft agar assays (P=0.001). Treatment with GSI-18 at a concentration of 5µM led to a more than seven-fold reduction in colony numbers in empty vector-transfected PANC-1 cells (P<0.001), while only minimal reduction in colony formation was observed in PANC-1-N1ICD cells (Figures 5B and C). Thus, enforced expression of N1ICD is able to rescue PANC-1 cells from the effects of GSI-18, underscoring the relative “on-target” effects of this small molecule inhibitor.
Figure 5. Enforced expression of Notch 1 intracytoplasmic domain (N1ICD) rescues anchorage independent growth phenotype of PANC-1 cells treated with GSI-18.
Enforced expression of N1ICD in PANC-1 cells (A) led to increased colony formation and anchorage independent growth in soft agar as compared to empty vector transfected cells. Treatment with GSI-18 (5 µM) significantly reduced colony formation of empty vector transfected cells, but no significant effects were observed in the N1ICD-overexpressing cells (B). Colony counts are provided in (C).
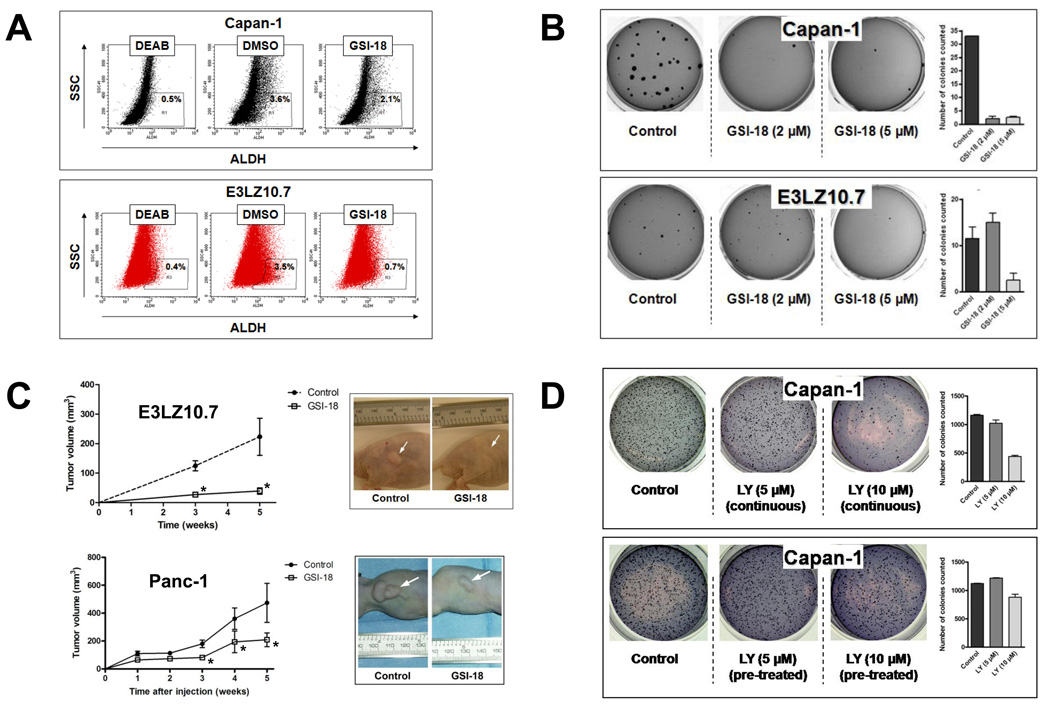
Transient Notch pathway inhibition eliminates a subpopulation of ALDH “bright” cells with tumor initiating properties in pancreatic cancer
Emerging lines of evidence in solid cancers suggest that a subpopulation of cells with tumor-initiating properties (so-called “cancer stem cells”) can be identified by elevated expression of the enzyme aldehyde dehydrogenase (ALDH) (5, 24, 28). We have recently identified ALDH “bright” cells in pancreatic cancer that are highly sensitive to Hedgehog pathway blockade with cyclopamine or related small molecule inhibitors (5, 6). We have also shown that selective elimination of these ALDH “bright” cells by transient pre-treatment with Hedgehog inhibitors inhibits subsequent tumor initiation (engraftment) in xenograft models (6). In order to determine whether this putative tumor initiating population is also Notch pathway dependent, we treated CAPAN-1 and E3LZ10.7 cells with GSI-18 in vitro for 24 hours. These two cell lines have been documented to have robust ALDH ‘bright” cells detectable by the Aldefluor assay (6). We observed a selective depletion of this subpopulation with transient GSI-18 exposure in both CAPAN-1 and E3LZ10.7 cells (Figure 6A). Upon subsequent plating in soft agar, these transiently pre-treated cells also demonstrated profound inhibition of anchorage independent growth (Figure 6B). Further, when equal numbers of viable E3LZ10.7 or PANC-1 cells, which had been transiently exposed to either GSI-18 or vehicle for 24 hours, respectively, were injected subcutaneously in athymic mice, a significant blockade of xenograft engraftment was observed in both sets of treated cell lines, during 5 weeks of follow up (Figure 6C). These findings underscore the importance of sustained Notch signaling in maintaining the viability of tumor-initiating ALDH “bright” cells in pancreatic cancer, and demonstrate that even transient exposure to Notch antagonists has deleterious effects on tumor engraftment in vivo.
