Abstract
The hypothesis-driven fMRI data analysis methods, represented by the conventional general linear model (GLM), have a strictly defined statistical framework for assessing regionally specific activations but require prior brain response modeling which is hard to be accurate. On the contrary, exploratory methods, like the support vector machine, are independent of prior hemodynamic response function (HRF), but generally lack a statistical inference framework. To take the advantages of both kinds of methods, this paper presents a composite approach through combining conventional GLM with SVM. This hybrid SVM-GLM concept is to use the power of SVM to obtain a data-derived reference function and enter it into the conventional GLM for statistical inference. A strategy is also presented to extract the temporal profile from the SVM classifier to be used as the data-derived regressor in SVM-GLM. In simulations with synthetic fMRI data, SVM-GLM demonstrated a better sensitivity and specificity performance for detecting the synthetic activations, as compared to the conventional GLM. With real fMRI data, SVM-GLM showed better sensitivity than regular GLM for detecting the sensorimotor activations.
Keywords: support vector machine, general linear model, spatiotemporal processing, fMRI, group analysis, random effect analysis
Introduction
Functional MRI (fMRI) data analysis methods can be roughly divided into two main categories: the hypothesis-driven methods and the exploratory methods. The first type of methods, represented by the univariate general linear model (GLM) based method (Bandettini et al., 1993; Friston et al., 1995a,b; Worsley and Friston, 1995), gains their popularity due to the easiness of result interpretation and function localization. However, they have been criticized for using a canonical hemodynamic response function (HRF) while the actual shape of HRF may differ significantly in different populations and may differ markedly from subject to subject (Aguirre et al., 1998). The second type of fMRI data analysis methods are HRF model-free and data driven (or semi-data driven), therefore are more flexible than hypothesis-driven approaches for analyzing fMRI data with various experimental designs or even resting fMRI data. However, by extracting the activation patterns as a whole, the exploratory methods lose the specificity of function localization. Without prior hypothesis, the activation patterns may also not lead to a meaningful neurophysiological interpretation. Moreover, a statistical framework for assessing the analysis results of the exploratory methods is generally not available. A combination of the hypothesis-driven approach and the exploratory approach could then provide a good solution to these issues of both types of methods via taking the advantages of both of them.
This paper presents a hybrid exploratory and hypothesis-driven fMRI data analysis method through combining conventional GLM with the support vector machine (SVM) (Vapnik, 1995; Burges, 1998). SVM is a machine learning-based auto-classification method which has been demonstrated to be useful for analyzing neuroimaging data in many applications (Cox and Savoy, 2003; Wang et al., 2003; Davatzikos et al., 2005; Mitchell et al., 2004; Wang et al., 2008b; Zhang et al., 2005b; Fan et al., 2007; LaConte et al., 2005; Mourão-Miranda et al., 2005; Wang et al., 2006, 2007a,b). The idea of a hybrid SVM-GLM is to use the power of SVM to obtain a data-derived reference function and enter it into the conventional GLM for statistical inference. As SVM has shown good promise for exploring the spatial brain discriminance patterns (SDP) between different populations or between different brain states (Cox and Savoy, 2003; Wang et al., 2003; Davatzikos et al., 2005; Mitchell et al., 2004; Wang et al., 2008b; Zhang et al., 2005b; Fan et al., 2007; LaConte et al., 2005; Mourão-Miranda et al., 2005; Wang et al., 2006, 2007a,b), it is also desirable to use it to assess the temporal fluctuations of brain activations. The temporal profile of SDP (SDPtp) itself reflects the task induced hemodynamic changes, and could then be imported into the standard GLM as a data-derived reference function. Mourão-Miranda et al. (Mourão-Miranda et al., 2006a) proposed a way to examine SDPtp by incorporating the temporal information into the SVM training process as done by Zhang et al. (Zhang et al., 2005a). By treating the images from each block repetition as a single 4D training sample, their method assumed that the temporal profile did not change across different repetitions and could then only assess the average temporal variation of SDP within the contrasted functional conditions. A more general approach is required to extract the entire SDPtp either for monitoring the entire temporal fluctuations of SDP or for the hybrid SVM-GLM.
A strategy to extract the entire SDPtp is presented in this paper. SDP was obtained through estimating the whole brain spatial discriminance map (SDM) (LaConte et al., 2005; Mourão-Miranda et al., 2005; Wang et al., 2007a) from the intrasubject SVM classifier as described in previous work (Wang et al., 2007a) (a detailed definition of SDP and SDM were given in Theory); SDPtp was then extracted via calculating the distance between SDM and an fMRI image at each time point. Providing a reference function for the hybrid SVM-GLM aside, SDPtp gives a way to assess how the spatial brain activity patterns vary along the time. Additionally a statistical inference for the entire SDP can be obtained by correlating SDPtp to the design paradigm. Both synthetic activation data and two fMRI data with a well-characterized sensorimotor task were used to evaluate the proposed SVM-GLM with a comparison to conventional GLM.
