Abstract
We demonstrate for the first time, to our knowledge, a unique gene expression assay by surface-enhanced Raman scattering (SERS) using nonfluorescent Raman labels to quantify gene expression at the resolution of alternative splicing using RNA extracted from cancer cells without any amplification steps. Our approach capitalizes on the inherent plasmon-phonon mode of SERS substrates as a self-referencing standard for the detection and quantification of genetic materials. A strategy integrating S1 nuclease digestion with SERS detection was developed to quantify the expression levels of splice junction Δ(9,10), a segment of the breast cancer susceptibility gene 1 (BRCA1) from MCF-7 and MDA-MB-231 cells. Quantification results were cross-validated using two Raman tags and qualitatively confirmed by RT-PCR. Our methodology based on SERS technology provides reliable gene expression data with high sensitivity, bypassing the intricacies involved in fabricating a consistent SERS substrate.
Introduction
Novel analytical techniques for efficient and reliable gene expression screening are highly desirable in the new genetic era. Commonly used Northern blots, conventional reverse transcription polymerase chain reaction (RT-PCR), and quantitative nuclease protection assays for gene expression studies are either semiquantitative or not sensitive enough (1). Although it is known to be a very sensitive method, quantitative real-time RT-PCR is not always appropriate for studying gene expression, especially when high-amplification efficiency is not possible or available (2). Therefore, methods that can provide quantitative information, monitor low-abundant genetic molecules, and do not require an amplification step are extremely promising.
With its superb multiplexing capability attributed to the intrinsic sharp fingerprints of Raman tags (3,4), single-molecule sensitivity (5,6), photostability, and a wide choice of labels, surface-enhanced Raman spectroscopy (SERS) could be an excellent choice for gene expression screening. However, due to the lack of signal reproducibility or variability in surface enhancement, most SERS applications, spanning the areas of molecular imaging (7–9), molecular diagnostics (10–12), genotyping (13,14), and several others (15,16), are qualitative or at best semiquantitative in nature. To date, the use of SERS for quantification of genetic materials has been demonstrated only for a dilution series of dye-labeled DNA sequences (17,18). If harnessed properly, a reliable SERS quantification approach could possess unique advantages due to its high sensitivity, multiplexing ability, and high-throughput screening capability.
Although several factors contribute to the variation in SERS (19), it is known that heterogeneity of enhancement is a major hurdle for SERS-based quantification. Great strides have been made in fabricating tunable and reproducible substrates for SERS studies (20–28), which often require advanced lithographic techniques (29–31). However, uniform enhancement, which is critical for quantification, remains a challenge. The isotope-edited internal standard method has been suggested (32); however, this method is technically difficult because the corresponding isotopes need to be synthesized. Although it is known that the sensitivity of SERS can rival that of fluorescence (33), due to the probabilistic nature of SERS enhancement or the complexity of substrate synthesis, a consistent SERS quantification strategy in cell/molecular biology is still a challenge.
In this study, a quantification method that takes advantage of the phonon-plasmon Raman band around 180–280 cm−1 inherent to metallic nanostructures (34) as a self-referencing standard is proposed for gene quantification using nonfluorescent Raman labels. We report for the first time, to our knowledge, an SERS approach to quantify gene expression at the resolution of alternative splicing, which is a fundamental mechanism of gene expression and a potential marker for cancer diagnosis. In our previous work (35), we demonstrated the concept of an array-format SERS sandwich assay for multiplex detection of DNA targets using fabricated DNA molecules. In the study presented here, we integrate S1 nuclease digestion with the SERS sandwich assay to quantify splice variants obtained directly from cells without amplification. The breast cancer susceptibility gene 1 (BRCA1) was chosen as a model gene to illustrate the validity of our approach. Expression of the splice junction variant Δ(9, 10) (skipping exons 9 and 10) of BRCA1 was compared between two breast cancer cell lines, MCF-7 and MDA-MB-231, and the quantification results were cross-confirmed using two different Raman labels and validated by RT-PCR. The appeal of the proposed methodology is its ability to estimate an unknown amount of genetic material extracted from cells without any amplification steps and with excellent sensitivity, paving the way to a possible quantification of even lowly abundant variants, the potential to detect multiple variants, the striking specificity intrinsic to Raman spectroscopy, and finally, reduced costs of detection due to the use of nonfluorescent labels.
Materials and Methods
Instrumentation
A SENTERRA confocal Raman system (Bruker Optics, Billerica, MA) fitted with a 785 nm laser, and a 20× air objective (N.A. 0.7) was used to investigate SERS substrates. An integration time of 20 s and a laser power of 10 mW at the laser source were used to examine the mode shift of naked materials. For studying the band shift with changing surface modifications, 1 mW of laser power at the source was found to be sufficient. Further evaluation and characterization of the band was done using the SENTERRA Raman system (830 nm laser line, 20 s integration time, and 100 mW of power) fitted with a darkfield illumination module (Cytoviva, Auburn, AL) for simultaneous Raman spectroscopy and darkfield imaging, and the same sample was interrogated by scanning electron microscopy (SEM) using an S-4800 UHR FE-SEM system (Hitachi, Pleasanton, CA). All Raman spectra for DNA detection were acquired using a 785 nm laser with a power of 10 mW at the source, a 50× air objective (N.A. 0.7), and an integration time of 20 s.