Figure 6. Transient ex vivo exposure of pancreatic cancer cells to Notch inhibitors depletes ALDH “bright” cells and impedes tumor initiation in vivo.
(A) Transient incubation of CAPAN-1 and E3LZ10.7 cells with GSI-18 (5 µM) for 24 hours reproducibly diminished the fraction of ALDH “bright” cells as determined by the Aldefluor assay (3.6% to 2.1% for CAPAN-1 and 3.5% to 0.7% for E3LZ10.7). The ALDH inhibitor DEAB was used as negative control in the assay.
(B) CAPAN-1 and E3LZ10.7 cells were incubated with 2 and 5µM doses of GSI-18 for 24 hours, followed by full serum recovery for an additional 24 hours, and plating in soft agar for colony assays. No further GSI-18 exposure was administered to the plated cells. Compared to equal numbers of viable plated cells in the DMSO-treated group, reduction in colony formation is observed at 2 and 5µM doses for CAPAN-1 cells, and at 5µM dose for E3LZ10.7 cells.
(C) E3LZ10.7 or PANC-1 cells were incubated with GSI-18 at a concentration of 5 µM for 24 hours, followed by full serum recovery for an additional 24 hours, and injection of 5×106 cells in the subcutaneous milieu of athymic mice. No further in vivo treatment was performed. Compared to equal numbers of viable injected cells in the DMSO-treated group, significant reduction in size of the engrafted tumors is seen with transient GSI-18 exposure in both sets of cell lines, beginning at 3 weeks post-injection and persisting at 5 weeks (asterisks).
(D) In contrast to the phenotype observed with GSI-18 pre-treatment, no effects of transient exposure are seen with LY294002, a small molecule inhibitor of the Akt signaling pathway, in CAPAN-1 cells. Specifically, CAPAN-1 cells were exposed to two doses (5 and 10µM) of LY294002 in one of two modes: “continuous”, wherein cells were incubated with continuous exposure to the drug, as in a conventional colony assay, and “pre-treatment”, mimicking the transient pre-treatment exposure for 24 hours performed with GSI-18.
One potential pitfall of the “pre-treatment” strategy is the possibility that overall cellular function is sufficiently compromised by the transient exposure to GSI-18 that colony formation and engraftment in nude mice are inhibited, irrespective of any specific impact on the tumorigenic population of cells. To exclude this possibility, we performed a parallel series of colony assays in soft agar, wherein CAPAN-1 cells were either “pre-treated” transiently with LY294002, an antagonist of Akt/PI-3-kinase pathway, prior to plating, or exposed continuously to the drug in a more conventional colony assay format. In contrast to our observations with GSI-18, transient pre-treatment has no effect on anchorage independent growth, while the conventional colony assays demonstrate the expected reduction in colonies at 2 weeks (Figure 6D). This provides additional confirmation that the loss of tumorigenic phenotype observed with transient Notch inhibition is unlikely to be a non-specific deleterious effect on cellular function.
Discussion
The Notch signaling pathway plays a critical role in pancreatic development and in the homeostasis of mature pancreatic tissues (9, 29, 30). In the adult pancreas, we and others have shown that Notch activation is predominantly restricted to the centroacinar cells within the exocrine compartment (13, 31). It is believed that these cells represent a persistent pool of progenitor-type cells in the adult pancreas, and that the Notch pathway is a sine qua non for maintaining the undifferentiated state of these cells. An abnormal expansion of Notch-expressing cells is observed in states of exocrine injury, while abrogation of Notch signaling impairs subsequent epithelial regeneration, underscoring the importance of this pathway to tissue homeostasis in the pancreas (32–34). A role for aberrant Notch signaling in pancreatic cancer has emerged from studies conducted in human and animal models of this disease (13–17, 35). For example, the basic helix-loop-helix (bHLH) transcription factor Hes-1 is a prototypal Notch gene target (36), and Hes-1 upregulation is observed at the earliest, non-invasive stages of human and mouse pancreatic cancer (13, 35).