Theory
Spatial discriminating patterns
To be self-contained, a brief introduction to SDP extraction was given in this subsection, more details could be found in (LaConte et al., 2005; Mourão-Miranda et al., 2005; Wang et al., 2007a).
For SDP extraction, all the acquired fMRI data are included in the training process. The major steps are: 1) fMRI data preprocessing, 2) data restacking into a big data matrix with one volume per column and one voxel per row, 3) spatial dimension reduction and eigenvector-based data representation for the big data matrix using standard principal component analysis (PCA), 4) linear SVM classifier training using all acquired fMRI data projected into the eigenspace, 5) SDP extraction through projecting the normal weight of the separating hyperplane of the trained SVM classifier into the original image space.
Suppose we have an fMRI data matrix SN×L with one volume per column and one voxel per row. Here N and L are the number of intracranial voxels and the number of timepoints. Since N≫L, the maximum rank of S is L, which means S only has L nonzero eigenvalue associated principal vectors (eigenvectors) E = (e1, e2, ⋯ ,eL), ei = (ei,1, ei,2, ⋯ , ei,N)T (Vetterling and Flannery, 2002). Using standard principal component analysis, S can be loss-lessly compressed into a L × L matrix X = ETS, with one representation coefficient vector xi = (x 1,i, x2,i, ⋯, xL, i)T per column corresponding to one original image volume. The purpose of this preprocessing is to reduce the computational burden of SVM classification. And without any ambiguity, the real SVM classifier is trained with the eigenvectors based representation coefficient vector series X = (x1, x2,⋯ , xL).
For the classical 2-class (2 contrasting experimental conditions in fMRI) classification (Multi-class classification (representing multi-condition fMRI data classification) can be extended from the 2-class case (Burges, 1998)), SVM pursues an optimal hyperplane to separate the samples from different classes with a maximal margin between the two boundaries (Vapnik, 1995; Burges, 1998). With the training samples {xi, yi}, i = 1,⋯ ,L,yi ∈ {−1,+1} from different classes (−1 indicates condition A, +1 indicates condition B), a linear SVM is to seek the optimal separating hyperplane defined by w · x + b = 0 in the feature space ℝ L. Here, w is the weight vector normal to the hyperplane, and ‖w‖ is the Euclidean norm of w. b is the offset, and |b|/‖w‖ is the perpendicular distance from the hyperplane to the origin. A simple diagram of a linear SVM classifier for two dimensional data points was shown in Fig. 1A. The final solution of w represents the direction along which the training samples of both classes differ most. Projected back to the image space through Ew, we can then get the SDM. Without ambiguity, SDM in this paper referred to the map which is an image volume in the case of linear SVM-based classification; SDP referred to the patterns carried in SDM.
Figure 1.
SVM-based classification and temporal profile extraction. A) 2-dimensional data classification using a linear SVM, B) the temporal profile extracted from the distance of each sample to the separating plane. Green and red colors indicate the two different categories. The thick dot line represents the separating hyperplane, and the two dash dot lines indicate the boundaries with the maximal margin determined by a linear SVM. The circled symbols are the support vectors.
SDP temporal profile extraction
As shown in Fig. 1, a natural measure for the temporal fluctuations of SDP is the distance of each sample to the separating hyperplane. To indicate the side of the hyperplane which an image sample xi is on, we used the signed distance in this paper:
| (1) |
Fig. 1 gives a geometric illustration for the distance-based SDPtp extraction, where the distance of each sample point to the hyperplane is measured with the length of the perpendicular (red and green lines) from that point to the hyperplane. The sign of the distance is indicated by the color, which corresponds to the side of the hyperplane. As shown in Fig. 1B, the final SDPtp was obtained after arranging these signed distances according to the acquisition time of the corresponding samples.
Statistical inference on SDPtp
Since the experimental design information is used to label functional images before SVM classification, a statistical inference on SDPtp and SDM can be obtained through the correlation between SDPtp and the design paradigm. In another words, if the P value of this correlation is less than a certain threshold (like 0.05) then we can say that the extracted SDPtp is significantly correlated to the experiment design and the SDP (carried in SDM) are significantly related to the administered functional task.
A permutation testing as stated in (Mourão-Miranda et al., 2005) can be also conducted for an alternative statistical inference. By randomly shuffling the training samples and extracting SDPtp for each permutation, a probability P can be calculated as the proportion of the correlation coefficients of the pseudo SDPtps that are no less than the coefficient of the original SDPtp. If the P value is less than a certain threshold, e.g., P < 0.05, then there is a far from few chance that the null hypothesis is true that the observed correlation between the extracted SDPtp to the experimental paradigm is an effect due to the prior labeling of each fMRI image to be within different conditions.
The hybrid SVM-GLM
Without considering the temporal autocorrelation, a general linear model for fMRI data can be described as y = Gβ + e, where y is the observed time series, G is the regressor matrix, β is the fitting parameter vector, and e is the residual error vector. In standard GLM-based fMRI data analysis, the main regressor of interest in G is generated by convolving the experimental design function with a canonical HRF function. In SVM-GLM, this main reference function is replaced by SDPtp defined in Eq. 1 to add a semi-data driven property to the standard GLM. A parametric map can be then obtained by running SVM-GLM and collecting the fitting parameter value at each voxel.