Preparation of SERS substrates
Gold (Au) nanoparticles were synthesized by reduction of gold (III) chloride with sodium citrate, and silver (Ag) nanoparticles were synthesized by citrate reduction of silver nitrate. To form Au and Ag nanoparticle films, glass slides were first cleaned with piranha solution for 30 min, rinsed with Nanopure water, and dried in nitrogen. Next, the slides were immersed in an ethanol solution of 2% 3-aminopropyltrimethoxysilane (APTMS) overnight. APTMS-treated slides were then rinsed with ethanol and Nanopure water three times and dried in nitrogen. Amino-functionalized glass slides were immersed in Au or Ag nanoparticle solution for 5 h and washed copiously with Nanopure water. Finally, the slides were dried in nitrogen and kept ready for measurement.
SERS sandwich assay
The splice junction Δ(9, 10) of BRCA1 was studied in this research. Their sequences were obtained from GenBank (accession number U14680), maintained by the National Institutes of Health. DNA oligonucleotides of different compositions (Table 1) were purchased from IDT (Coralville, IA). Ten thymine bases were used as linkers in the capturing strands (CS) and probing strands (PS) to reduce nonspecific binding and improve hybridization efficiency, since thymine has the lowest affinity to gold compared to other nucleobases (36). Procedures described in Sun et al. (37) were followed to prepare DNA-AuP-RTag probes for SERS sandwich assays. Briefly, thiolated DNA oligonucleotides in a disulfide form were reduced using Reductacryl (EMD Biosciences, San Diego, CA) at a mass ratio of 1: 50. Probing DNA strands were then added to the precipitate obtained from 10 mL of Au nanoparticle solution to obtain a 1 mL solution with an oligonucleotide concentration of 1 μM. After 24 h, the solution was buffered at pH 7.5 (10 mM phosphate buffer with 0.01% Tween 20) and salted slowly with 4 M NaCl to reach a final salt concentration of 0.3 M. After “aging” for 40 h and washing, 1 mL of RTag solution (4-mercaptopyridine or 2-thiazoline-2-thiol) was added and incubated for 24 h. The resulting DNA-AuP-RTag probes were finally washed three times with 0.3 M PBS (pH 7.5). For the SERS sandwich assay, gold-coated glass slides were first treated with piranha solution for 1 h, and the slides were covered with a silicone mask with an array of holes (3 mm in diameter) to avoid cross-talk between neighboring spots. Ten microliters of CS, target strands (TS), and DNA-AuP-RTag probes were sequentially spotted on the slide and incubated in a humidity chamber at 37°C to form a sandwich structure. After a thorough washing protocol, a silver enhancement procedure, and a final nitrogen drying step were completed, the slide was taken for Raman studies as detailed in Sun et al. (35).
Table 1.
Oligonucleotide sequences used in experiments
| Oligonucleotide | Sequence (5′- 3′) |
|---|---|
| Δ(9, 10): TS | TCAGAAAATTCACAAGCAGCCAATTCAATGTAGACAGACG |
| Δ(9, 10): CS | GCTGCTTGTGAATTTTCTGATTTTTTTTTT-C3-SH |
| Δ(9, 10): PS | SH-C6-TTTTTTTTTTCGTCTGTCTACATTGAATTG |
| Δ(9, 10): sense primer | GCAGGAAACCAGTCTCAGTG |
| Δ(9, 10): anti-sense primer | TTCACAAGCAGCCAATTCAATG (amplicon size: 118 bp) |
| β-actin: sense primer | CATGTACGTTGCTATCCAGGC |
| β-actin: anti-sense primer | CTCCTTAATGTCACGCACGAT (amplicon size: 250 bp) |
TS: target strand; CS: capturing strand; PS: probing strand.
Preparation of DNA targets from cancer cells
MCF-7 cells were grown in Eagle's minimum essential medium (ATCC, Manassas, VA) supplemented with 0.01 mg/mL bovine insulin and 10% of FetalPlex animal serum complex (Gemini Bio-Products, West Sacramento, CA). MDA-MB-231 cells were grown in ATCC-formulated RPMI-1640 medium supplemented with 10% of FetalPlex animal serum. Total RNA was isolated from ∼107 cells with the use of an RNeasy Mini Kit (QIAGEN, Valencia, CA). Next, 30 μL of hybridization reaction containing 20 μL of RNA, 9 μL of 3× aqueous hybridization solution (3 M NaCl, 0.5 M HEPES (Invitrogen, Carlsbad, CA), 1 mM EDTA), and 1 μL of DNA oligonucleotides (0.3 ng) were heated to 75°C for 10 min and incubated overnight at 55°C. Then 270 μL of S1 nuclease mix were added to the tube, the mixture was incubated at 37°C for 45 min, and a hydrolysis solution (1.6 M NaOH, 135 mM EDTA) was added. The mixture was heated to 95°C for 15 min and a neutralizing solution (1 M HEPES, 1.6 M HCl, 6× SSC) was added. The unknown targets present in the resulting solution were then used for SERS detection.