In mammalian cells, the canonical Notch pathway includes four distinct Notch receptors, NOTCH1-4. Previous reports have elucidated context-dependent and cancer-type specific effects of the Notch receptors on carcinogenesis. Thus, NOTCH1 is oncogenic in T-cell leukemia and in breast cancers (18, 37), while loss of NOTCH1 function promotes tumorigenesis in medulloblastoma and in skin cancers (21, 38). In medulloblastoma, by contrast, NOTCH2 appears to be the dominant oncogenic receptor (21). Sarkar et al have demonstrated the primacy of NOTCH1 as the Notch pathway receptor responsible for tumor maintenance in pancreatic cancer (14–17). Genetic or pharmacological inhibition of NOTCH1 activity in pancreatic cancer has profound deleterious effects on cell growth, cell survival, and invasion, through downregulation of critical signaling moieties like NFκB, vascular endothelial growth factor (VEGF) and matrix metalloproteinases (MMPs) (14–17). These existing reports provided the seedbed for our current study exploring the mechanisms of Notch activation in pancreatic cancer, and an assessment of the effects of Notch inhibition on the putative tumor-initiating compartment in this malignancy.
The Notch pathway is activated through somatic mutations of NOTCH1 in approximately 50% of T-cell leukemias (18), and in a minor subset (<5% by conservative estimates) of other solid cancers like breast, lung and colon cancer (39). The Catalog of Somatic Mutations in Cancer (COSMIC) database (http://www.sanger.ac.uk/genetics/CGP/cosmic/) provides an online compendium of these mutations. Sequencing the complete coding regions of NOTCH1 and NOTCH2 genes in 42 pancreatic cancer samples (20 cell lines and 22 xenografts) failed to elicit evidence of activating non-synonymous alterations. A single hemizygous L2458V PEST domain alteration was identified in the MIAPaCa-2 cell line; however, in the absence of functional correlates of pathway activation (as gauged by Notch reporter and target gene analysis), the significance of this change remains uncertain. Of note, a recent large scale sequencing effort of the pancreatic cancer genome also failed to identify somatic point mutations in NOTCH1 or NOTCH2, as well as within any of the genes encoding Notch ligands (40). These results reiterate that mutational activation of the Notch pathway in pancreatic cancer is rare.
In mammalian cells, at least five distinct ligands (JAGGED-1, JAGGED-2, DLL-1, DLL-3, and DLL-4) initiate Notch signaling upon binding to the cognate receptors. We found evidence for striking overexpression of Notch ligand transcripts, especially JAGGED-2 and DLL-4, in the majority of pancreatic cancer cell lines. Thus, as many as 18 of 20 (90%) of the cell lines examined in our panel demonstrated JAGGED-2 upregulation compared to hTERT-HPNE cells, with the majority having >50-fold relative expression levels. JAGGED-2 expression was mirrored by, and correlated with, mRNA expression of the Hairy enhance of split family of bHLH transcription factors recognized as Notch gene targets, underscoring the functional relevance of ligand-dependent activation. Ligand-dependent activation of embryonic signaling pathways is not unique to Notch, as we and others have described the existence of an analogous mechanism for both Hedgehog and wnt pathways, respectively, in pancreatic cancer (41, 42). In both instances, somatic mutations in downstream components (for example, GLI1 or CTNNB1) are rare to absent, accompanied by endogenous overexpression of stimulatory ligand. An enigmatic question pertains to the upstream genetic influence(s) promoting such profound Notch ligand expression in pancreatic cancer cells. In the context of Hedgehog signaling, we and others have shown that mutant Kras upregulates the transcription of endogenous Hedgehog ligand in pancreatic cancer cells (43, 44). Whether parallel mechanisms are in place for the Notch pathway remains a matter of investigation. In light of the prior observations by Miele and colleagues pertaining to the absolute requirement of Notch signaling for maintaining the neoplastic phenotype of human Ras-transformed cells (45), and the demonstration of Notch activation in pancreatic ductal lesions arising in Kras-driven genetically engineered mouse models of pancreatic cancer (35), the existence of such an axis is not beyond the realm of speculation. In passing, we should add that in a minority of instances, Notch activation appears to be a consequence of genomic copy number alterations at chromosome 19q13, the DLL3 gene locus (19, 26, 27). We have confirmed the existence of increased gene dosage, and associated DLL3 transcript overexpression, in two cell lines, and shown that knockdown of DLL3 by RNAi can inhibit anchorage independent growth in the SU86.86 cell line. Curiously, DLL3 RNAi in PANC-1 cells did not exhibit a discernible growth phenotype, suggesting that redundant ligand-driven activation can bypass the blockade of any one single Notch ligand, and underscores the need for targeting downstream elements of this pathway in cancer therapy.