Materials and methods
Imaging parameters
Imaging experiments were performed on a 3T Siemens Trio whole body MR scanner with a standard transmit/receive (Tx/Rx) head coil (Bruker BioSpin, USA). High resolution 3D T1-weighted anatomical images using the MPRAGE (TR/TE/TI = 1630/3/1100msec) sequence were obtained for each subject for spatial image normalization.
Resting and sensorimotor blood-oxygen-level-dependent (BOLD) fMRI scan
A gradient echo planar BOLD fMRI sequence was used to acquire 1) a 6 min resting BOLD fMRI scan, and 2) a 8 min sensorimotor BOLD fMRI scan. Nine young healthy subjects (6 males, 3 females) were scanned with written informed consent obtained before scanning following an Institutional Review Board approved protocol. The acquisition parameters were: 30 interleaving slices, slice thickness=3 mm, flip angle=75°, TR/TE=3000/30 msec, in plane FOV=220 mm. The resting scan was performed first and the subject was asked to stay still and do nothing in the scanner with eyes opened. During the sensorimotor fMRI session, visual stimuli with an 8.3 Hz reversing black and white checkerboard were presented for 5 times each lasting 48 sec interleaved with a 48 sec black screen block (baseline). Each subject was also asked to perform a self-paced left hand only fingertapping task during the visual stimuli.
Data preprocessing and GLM analysis
All data preprocessing was performed in batch mode with SPM5 software (Wellcome Department of Cognitive Neurology, London, UK, http://www.fil.ion.ucl.ac.uk) based batch scripts (Wang et al., 2008a). Functional images of each subject were motion corrected, coregistered with the structural images, and smoothed with an isotropic Gaussian filter (FWHM=6 mm) using the SPM5 software. The structural image was normalized to the Montreal Neurological Institute/International Consortium for Brain Mapping (MNI/ICBM) 152 standard brain using SPM5.
Voxel-based statistical analysis was performed on the spatially smoothed sensorimotor fMRI data using the univariate GLM approach implemented in SPM5. The design paradigm convolved with the canonical HRF function was used as the reference function in the GLM. The temporal autocorrelated noise was considered using the AR(1) model (Friston et al., 1995a; Bullmore et al, 1996), and the high frequency noise was lowpassed with a cutoff of 1/128 Hz. Statistical parametric maps (SPMs) of the task-baseline contrast were collected at each voxel using a standard t-test, which were subsequently normalized to the MNI space using the same normalization parameters estimated from the T1 images and finally a group analysis was performed by running a one-sample t-test on these spatially normalized parametric maps.
Synthetic data generations
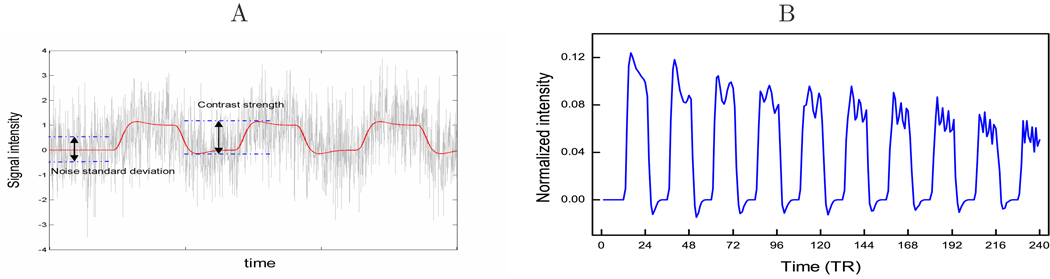
Eight subjects’ motion corrected and smoothed resting fMRI data were used to generate synthetic activation data. Only the central 9 slices were used for simulations to reduce the computational burden. The 120 resting fMRI images were high-pass filtered using SPM5 and randomly permuted to minimize the effects of any possible pseudo-activations. Synthetic activations were then inserted into 40 voxels in grey matter consisting of two 20-voxel clusters using a block-wise baseline/task paradigm. Five cycles were simulated; each block consists of 12 images. Five contrast-to-noise-ratios (CNRs, as defined in Fig. 2A), 0.01, 0.05, 0.1, 0.5, and 1 were used to simulate different activation strengths. The artificial brain activation time course was generated from the canonical HRF convolved boxcar function (the design function) with additional modulations from an exponentially decaying function f(t) = (1 −t/240)0.5 and a nonstable function f(t) = sin(1/4πt2)/9. The decay was introduced to simulate a possible habituation process, and the nonstable component was used to add a nonlinear and nonstable feature to the synthetic data. Fig. 2B shows an example of the activation time course when CNR=1.0. Random Gaussian noise was added to the rest of the brain. The simulated functional image series were then processed through data reduction, SVM training, SDPtp extraction and SVM-GLM. Voxel based GLM was conducted as a comparison benchmark.