RT-PCR
One microgram of RNA was first treated with DNase I (New England BioLabs, Ipswich, MA). Then an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) was used to synthesize cDNA according to the manufacturer's instructions. Taq DNA polymerase (Fisher Scientific, Fair Lawn, NJ) and primers (Table 1) were subsequently added for PCR using a DNA Engine (PTC-220) Peltier Thermal Cycler (MJ Research, Waltham, MA). Finally, 3% of agarose gel prestained with ethidium bromide (Fisher Scientific) was used to analyze the PCR products.
Results and Discussion
Phonon-plasmon band as a self-referencing standard
Since it is very difficult to obtain uniform surface enhancement, it would be advantageous if surface characteristics related to enhancement of an SERS substrate could be defined by a simple criterion and taken into account in a Raman measurement. In this section, we will demonstrate that the band around 180–280 cm−1 from nanoparticle SERS substrates can be used as a self-referencing standard for Raman-based quantification. Raman spectra from two widely used SERS substrates (Au and Ag nanoparticles) on both glass and gold-coated glass slides were examined first. An intermediary substrate consisting of an Au-Ag composite was also evaluated, since Ag deposition on gold nanoparticles is commonly applied in SERS detection for further surface enhancement (13). A Raman band around 230–260 cm−1 appeared when nanoparticles were present, but this band was not noted when smooth surfaces were used (Fig. 1, a–h), suggesting a morphology dependence. A consistent left-shift in this band was noted during the transition from Au to Ag, and a similar trend in band position was observed for Au and Ag nanoparticles on both glass and gold-coated glass slides, indicating that this band is material-dependent. This low-frequency Raman band can be assigned to the plasmon-phonon coupling resulting from acoustic vibrations of surface characteristics selected by the resonant excitation of localized plasmons (38–40). It is well known that surface enhancement depends on the morphology and material type of metal nanostructures. Hence, it is reasonable to expect that this plasmon-phonon band can depict the local enhancement of specific SERS substrates. The plasmon-phonon band was also sensitive to surface modifications (Fig. 1, i–l). A shift in the band was noted as expected upon subsequent modification of the particles with thiolated DNA (DNA-AuP) and nonfluorescent Raman tags (DNA-AuP-RTag). Nonfluorescent labels, 4-mercaptopyridine and 2-thiazoline-2-thiol, were used as Raman Tag 1 (RTag-1) and Raman Tag 2 (RTag-2), respectively. It is interesting to note that there was another intense band below 100 cm−1 that appeared in the presence of metal (both metallic nanostructures and smooth metal surface (Fig. 1 e)) and disappeared when bare glass slides were examined (Fig. 1 a). This band could be attributed to backscattering.
Figure 1.
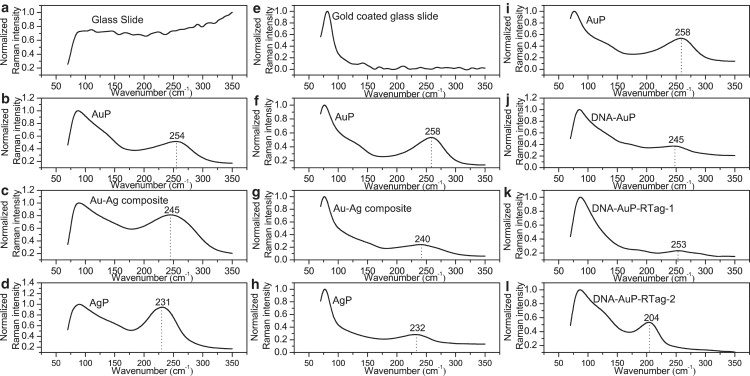
Raman spectra of different SERS substrates and Au nanoparticles with different surface modifications. (a–d) Au and Ag nanoparticles and Au-Ag composite on a glass slide. (e–h) Au and Ag nanoparticles and Au-Ag composite on a gold-coated glass slide. (i–l) Surface modifications of Au nanoparticles using DNA and Raman tags on a gold-coated glass slide.
As a next step, we sought to demonstrate the relevance of the plasmon-phonon band for surface enhancement at the measurement location, to show that the plasmon-phonon mode can be used as a self-referencing standard to correct for heterogeneous enhancement and ensure reliable quantification. A drop of Au colloids was dried on a glass slide. Three locations with significantly different particle densities were identified and the phonon-plasmon mode intensity was measured. Surface enhancement could potentially be higher with a larger number of particle assemblies, which could include more hot areas. A SENTERRA Raman workstation equipped with a darkfield imaging system was used to evaluate the chosen locations. Coupled darkfield and SEM images were obtained at exactly the same positions via precise stage positioning using a coordinate measurement method based on reference markers. Darkfield images are shown in Fig. 2 a, with the corresponding brightness indicating the respective particle density (from left to right (positions 1–3): dense particles, few particles, and no particles). The drastic difference in particle densities is clearly seen in the corresponding SEM images (Fig. 2 b) representing the center of an excitation laser spot. The highest particle density suggests the strongest enhancement at position 1. The corresponding Raman spectra from the spots corresponding to the three positions (Fig. 2 c) clearly point to the fact that the Raman intensity of the plasmon-phonon band is correlated to enhancement, with increasing intensity depicting an increase in enhancement. No particles were present at position 3, and the plasmon-phonon band was consequently not present, which supports our hypothesis that the plasmon-phonon mode is characteristic of the surface structure itself. For further validation, we applied coupled darkfield imaging and SERS measurement to Ag nanoparticles, a gold nanoparticle film, and a silver nanoparticle film. Similar results (see Fig. S1 in the Supporting Material) were obtained, reinforcing our hypothesis that the plasmon-phonon mode inherent to metallic structures has an increasing Raman intensity to correspond with increasing surface enhancement. For a metal nanoparticle substrate, a drop of metal colloids was dried on a glass slide to form a circular spot with higher particle density at the rim and lower particle density at the center. For a metal nanoparticle film substrate, a layer of nanoparticles was attached to a 3-aminopropyltrimethoxysilane (APTMS)-treated glass slide. These substrates are the most commonly used in SERS-based detection, which indicates that the plasmon-phonon band can be utilized in many SERS applications. Thus a preliminary confirmation of the use of the plasmon-phonon band as a self-referencing standard was obtained, paving the way to a potentially more robust SERS quantification methodology. This method circumvents the intricacies involved in fabricating reproducible SERS substrates. It does not require any secondary technique or any additional step to characterize the SERS substrate, and information on local enhancement can be acquired simultaneously with the detection of Raman labels by a simple Raman measurement.