In addition to exploring the mechanisms of Notch activation in pancreatic cancer, we also assessed the potential of Notch as a therapeutic target in pancreatic cancer, and in particular, whether Notch inhibition has a preferential deleterious effect on the tumor-initiating (“cancer stem cell”) compartment. In light of the prior series of studies by Sarkar and colleagues (14–17), our findings on Notch inhibition and pancreatic cancer maintenance are mainly confirmatory in nature. Nevertheless, these studies expand the repertoire of pancreatic cancer cell line models in which the anti-cancer effects of Notch inhibition, either by genetic or pharmacological means, are evident. Further, our findings confirm that NOTCH1 is possibly the dominant oncogenic Notch receptor in this malignancy, and that gamma secretase inhibitors like GSI-18, or other comparable small molecules (7, 46), warrant further preclinical evaluation in pancreatic cancer. In contrast to genistein and curcumin, two previously reported Notch inhibitors that are natural plant polyphenols (14, 16, 17), the synthetic gamma secretase inhibitors are likely to have a more limited repertoire of targeted intracellular effects. Gamma secretase inhibitors are currently in advanced phase clinical trials for Alzheimer disease, having demonstrated favorable toxicity profiles in healthy volunteers (47, 48), and therefore, the transition to being utilized as an anti-cancer agent may be an option in not too distant a future. Besides pancreatic cancer maintenance, however, a novel finding of our study has been the demonstration that even transient ex vivo pharmacological Notch inhibition depletes the putative tumor-initiating population in pancreatic cancer. We and others have recently identified ALDH “bright” cells detectable by Aldefluor assay as an enriched cancer stem cell compartment in a variety of solid cancers, including pancreatic cancer (5, 6, 24, 28). The ALDH “bright” cells are eliminated upon systemic Hedgehog inhibitor therapy, and correlate with abrogation of metastases in orthotopic xenograft models of pancreatic cancer (5, 6). Here we have demonstrated that transient ex vivo treatment with GSI-18 depletes the ALDH “bright” population in pancreatic cancer cell lines, and this is paralleled by a significant reduction in anchorage independent growth and xenograft engraftment in athymic mice. Due to limited drug availability, we were unable to perform systemic trials in orthotopic xenograft models, but our results lay the groundwork for such analyses in the future. The observation that the ALDH “bright” cells are both Hedgehog and Notch dependent for their viability suggests that combinatorial therapy with small molecule inhibitors against both pathways might have even more potent effects in vivo than single agent treatment. Further, our findings reiterate our previously stated postulate that effective therapy of pancreatic cancer will likely require targeting both the “bulk” tumor cells with a conventional anti-metabolite like gemcitabine, as well as a stem cell directed therapy like Notch or Hedgehog inhibitor to eliminate the cells responsible for metastases and disease recurrence.
In conclusion, we demonstrate that ligand-dependent activation of the Notch signaling pathway is common in pancreatic cancer. Pharmacological inhibition of Notch signaling is a valid therapeutic strategy in this malignancy, based on the requirement of sustained Notch activation for tumor initiation as well as for tumor maintenance of pancreatic cancer.
Supplementary Material
Acknowledgement
A.M. was supported by the Sol Goldman Pancreatic Cancer Research Center, the Michael Rolfe Foundation, NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer P50CA62924 and NIH R01CA113669. G. F. was supported by a fellowship grant within the postdoctoral program of the German Academic Exchange Service (DAAD). C.G.E. was supported by R01NS055089. X.F. was supported by the American Brain Tumor Association (ABTA) and the Accelerate Brain Cancer Cure (ABC2) foundation. J-B.K. was supported by the Netherlands Cancer Research Foundation and the International Exchange Program grant provided by the University Medical Center Utrecht.