Figure 2.
A) Definition of CNR, B) the artificial brain activation time course for simulations with CNR=1.0.
ROC analysis
The receiver operator characteristic (ROC) method (Metz, 1978) was used to assess the performance and efficacy of SVM-GLM. To calculate the ROC curves, the voxel value of each subject’s GLM parametric map and SVM-GLM parametric map were sorted in descending order. ROC curves were generated based on the true positive (activation) rate vs. the false positive (activation) rate throughout the range of sorted maps, by comparing the locations of the predetermined ROIs. The area under curve (AUC) was used as the surrogate score for the efficacy of each method.
SDPtp extraction and SVM-GLM for the sensorimotor BOLD fMRI data
To reduce computational burden, out-of-brain voxels were first excluded using a mask generated from the motion corrected functional images. The remained voxels were then stacked into a column vector, and all column vectors were grouped into a data matrix. All nonzero eigen-values associated eigenvectors were extracted using the same method as in (Mourão-Miranda et al., 2005; Wang et al., 2007a). Each image vector was finally projected into the eigen space spanned by the extracted eigenvectors, yielding a representing coefficient vector. The dimension reduced whole brain data based SVM classification was performed using SVMlight software (Joachim, 1999). The regularization parameter that trades off the margin and training error was set to to avoid an overfitting issue (Joachims, 2002). Eq. 1 was used to extract SDPtp, followed by a correlation analysis and a permutation testing for a statistical inference. SVM-GLM was then performed on the preprocessed fMRI image series (the motion corrected and smoothed images) using the batch mode SPM scripts by replacing the HRF convolved experiment design function with the extracted SDPtp. The statistical parametric map of each individual subject’s SVM-GLM analysis was then spatially normalized to the MNI space using the transformation information obtained with the T1 images, and the group analysis was performed using a one-sample t-test.
Histograms of t-scores within the visual cortex and motor cortex
To further assess the performance difference of regular GLM and SVM-GLM, two large regions-of-interest (ROIs) were defined in the visual cortex and motor cortex, respectively. Each ROI was obtained from the suprathreshold clusters of the group analysis in the visual cortex and motor cortex using a thresh of t > 1.5. The suprathreshold clusters from the regular GLM analysis and SVM-GLM based group analyses were combined into a big cluster; a sphere centered around the peak t spot covering the whole combined cluster was then defined as the final ROI for extracting t-scores from the group level statistical parametric maps. The same process was conducted for defining the visual cortex (VC) ROI and the motor cortex (MC) ROI separately. A histogram for each ROI was generated by counting the number of voxels presenting the same t-scores.
SVM-GLM evaluation with arterial spin labeled (ASL) perfusion fMRI data
Previously published ASL perfusion fMRI data (Wang et al., 2007a) were also used to evaluate SVM-GLM for assessing brain activations in an environment with even lower signal-to-noise ratio than BOLD fMRI (Wong, 1999). The functional stimuli were similar to those used in the BOLD fMRI experiment, but the participants were asked to perform right hand only self-paced finger tapping whenever they saw the flashing checker-board. Ten subjects were included in the individual level analysis and the group level analysis. A detailed description of the preprocessing steps for this data set could be found in (Wang et al., 2007a). Briefly, cerebral blood flow (CBF) image series were generated for each subject after applying motion correction to the raw ASL data. GLM and SVM-GLM were then performed on the spatially smoothed (3D isotropic Gaussian kernel with FWHM=6 mm) CBF series. Additionally acquired T1 images were used to normalize the individual level statistical parametric maps into the MNI space for group analysis.
Results
SDPtp extraction and statistical inferences
The correlation coefficient between SDPtp and the experimental design function was greater than 0.85 for all 9 subjects. With 1000 permutations, the probability was P=1/1001 for every subject’s data for testing the null hypothesis that the correlation between the extracted SDPtp and the design paradigm is due to the prior labeling of the acquired images. Fig. 3A shows a typical SDPtp extracted from a representative subject’s sensorimotor BOLD fMRI data, which was significantly (r=0.89, P=9.51e-56) correlated to boxcar function as shown in Fig. 3B (experimental design function). The correlation coefficient between SDPtp and the fMRI time series extracted from a motor cortex ROI was 0.71 (P=2.7e-26); while the correlation coefficient between the HRF convolved reference function (Fig. 3B) and the fMRI time series was 0.70 (P=4.2e-25).
Figure 3.
An SDPtp extracted from a representative subject’s sensorimotor BOLD fMRI data. A) the extracted SDPtp, B) experimental design function (the thin black line) and the canonical HRF convolved version (the thick blue line), C) fMRI time course from ROI defined in the motor cortex. The green bars indicate the baseline (off) condition; and the red bars indicate the task condition (on).
