Abstract
We present molecular dynamics simulations of a multicomponent, asymmetric bilayer in mixed aqueous solutions of sodium and potassium chloride. Because of the geometry of the system, there are two aqueous solution regions in our simulations: one mimics the intracellular region, and one mimics the extracellular region. Ion-specific effects are evident at the membrane/aqueous solution interface. Namely, at equal concentrations of sodium and potassium, sodium ions are more strongly adsorbed to carbonyl groups of the lipid headgroups. A significant concentration excess of potassium is needed for this ion to overwhelm the sodium abundance at the membrane. Ion-membrane interactions also lead to concentration-dependent and cation-specific behavior of the electrostatic potential in the intracellular region because of the negative charge on the inner leaflet. In addition, water permeation across the membrane was observed on a timescale of ∼100 ns. This study represents a step toward the modeling of realistic biological membranes at physiological conditions in intracellular and extracellular environments.
Introduction
Membranes are important components of biological cells because they separate the content of cells and organelles from the surrounding fluids. At the same time, membranes play an active role in regulating traffic across these open systems, and regulate communications among cells. Nature accomplishes all these tasks by designing membranes in the form of sophisticated bilayers containing mixtures of different lipids and proteins (1). The properties of lipids and of proteins in membranes are mutually adjusted to perform specific functions. In view of the enormous number of functions to be performed in different cells, it is not surprising that a wide variety of lipids and proteins can be found in membranes. This variety of proteins and especially lipids provides biological membranes with flexible electrostatic and elastic properties. By mixing lipids with different headgroups and different tails, and by building bilayers asymmetrically from two monolayers of different compositions, membranes can adjust their thickness to adopt proteins and exclude or include them, can change the membrane curvature, and can turn on or off electrostatic interactions (2).
A prime example of how nature can use variable properties of lipids in membranes involves lipid raft domains, which contain an enhanced number of sphingomyelin molecules and cholesterol (3,4). The roles of proteins in rafts for biological membranes, and even more fundamental issues such as the equilibrium or nonequilibrium nature of rafts, remain to be clarified (5). Another interesting issue related to rafts concerns the asymmetry of membranes (6). Although sphingomyelin is prevalent in the outer leaflet of a plasma membrane, it is unclear what lipid can play its role in the creation of rafts in the inner leaflet.
The distribution of salt ions inside and outside cells represents another pronounced asymmetry. While the dominant cations inside the cellular media are K+, Na+ ions are mostly located outside (1). This nonequilibrium situation is primarily maintained by an intricate system of ion pumps and channels, but a question arises over whether the asymmetry of the membrane and the fact that the inner leaflet bears a negative charge are also related to this ionic selectivity. Recent simulations and experiments show that ions display selective adsorptive properties toward lipid bilayers (7–14). What is the role of bilayer asymmetry and selective adsorption properties on the total electrostatic properties of membranes? How can membrane asymmetry influence membrane/peptide interactions that are screened by salt? To address some of the issues raised by these questions, we performed computer simulations of asymmetric model biological membranes in aqueous solutions containing KCl and NaCl salts, and we report on the results of these simulations here.
Systems and Methods
System composition
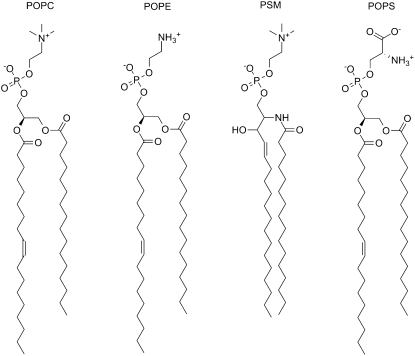
As mentioned in the Introduction, the composition of membranes found in biological cells is variable, depending on the biological species and type of organelle the membrane encloses. For example, the plasma membrane of the human erythrocyte is strongly asymmetric, i.e., the composition of the two leaflets in the bilayer is different. The detailed composition of the leaflets for this membrane was investigated, and the results are available elsewhere (15,16). To create a more biologically realistic model of an asymmetric membrane in our simulations, we chose to mimic the plasma membrane of the human erythrocyte. To this end, we constructed an asymmetric membrane containing four different lipids, three of them zwitterionic: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and N-palmitoyl sphingomyelin (PSM). The fourth lipid was anionic: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS). The chemical structures of these four mentioned phospholipids are depicted in Fig. 1.