references
- 1.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher FC, Fennelly D, Rafferty M. Common critical pathways in embryogenesis and cancer. Acta Oncol. 2006;45:375–388. doi: 10.1080/02841860600602946. [DOI] [PubMed] [Google Scholar]
- 4.Ball DW, Leach SD. Notch in malignancy. Cancer Treat Res. 2003;115:95–121. doi: 10.1007/0-306-48158-8_4. [DOI] [PubMed] [Google Scholar]
- 5.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer research. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldmann G, Fendrich V, McGovern K. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725–2735. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science (New York, NY. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 9.Leach SD. Epithelial differentiation in pancreatic development and neoplasia: new niches for nestin and Notch. J Clin Gastroenterol. 2005;39:S78–S82. doi: 10.1097/01.mcg.0000155547.83901.a3. [DOI] [PubMed] [Google Scholar]
- 10.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 12.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto Y, Maitra A, Ghosh B, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer research. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Inhibition of nuclear factor kappab activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer. 2006;118:1930–1936. doi: 10.1002/ijc.21589. [DOI] [PubMed] [Google Scholar]
- 18.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (New York, NY. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 19.Calhoun ES, Hucl T, Gallmeier E, et al. Identifying Allelic Loss and Homozygous Deletions in Pancreatic Cancer without Matched Normals Using High-Density Single-Nucleotide Polymorphism Arrays. Cancer research. 2006;66:7920–7928. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 20.Lawson T, Ouellette M, Kolar C, Hollingsworth M. Culture and immortalization of pancreatic ductal epithelial cells. Methods Mol Med. 2005;103:113–122. doi: 10.1385/1-59259-780-7:113. [DOI] [PubMed] [Google Scholar]
- 21.Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer research. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer research. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 23.Lewis SJ, Smith AL, Neduvelil JG, et al. A novel series of potent gamma-secretase inhibitors based on a benzobicyclo[4.2.1]nonane core. Bioorg Med Chem Lett. 2005;15:373–378. doi: 10.1016/j.bmcl.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 24.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Nowak NJ, Gaile D, Conroy JM, et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2005;161:36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Bashyam MD, Bair R, Kim YH, et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakhai H, Siveke JT, Klein B, et al. Conditional ablation of Notch signaling in pancreatic development. Development. 2008 doi: 10.1242/dev.013722. [DOI] [PubMed] [Google Scholar]
- 31.Stanger BZ, Dor Y. Dissecting the cellular origins of pancreatic cancer. Cell cycle (Georgetown, Tex. 2006;5:43–46. doi: 10.4161/cc.5.1.2291. [DOI] [PubMed] [Google Scholar]
- 32.Jensen JN, Cameron E, Garay MV, Starkey TW, Gianani R, Jensen J. Recapitulation of elements of embryonic development in adult mouse pancreatic regeneration. Gastroenterology. 2005;128:728–741. doi: 10.1053/j.gastro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Su Y, Buchler P, Gazdhar A, et al. Pancreatic regeneration in chronic pancreatitis requires activation of the notch signaling pathway. J Gastrointest Surg. 2006;10:1230–1241. doi: 10.1016/j.gassur.2006.08.017. discussion 42. [DOI] [PubMed] [Google Scholar]
- 34.Siveke JT, Lubeseder-Martellato C, Lee M, et al. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134:544–555. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 36.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 37.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer research. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas M, Wolfer A, Raj K, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, Jeong EG, Yoo NJ, Lee SH. Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias. Apmis. 2007;115:1357–1363. doi: 10.1111/j.1600-0463.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones S, Zhang X, Parsons DW, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science (New York, NY. 2008 doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 42.Pasca di Magliano M, Biankin AV, Heiser PW, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS ONE. 2007;2:e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldmann G, Habbe N, Dhara S, et al. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008 doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. The Journal of biological chemistry. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 45.Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature medicine. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 46.Best JD, Smith DW, Reilly MA, et al. The novel gamma secretase inhibitor N-[cis-4-[(4-chlorophenyl)sulfonyl]-4-(2,5-difluorophenyl)cyclohexyl]-1,1, 1-trifluoromethanesulfonamide (MRK-560) reduces amyloid plaque deposition without evidence of notch-related pathology in the Tg2576 mouse. J Pharmacol Exp Ther. 2007;320:552–558. doi: 10.1124/jpet.106.114330. [DOI] [PubMed] [Google Scholar]
- 47.Siemers ER, Dean RA, Friedrich S, et al. Safety, tolerability, and effects on plasma and cerebrospinal fluid amyloid-beta after inhibition of gamma-secretase. Clin Neuropharmacol. 2007;30:317–325. doi: 10.1097/WNF.0b013e31805b7660. [DOI] [PubMed] [Google Scholar]
- 48.Siemers E, Skinner M, Dean RA, et al. Safety, tolerability, and changes in amyloid beta concentrations after administration of a gamma-secretase inhibitor in volunteers. Clin Neuropharmacol. 2005;28:126–132. doi: 10.1097/01.wnf.0000167360.27670.29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.