ROC analysis
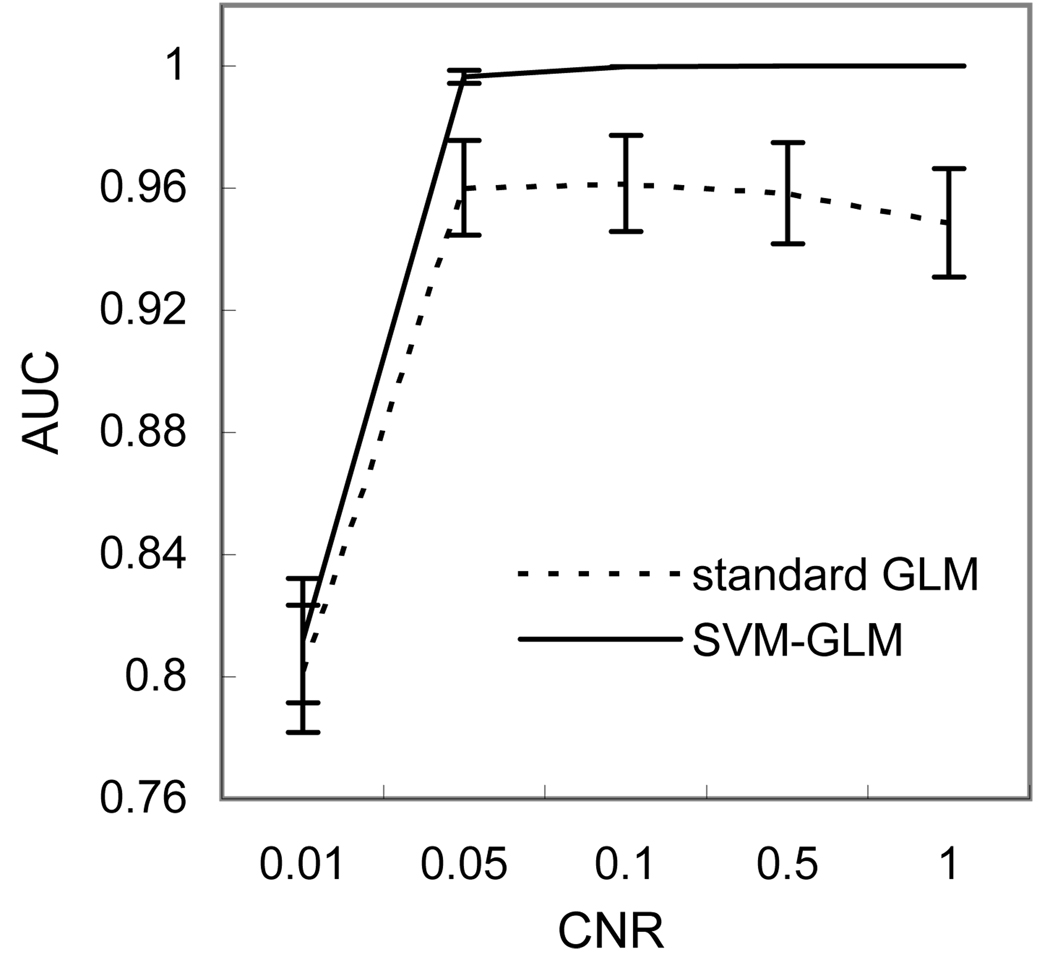
Fig. 4 shows the ROC analysis results of applying standard GLM (the dashed curve) and SVM-GLM (the solid curve) on the synthetic fMRI data with different CNR. No performance difference was observed between the AUC curves of standard GLM and SVM-GLM when the CNR was 0.01. SVM-GLM demonstrated a significantly better sensitivity/specificity performance after CNR was increased to 0.05 and above.
Figure 4.
Averaged (n=8) AUCs of regular GLM and SVM-GLM on the synthetic data generated with different activation CNRs. The error bars mean the standard deviations.