Figure 2.
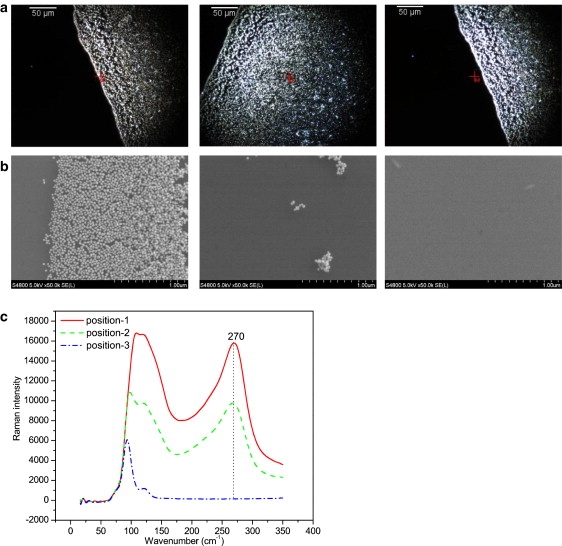
The plasmon-phonon mode of Au nanoparticles. (a) Darkfield images of Au nanoparticles with different densities. From left to right (positions 1–3): dense particles, few particles, and no particles. (b) SEM images of the corresponding positions. (c) Raman spectra of positions 1–3.
SERS-based quantification of BRCA1 splice variants from cancer cells
The concept of utilizing the plasmon-phonon band as a self-referencing standard was demonstrated to quantify splice variants obtained from cancer cells. First, a calibration curve was constructed for quantitative studies based on an SERS sandwich assay to detect target DNA strands specific to the splice junction Δ(9, 10) of the BRCA1 gene using fabricated probes. Then RNA isolation, S1 nuclease digestion, alkaline hydrolysis, and an SERS sandwich assay were performed to quantify an unknown concentration of the splice junction Δ(9, 10) of the BRCA1 gene obtained from breast cancer cell lines, MCF-7 and MDA-MB-231, as shown in Fig. 3. The detailed procedures will be described later.
Figure 3.
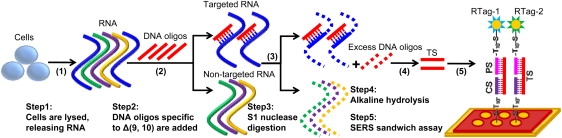
Detection schematic integrating S1 nuclease digestion and an SERS sandwich assay to quantify splice variants from targets obtained directly from cancer cells.
To establish the calibration, a gold-coated glass slide covered with a silicone mask with an array of holes was used as a detection platform. The CS with the sequence spanning the downstream half of the exon/exon junction with 3′ thiol modification was immobilized on the gold surface via S-Au interaction. Then the TS, which contained the sequence complementary to the exon/exon junction, was allowed to hybridize with the CS and was tested for a series of concentrations ranging from 1 fM to 500 nM. Finally, the DNA-AuP-RTag probe containing a PS sequence to mimic the upstream half of the exon/exon junction with 5′ thiol modification was hybridized to the overhanging part of the TS. Two nonfluorescent Raman tags (RTag-1 and RTag-2) identified in our earlier work (35) were used independently, and 1091 cm−1 and 1349 cm−1 were chosen as the characteristic peaks for RTag-1 and RTag-2, respectively. To utilize the plasmon-phonon band as a self-referencing standard, the baseline magnitude chosen according to the characteristic peaks was first subtracted from both the characteristic peak and the plasmon-phonon band to obtain corrected Raman intensities (denoted as I and , respectively), and the characteristic band was normalized against the plasmon-phonon band (). The normalization process, shown in Fig. 4, was performed for each sample for a range of TS concentrations and for the control sample in which no TS was present. The thus obtained for the control sample was the nonspecific signal, which was subtracted from the obtained from other samples for calibration purposes. As expected, Raman signals from the control samples were very weak, demonstrating that nonspecific binding was trivial and not statistically significant. All of the points on the calibration plot were based on an average of three measurements from positions randomly chosen from the sample spot, and normalization was performed independently for each of these measurements. The data points were fitted using MATLAB R2008a (The MathWorks, Natick, MA) by a power relationship with R2 of 0.89 and 0.93, respectively, for RTag-1 and RTag-2 (Fig. 5). It is worth noting that the calibration performed with the proposed approach is valid over a rather wide range of concentrations that are rarely reported in the literature, yielding an extensive dynamic range for the assay. The calibration established in this work is based on a sandwich assay, and hence all of the factors that affect detection are compensated for inherently in the calibration. To show the effectiveness of normalization to the self-referencing standard, measurements at three concentrations (1 nM, 1 pM, and 1 fM) using both Raman tags were compared before (Fig. 6 a) and after normalization (Fig. 6 b). A significant reduction in measurement error was obtained after normalization. The average relative standard deviation (RSD, standard deviation divided by the mean) for all of the available measurements before normalization was 40% for both tags, whereas after normalization the average RSD was reduced to 20% for both tags. It was observed that before normalization there was no reliable correlation between the Raman intensity and target concentration for either Raman tag.