SVM-GLM with the sensorimotor BOLD and ASL perfusion fMRI data
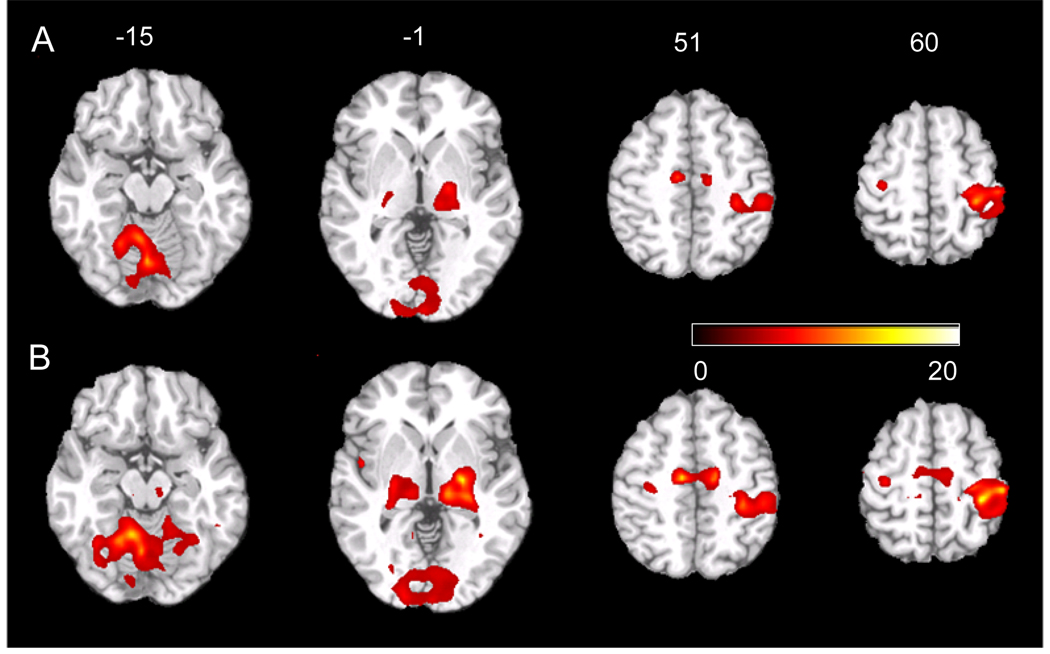
Fig. 5 shows several axial slices of the statistical parametric map (t-map) of the group level analysis for the 9 subjects’ sensorimotor BOLD fMRI data. At the same significance level P≤ 0.0005 (uncorrected), GLM (Fig. 5A) and SVM-GLM (Fig. 5B) revealed similar activation patterns in cerebellum, visual cortex, thalamus, right primary motor cortex, supplementary motor area, and the left primary motor cortex, while SVM-GLM demonstrated higher peak t value and larger suprathresholded clusters than GLM.
Figure 5.
Group level statistical analysis results of the left-hand sensorimotor BOLD fMRI data. The individual level analysis was conducted using A) regular univariate GLM and B) SVM-GLM. The t-maps were thresholded at t≥ 5.04 (corresponding to P≤ 0.0005, uncorrected). The number above each slice indicates the spatial location of each slice in the z axis of the MNI space.
The histograms of the group level t-scores extracted from the VC ROI and MC ROI were shown in Fig. 6. Within the VC and MC ROIs, SVM-GLM yielded a maximum t of 23.32 and 20.98 in VC and MC, respectively; GLM had a maximum of 11.42 and 12.64 in VC and MC, respectively. To be convenient for visual comparison, only the major part of the distributions between 0–12 were shown here. From these figures, we can see that SVM-GLM yielded more voxels with a t-score between 1–8 in both VC and MC ROIs than regular GLM.
Figure 6.
T-score histograms of group level analysis based on individual GLM and SVM-GLM results of the same fingertapping data. The horizontal axis is t-score, and the vertical axis is the number of voxels.
Fig. 7 shows the group level statistical analysis results for the previously published 10 subjects’ sensorimotor ASL fMRI data (Wang et al., 2007a). At the same significance level P≤ 0.001 (uncorrected), GLM (Fig. 5A) and SVM-GLM (Fig. 5B) revealed similar activation patterns in visual cortex, left primary motor cortex, and supplementary motor area. SVM-GLM presented higher sensitivity for activations in the left and right motor cortex. In visual cortex, SVM-GLM showed slightly increased peak t-value, while GLM demonstrated better sensitivity in the fusiform.
Figure 7.
Group level statistical analysis results of the right-hand sensorimotor ASL perfusion fMRI data. The individual level analysis was conducted using A) regular univariate GLM and B) SVM-GLM. The t-maps were thresholded at t≥ 4.3 (corresponding to P≤ 0.001, uncorrected). The number above each slice indicates the spatial location of each slice in the z axis of the MNI space.
Discussion
Exploratory methods, like the independent component analysis (ICA) (Bell and Sejnowski, 1995; Hyvärinen, 1999), has been incorporated into the regular GLM-based fMRI data analysis to improve the accuracy of hemodynamic response modeling (McKeown, 2000; Beckmann et al., 2000; Hu et al., 2005). In this paper, a new combination of exploratory method and GLM is proposed through replacing the regular prior-defined reference function of GLM with SDPtp extracted by SVM. The reason for choosing SVM to extract the data-derived GLM reference function is due to its successful performance for assessing the spatial discriminating patterns in fMRI. Moreover, SVM yields a unique discriminating map rather than multiple component maps as ICA does, so that it does not have the problem of picking up the interested components from a large number of output components, which could even be randomly ordered for different subjects as in ICA based fMRI data analysis.
To derive the reference function for the hybrid SVM-GLM, a new strategy is presented to extract the entire SDPtp from the trained SVM classifier using the signed distance between each image and the separating hyperplane. Aside from giving a way to examine how the SDP vary along time, SDPtp can be used to statistically infer SDP through checking the correlation between SDPtp and the experimental design function. For all 9 subjects recruited in this paper, the extracted SDPtps of their sensorimotor BOLD fMRI data showed significant correlations to the experiment design function, meaning the extracted SDP were significantly related to the functional task.
SDPtp showed a higher correlation than the canonical HRF convolved experimental design paradigm to the mean fMRI time series within the functional ROI (the motor cortex), suggesting that SDPtp more accurately reflected the brain hemodynamic response of the task-induced behavioral state than the canonical HRF convolved experimental design function. As a result, using SDPtp as the regressor in GLM analysis should provide a higher sensitivity for detecting the underlying activations. The ROC analysis using fMRI data with synthetic activations showed that SVM-GLM yielded a better sensitivity/specificity performance than conventional GLM. Applied to a BOLD fMRI dataset, SVM-GLM revealed similar activation patterns but with higher significance level than regular GLM for detecting the sensorimotor activations. Similar improvement was also observed in application to an ASL perfusion fMRI dataset, though regular GLM showed a slightly better performance for revealing the activation in the fusiform. These results demonstrated that SVM could yield a more data adapted reference function than the canonical HRF convolved one for analyzing fMRI data, and therefore increase the activation detection sensitivity.
While these results showed a promising fMRI data analysis using SVM-GLM, one caveat of this combination is that SVM-GLM could be less sensitive to the spatially incoherent activations than conventional GLM. This is because the underlying SVM-based fMRI data classification is tuned more effectively by the spatially coherent brain activations and less by the incoherent ones (Wang et al., 2007a); and the associated SDPtp is subsequently more adapted to the spatially coherent activations. This could explain the discrepancy between the suprathreshold clusters of GLM and SVM-GLM in fusiform and the visual cortex. From another point of view, a larger smoothing kernel could potentially further increase the sensitivity of SVM-GLM for the spatially coherent activations. Another limit of the proposed SDPtp extraction approach and SVM-GLM is that they can not be applied for fMRI data with multiple conditions/states especially due to the use of the signed distance. The data with more conditions/states will have to be split into several sub-series, each composing of two interested contrasting conditions/states. Additionally, the temporal compression method (Mourão-Miranda et al., 2006b) can not be incorporated into the SDPtp extraction, though it has been demonstrated to be helpful for improving the classification precision.
Although a linear SVM classifier is widely used in fMRI data analysis (LaConte et al., 2005; Wang et al., 2007a) as well as in this paper, it is worth to note that the concept of SDPtp is not limited to linear SVM-based classification. It also applies to a nonlinear SVM classifier since a distance can be always calculated between the sample points and the SVM separating hyperplane.
A final issue should be addressed is the overfitting problem of the SVM classifier training process. Overfitting is a common problem for supervised machine learning, meaning that the trained classifier is heavily fitted to the training samples which are generally contaminated by noise and consequently the classifier is less sensitive to the new input data. In the applications to neuroimaging including fMRI data, SVM has repeatedly demonstrated a high prediction accuracy (Cox and Savoy, 2003; Wang et al., 2003; Mitchell et al., 2004; Zhang et al., 2005b; LaConte et al., 2005; Davatzikos et al., 2005; Mourão-Miranda et al., 2005; Wang et al., 2007a), indicating that overfitting is a less problematic issue. Actually, a key point of SVM is to avoid overfitting through maximizing the margin. In our SVM-based SDP and the SDPtp extraction, The risk of overfitting is further minimized because: 1) a linear SVM model could not be trained to fit every training sample, and 2) a small value (, generally less than 1) was used for the regularization parameter to prevent overfitting during the learning process.
Acknowledgements
This research was supported by NIH/NIDA grant R03DA023496.
References
- Aguirre GK, Zarahn E, D’Esposito M. The variability of human BOLD hemodynamic responses. Neuroimage. 1998;8:360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional mri of the human brain. Magn Reson Med. 1993;30:161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Tracey I, Noble JA, Smith SM. Combining ICA and GLM: a hybrid approach to FMRI analysis. NeuroImage. 2000;11(5):s643. [Google Scholar]
- Bell A, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Computation. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Brammer M, Williams S, Rabe-Hesketh S, Janot N, David A, Mellers J, Howard R, Sham P. Statistical methods of estimation and inference for functional MR image analysis. Magnetic Resonance in Medicine. 1996;35(2):261–277. doi: 10.1002/mrm.1910350219. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Mining and Knowledge Discovery. 1998;2:121–167. [Google Scholar]