Figure 4.
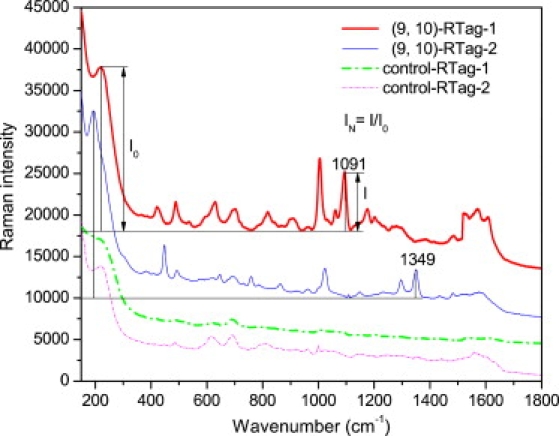
The normalization method using the plasmon-phonon band as a self-referencing standard.
Figure 5.
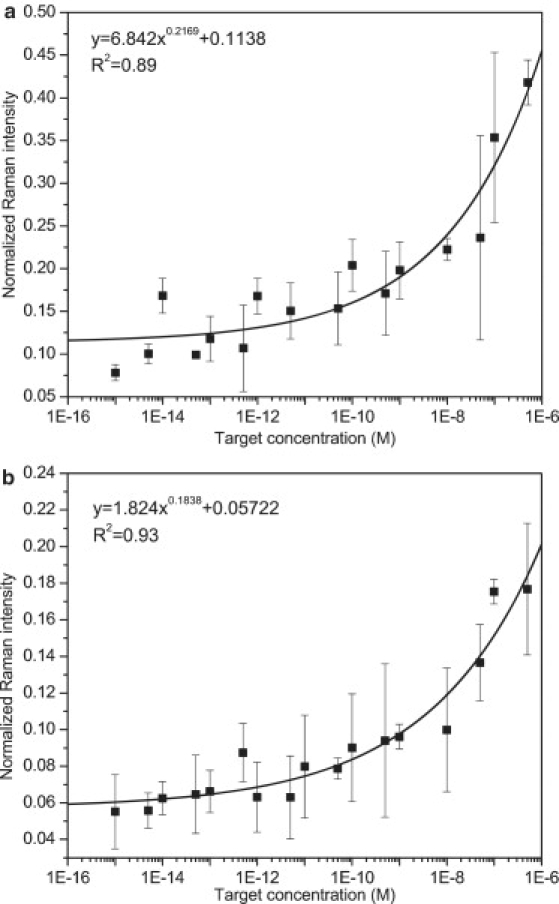
Calibration curves for detection of Δ(9, 10) in the concentration range between 1 fM and 500 nM using (a) RTag-1 and (b) RTag-2.
Figure 6.
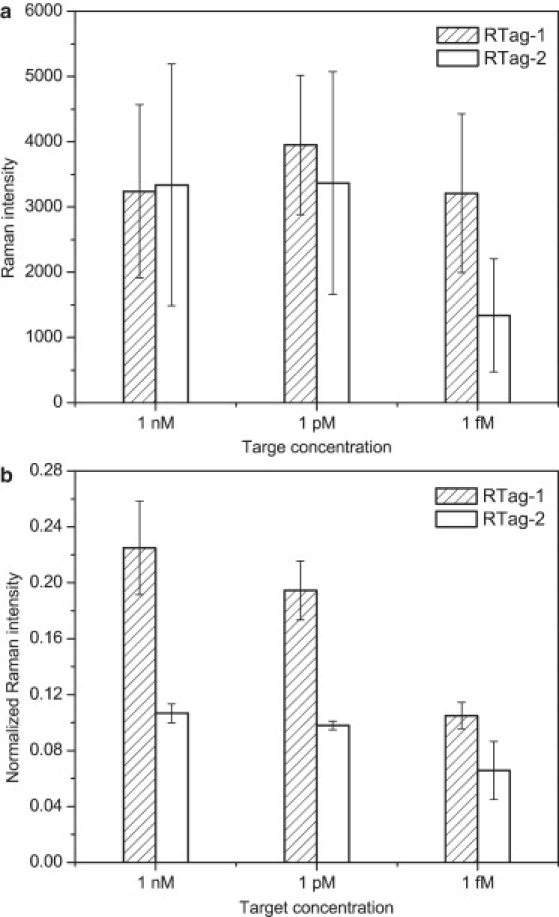
Comparison of Raman signals (a) before and (b) after normalization.
To demonstrate the ability of the SERS methodology to quantify gene expression, the splice junction Δ(9, 10) of BRCA1 gene obtained from two breast cancer cell lines, MCF-7 and MDA-MB-231, were analyzed. It should be noted that any gene segment could be quantified using the same procedures. As shown in Fig. 3, cells were first lysed to release total RNA. Then DNA oligonucleotides complementary to the splice junction Δ(9, 10) (40 bp, the same sequence of TS) were hybridized to mRNA, and S1 nuclease was sequentially applied to cleave all the single-stranded nucleic acids. As a result, the nontargeted RNA molecules and the single-stranded parts of the targeted RNA molecules, as well as the excess DNA oligonucleotides, were completely digested, leaving only the specific DNA-RNA duplexes intact. S1 nuclease was chosen because this endonuclease can remove heteroduplexes containing mismatched regions, such as hairpin loop structures, to result in specific DNA-RNA duplexes. Next, an alkaline hydrolysis step was introduced to inactivate the S1 nuclease and destroy the RNA component of the DNA-RNA duplex. The DNA thus obtained is specific to Δ(9, 10) and in proportion to the amount of mRNA containing the Δ(9, 10) variant in cancer cells. Finally, the DNA targets were tested by the SERS sandwich assay, and the resulting spectra were normalized as discussed above. It should be noted that a cocktail containing all of the components in the aforementioned strategy except for the DNA oligonucleotides was used as a control to ensure accurate assessment of the nonspecific signal. The nonspecific signal thus obtained was first subtracted from the sample signal, and the resulting signal intensity was used in the calibration equation to calculate the expression level.
Table 2 presents the quantification results from three independent SERS sandwich assays using the same target DNA obtained from MCF-7 and MDA-MB-231 cells. The determined concentrations of target DNA specific to the splice variant Δ(9, 10) were 29 pM and 1.40 pM, and were reproducible with RSDs of 20% and 5% for MCF-7 and MDA-MB-231 cells, respectively. Dual labels (RTag-1 and RTag-2) were used to analyze the same target, and the results are presented in Table 3. The matching results from the two RTags cross-validate the quantification results to affirm that the newly proposed SERS strategy is a robust and reliable method for quantifying genetic components at the splicing variant resolution. To further validate these results, a conventional RT-PCR method was used to reveal the expression level of the splice junction Δ(9, 10) qualitatively. cDNA was first synthesized from the isolated total RNA using primers designed to cross the exon/exon junction for PCR. Here β-actin, a housekeeping gene, was used as an internal control. Fig. 7 clearly shows that the splice variant Δ(9, 10) was expressed more in MCF-7 cells than in MDA-MB-231 cells, which agrees well with our SERS results and the literature (1). Because of the intrinsic quantification design, the proposed approach could be expanded to quantify any segment of the genome without any amplification steps, ensuring an accurate assessment since the variability caused by amplification could be avoided.
Table 2.
Quantification of the splice junction Δ(9, 10) from MCF-7 and MDA-MB-231 cells
| MCF-7 | MDA-MB-231 | |
|---|---|---|
| Test 1 | 34 pM | 1.46 pM |
| Test 2 | 26 pM | 1.43 pM |
| Test 3 | 26 pM | 1.32 pM |
| Average | 29 pM | 1.40 pM |
| SD | 5 pM | 0.07 pM |
| RSD | 20% | 5% |
Table 3.
Quantification of splice junction Δ(9, 10) from MCF-7 and MDA-MB-231 cells using two different nonfluorescent Raman tags
| MCF-7 | MDA-MB-231 | |
|---|---|---|
| RTag-1 | 31 ± 1 pM | 1.3 ± 0.2 pM |
| RTag-2 | 29 ± 5 pM | 1.4 ± 0.1 pM |
Figure 7.
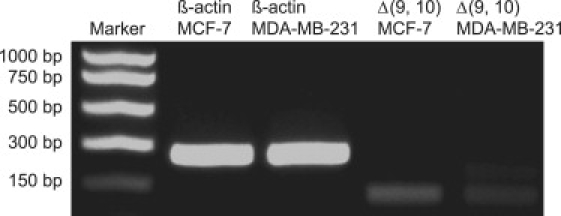
RT-PCR analysis of the splice junction Δ(9, 10) from MCF-7 and MDA-MB-231 cells.
Conclusions
To summarize, a self-referencing standard attributable to the plasmon-phonon mode of chosen metallic structures was shown to have significant potential for quantitative SERS. The position of the plasmon-phonon mode was shown to be dependent on morphology, material, and surface modification, and its intensity had a direct relevance to surface enhancement. SERS sandwich assays and signal normalization were performed over a wide dynamic range (1 fM to 500 nM) to quantify genetic materials extracted directly from cancer cells. A detection scheme combining S1 nuclease digestion and an SERS sandwich assay was developed to quantify the splice variant Δ(9, 10) from MCF-7 and MDA-MB-231 cells. The developed methodology was independently tested using two Raman labels and validated with RT-PCR. This first application (to our knowledge) of a novel SERS quantification strategy for gene expression studies is a significant step forward in enabling biological quantification of sparse molecules without the need for any amplification steps. Because of its intrinsic design, this methodology can be used to study any segment of the genome in a quantitative format. Broader applications in gene mapping and detection of epigenetic marks and mutations are possible to address fundamental disease-related questions using the developed chemistries in conjunction with the proposed analytical methodology.