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fmri activity in human visual cortex. NeuroImage. 2003;19(2):261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen D, Acharyya M, Loughead J, Gur R, Langleben D. Classifying spatial patterns of brain activity with machine learning methods: Application to lie detection. NeuroImage. 2005;28(3):663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Fan Y, Rao H, Hurt H, Giannetta J, Korczykowski M, Shera D, Avants BB, Gee JC, Wang J, Shen D. Multivariate examination of brain abnormality using both structural and functional mri. NeuroImage. 2007;36(4):1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Friston K, Holmes A, Poline J, Grasby PJ, Williams SCR, Frackowiak R, Turner R. Analysis of fmri time series revisited. NeuroImage. 1995a;2:45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- Friston K, Holmes A, Worsley K, Poline J, Frith C, Frackowiak R. Statistical parametric maps in functional imaging: A general linear approach. Human Brain Mapping. 1995b;2:189–210. [Google Scholar]
- Hu D, Yan L, Liu Y, Zhou Z, Friston KJ, Tan C, Wu D. Unified spmcica for fmri analysis. NeuroImage. 2005;25:746–755. doi: 10.1016/j.neuroimage.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. on Neural Network. 1999;10(3):626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- Joachim T. Making large-scale svm learning practical. In: Schölkopf B, Burges C, Smola A, editors. Advances in Kernel Methods-Support Vector Learning. Cambridge Boston: MIT Press; 1999. pp. 42–56. [Google Scholar]
- Joachims T. Learning to Classify Text Using Support Vector Machines. Norwell, MA: Kluwer Academic Publishers; 2002. [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Anderson J, Hu X. Support vector machines for temporal classification of block design fmri data. NeuroImage. 2005;26(2):317–329. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- McKeown MJ. Detection of consistently task-related activations in fMRI data with hybrid independent component analysis. NeuroImage. 2000;11:24–35. doi: 10.1006/nimg.1999.0518. [DOI] [PubMed] [Google Scholar]
- Metz CE. Basic principle of ROC analysis. Semin Nucl Med. 1978;VIII(4):283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- Mitchell TM, Hutchinson R, Niculescu RS, Pereira F, Wang X, Just M, Newman S. Learning to decode cognitive states from brain images. Machine Learning. 2004;57(1–2):145–175. [Google Scholar]
- Mourão-Miranda J, Bokde AL, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: Support vector machine on functional MRI data. NeuroImage. 2005;28(4):980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Friston KJ, Brammer M. Dynamic discrimination analysis: A spatial-temporal svm. NeuroImage. 2006a;36(1):88–99. doi: 10.1016/j.neuroimage.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Reynaud E, McGlone F, Calvert G, Brammer M. The impact of temporal compression and space selection on SVM analysis of single-subject and multi-subject fMRI data. NeuroImage. 2006b;33:1055–1065. doi: 10.1016/j.neuroimage.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Vapnik V. The Nature of Statistical Learning Theory. New York: Springer-Verlag; 1995. [Google Scholar]
- Vetterling WT, Flannery BP. Numerical Recipes in C++: The Art of Scientific Computing. 2nd edition. New York: Cambridge University Press; 2002. [Google Scholar]
- Wang X, Hutchinson R, Mitchell T. Training fMRI classifiers to discriminate cognitive states across multiple subjects. Proc. of 17th Annual Conference on Neural Information Processing Systems; Vancouver and Whistler, Canada. 2003. [Google Scholar]
- Wang Z, Aguirre GK, Rao H, Wang J, Fernández-Seara MA, Childress AR, Detre JA. Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx. Magnetic Resonance Imaging. 2008a;26(2):261–269. doi: 10.1016/j.mri.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Childress AR, Wang J, Detre JA. Support vector machine learning-based fMRI data group analysis. NeuroImage. 2007a;36(4):1139–1151. doi: 10.1016/j.neuroimage.2007.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Detre JA, Childres AR. Boost up the detection sensitivity of ASL perfusion fmri through support vector machine; Proceedings of the 28th IEEE EMBS Annual International Conference (EMBC06); 2006. pp. 1006–1009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Fernández-Seara MA, Alsop DC, Wang J, Liu W-C, Flax JF, Benasich AA, Detre JA. Assessment of functional development in normal infant brain using arterial spin labeled perfusion mri. NeuroImage. 2008b doi: 10.1016/j.neuroimage.2007.09.045. Epub ahead of print:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Ehrman R, Hole AV, MacDougall M, Jens W, Franklin T, Detre JA, O’Brien CP, Childress AR. A support vector machine (SVM) is superior to univariate glm for characterizing cue-induced limbic activation by arterial spin labeling (asl) perfusion fMRI; Annual Conference of Society for Neuroscience; 2007b. page online. [Google Scholar]
- Wong E. Potential and pitfalls of arterial spin labeling based perfusion imaging techniques for MRI. In: Moonen CW, Bandettini P, editors. Functional MRI, Medical Radiology:Diagnostic Imaging and Radiation Oncology. New York: Springer-Verlag; 1999. pp. 63–69. [Google Scholar]
- Worsley K, Friston K. Analysis of fmri time-series revisited-again. NeuroImage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- Zhang L, Samaras D, Tomasi D, Alia-Klein N, Cottone L, Leskovjan A, Volkow N, Goldstein R. Exploiting temporal information in functional magnetic resonance imaging brain data; Proc. of the Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv; 2005a. pp. 679–687. [DOI] [PubMed] [Google Scholar]
- Zhang L, Samaras D, Tomasi D, Volkow N, Goldstein R. Machine learning for clinical diagnosis from functional magnetic resonance imaging; IEEE International Conference Computer Vision and Pattern Recognition; 2005b. pp. 1211–1217. [Google Scholar]