Acknowledgments
We thank Leo Tom Varghese from the Birck Nanotechnology Center for assistance with SEM imaging, and Dr. Chungang Wang from the Bindley Bioscience Center for helpful discussions on Au/Ag films. The work was conducted at the Physiological Sensing Facility of Bindley Bioscience Center.
Funding from the National Cancer Institute, National Institutes of Health (5RO3CA121347-02), the Center for Food Safety Engineering, and the Bilsland Fellowship, Purdue University, is acknowledged.
Supporting Material
References
- 1.Orban T.I., Olah E. Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem. Biophys. Res. Commun. 2001;280:32–38. doi: 10.1006/bbrc.2000.4068. [DOI] [PubMed] [Google Scholar]
- 2.Faulds K., Smith W.E., Graham D. DNA detection by surface enhanced resonance Raman scattering (SERRS) Analyst. 2005;130:1125–1131. doi: 10.1039/b500248f. [DOI] [PubMed] [Google Scholar]
- 3.Ni J., Lipert R.J., Dawson G.B., Porter M.D. Immunoassay readout method using extrinsic Raman labels adsorbed on immunogold colloids. Anal. Chem. 1999;71:4903–4908. doi: 10.1021/ac990616a. [DOI] [PubMed] [Google Scholar]
- 4.Mulvaney S.P., Musick M.D., Keating C.D., Natan M.J. Glass-coated, analyte-tagged nanoparticles: a new tagging system based on detection with surface-enhanced Raman scattering. Langmuir. 2003;19:4784–4790. [Google Scholar]
- 5.Koo T.-W., Chan S., Sun L., Su X., Zhang J. Specific chemical effects on surface-enhanced Raman spectroscopy for ultra-sensitive detection of biomolecules. Appl. Spectrosc. 2004;58:1401–1407. doi: 10.1366/0003702042641227. [DOI] [PubMed] [Google Scholar]
- 6.Nie S., Emory S.R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 7.Qian X., Peng X.-H., Ansari D.O., Yin-Goen Q., Chen G.Z. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 8.Keren S., Zavaleta C., Cheng Z., de la Zerda A., Gheysens O. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc. Natl. Acad. Sci. USA. 2008;105:5844–5849. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chourpa I., Lei F.H., Dubois P., Manfait M., Sockalingum G.D. Intracellular applications of analytical SERS spectroscopy and multispectral imaging. Chem. Soc. Rev. 2008;37:993–1000. doi: 10.1039/b714732p. [DOI] [PubMed] [Google Scholar]
- 10.Wabuyele M.B., Vo-Dinh T. Detection of human immunodeficiency virus type 1 DNA sequence using plasmonics nanoprobes. Anal. Chem. 2005;77:7810–7815. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis R.M., Goodacre R. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev. 2008;37:931–936. doi: 10.1039/b705973f. [DOI] [PubMed] [Google Scholar]
- 12.Porter M.D., Lipert R.J., Siperko L.M., Wang G., Narayanan R. SERS as a bioassay platform: fundamentals, design, and applications. Chem. Soc. Rev. 2008;37:1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y.C., Jin R., Mirkin C.A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 14.Graham D., Mallinder B.J., Whitcombe D., Watson N.D., Smith W.E. Simple multiplex genotyping by surface-enhanced resonance Raman scattering. Anal. Chem. 2002;74:1069–1074. doi: 10.1021/ac0155456. [DOI] [PubMed] [Google Scholar]
- 15.Piorek B.D., Lee S.J., Santiago J.G., Moskovits M., Banerjee S. Free-surface microfluidic control of surface-enhanced Raman spectroscopy for the optimized detection of airborne molecules. Proc. Natl. Acad. Sci. USA. 2007;104:18898–18901. doi: 10.1073/pnas.0708596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murgida D.H., Hildebrandt P. Disentangling interfacial redox processes of proteins by SERR spectroscopy. Chem. Soc. Rev. 2008;37:937–945. doi: 10.1039/b705976k. [DOI] [PubMed] [Google Scholar]
- 17.Faulds K., Smith W.E., Graham D. Evaluation of surface-enhanced resonance Raman scattering for quantitative DNA analysis. Anal. Chem. 2004;76:412–417. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- 18.Stokes R.J., Macaskill A., Dougan J.A., Hargreaves P.G., Stanford H.M. Highly sensitive detection of dye-labelled DNA using nanostructured gold surfaces. Chem. Commun. 2007;27:2811–2813. doi: 10.1039/b705873j. [DOI] [PubMed] [Google Scholar]
- 19.Bell S.E.J., Sirimuthu N.M.S. Quantitative surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2008;37:1012–1024. doi: 10.1039/b705965p. [DOI] [PubMed] [Google Scholar]
- 20.Freeman R.G., Grabar K.C., Allison K.J., Bright R.M., Davis J.A. Self-assembled metal colloid monolayers: an approach to SERS substrates. Science. 1995;267:1629–1632. doi: 10.1126/science.267.5204.1629. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell D.J., Emory S.R., Nie S. Nanostructured thin-film materials with surface-enhanced optical properties. Chem. Mater. 2001;13:1082–1088. [Google Scholar]
- 22.Wang H., Levin C.S., Halas N.J. Nanosphere arrays with controlled sub-10-nm gaps as surface-enhanced Raman spectroscopy substrates. J. Am. Chem. Soc. 2005;127:14992–14993. doi: 10.1021/ja055633y. [DOI] [PubMed] [Google Scholar]
- 23.Baia M., Baia L., Astilean S. Gold nanostructured films deposited on polystyrene colloidal crystal templates for surface-enhanced Raman spectroscopy. Chem. Phys. Lett. 2005;404:3–8. [Google Scholar]
- 24.Kuncicky D.M., Prevo B.G., Velev O.D. Controlled assembly of SERS substrates templated by colloidal crystal films. J. Mater. Chem. 2006;16:1207–1211. [Google Scholar]
- 25.Sawai Y., Takimoto B., Nabika H., Ajito K., Murakoshi K. Observation of a small number of molecules at a metal nanogap arrayed on a solid surface using surface-enhanced Raman scattering. J. Am. Chem. Soc. 2007;129:1658–1662. doi: 10.1021/ja067034c. [DOI] [PubMed] [Google Scholar]
- 26.Qin L., Zou S., Xue C., Atkinson A., Schatz G.C. Designing, fabricating, and imaging Raman hot spots. Proc. Natl. Acad. Sci. USA. 2006;103:13300–13303. doi: 10.1073/pnas.0605889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y., Liu G.L., Kim J., Mejia Y.X., Lee L.P. Nanophotonic crescent moon structures with sharp edge for ultrasensitive biomolecular detection by local electromagnetic field enhancement effect. Nano Lett. 2005;5:119–124. doi: 10.1021/nl048232+. [DOI] [PubMed] [Google Scholar]
- 28.Lee S.J., Morrill A.R., Moskovits M. Hot spots in silver nanowire bundles for surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 2006;128:2200–2201. doi: 10.1021/ja0578350. [DOI] [PubMed] [Google Scholar]
- 29.Fromm D.P., Sundaramurthy A., Kinkhabwala A., Schuck P.J., Kino G.S. Exploring the chemical enhancement for surface-enhanced Raman scattering with Au bowtie nanoantennas. J. Chem. Phys. 2006;124:061101–061104. doi: 10.1063/1.2167649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Puebla R., Cui B., Bravo-Vasquez J.P., Veres T., Fenniri H. Nanoimprinted SERS-active substrates with tunable surface plasmon resonances. J. Phys. Chem. C. 2007;111:6720–6723. [Google Scholar]
- 31.McFarland A.D., Young M.A., Dieringer J.A., VanDuyne R.P. Wavelength-scanned surface-enhanced Raman excitation spectroscopy. J. Phys. Chem. B. 2005;109:11279–11285. doi: 10.1021/jp050508u. [DOI] [PubMed] [Google Scholar]
- 32.Zhang D., Xie Y., Deb S.K., Davison V.J., Ben-Amotz D. Isotope edited internal standard method for quantitative surface-enhanced Raman spectroscopy. Anal. Chem. 2005;77:3563–3569. doi: 10.1021/ac050338h. [DOI] [PubMed] [Google Scholar]
- 33.Graham D., Faulds K. Quantitative SERRS for DNA sequence analysis. Chem. Soc. Rev. 2008;37:1042–1051. doi: 10.1039/b707941a. [DOI] [PubMed] [Google Scholar]
- 34.Shamsaie A., Heim J., Yanik A.A., Irudayaraj J. Intracellular quantification by surface enhanced Raman spectroscopy. Chem. Phys. Lett. 2008;461:131–135. [Google Scholar]
- 35.Sun L., Yu C., Irudayaraj J. Raman multiplexers for alternative gene splicing. Anal. Chem. 2008;80:3342–3349. doi: 10.1021/ac702542n. [DOI] [PubMed] [Google Scholar]
- 36.Kimura-Suda H., Petrovykh D.Y., Tarlov M.J., Whitman L.J. Base-dependent competitive adsorption of single-stranded DNA on gold. J. Am. Chem. Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 37.Sun L., Yu C., Irudayaraj J. Surface-enhanced Raman scattering based nonfluorescent probe for multiplex DNA detection. Anal. Chem. 2007;79:3981–3988. doi: 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weitz D.A., Gramila T.J., Genack A.Z., Gersten J.I. Anomalous low-frequency Raman scattering from rough metal surfaces and the origin of surface-enhanced Raman scattering. Phys. Rev. Lett. 1980;45:355–358. [Google Scholar]
- 39.Kröger J. Phonons and electrons at metal surfaces. Appl. Phys. Mater. Sci. Process. 2007;87:345–350. [Google Scholar]
- 40.Nobile C., Fonoberov V.A., Kudera S., DellaTorre A., Ruffino A. Confined optical phonon modes in aligned nanorod arrays detected by resonant inelastic light scattering. Nano Lett. 2007;7:476–479. doi: 10.1021/nl062818+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