Figure 1.
Schematic drawings (molecular structures) of lipids POPC, POPE, PSM, and POPS.
The outer leaflet of such a membrane consists mainly of phosphocholine (PC) and sphingomyelin (SM) lipids with a ratio of 1:1, whereas the inner leaflet contains about one third of the charged phosphoserine (PS) lipids. The rest are zwitterionic lipids, with the main contribution coming from phosphosthanolamine (PE) phospholipids. Because we can simulate only a small patch of a membrane containing ∼60 lipid molecules in one leaflet, lipids that make minor contributions to the composition of the human erythrocyte membrane were not considered. Nor did we include cholesterol in our present model, postponing the study of its influence for future research.
The exact number of lipids in our simulations was determined by a careful matching of the inner and outer leaflet areas, to prevent any bending and undulation of the membrane. To this end, we initially performed simulations of symmetric membranes containing mixed leaflets of different compositions, and compared the resulting membrane areas. The initial guess for the areas and numbers of lipids was based on the data for the area per lipid obtained from previous simulations (17–22). The best match was obtained for a bilayer composed of 30 POPC and 30 PSM lipids with an area of 32.7 nm2, and the other bilayer contained 44 POPE and 20 POPS lipids with an area of 32.6 nm2. Therefore, these compositions were adopted as the leaflet compositions of our model asymmetric membrane. A somewhat inferior match was obtained for another tested membrane that contained 43 POPE and 20 POPS lipids, and had an area of 31.9 nm2.
The unit cell in these simulations contained two such bilayers in a symmetric arrangement (Fig. 2). Fig. 2 shows two compositionally asymmetric bilayers oriented with their inner leaflets toward the box center, separated by aqueous solution. We denote the central region between the inner leaflets as the interior, which models the solution in the cell cytoplasm, whereas the extracellular solution is represented by an exterior region between the outer leaflets. The outer region is also continuous, because of the applied periodic boundary conditions.
Figure 2.
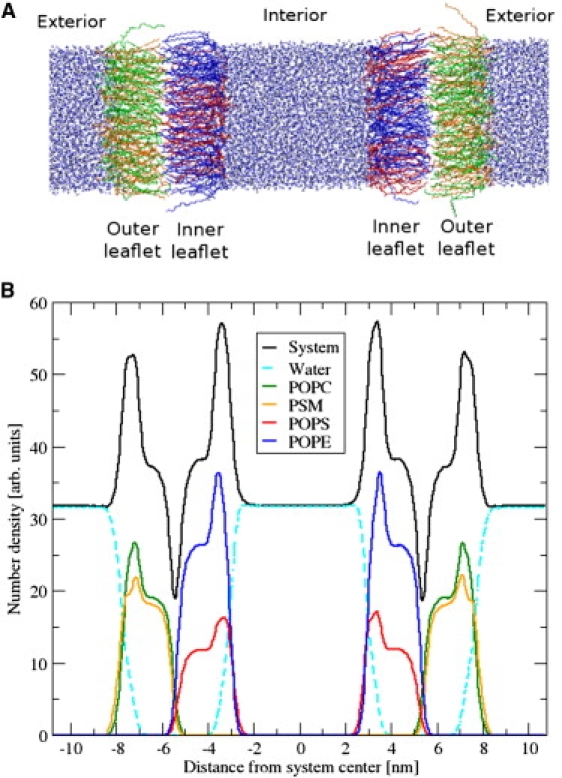
(A) System snapshot of double asymmetric membrane with separated interior and exterior solutions. (B) Number density profiles of our system as a function of distance perpendicular to the membrane surface. Data were averaged over 100 ns.
We investigated two different systems, and the compositions of the interior and exterior regions are summarized in Table 1. The first system, denoted as NaK-NaK, contained equal numbers of Na+ and K+ ions in both the interior and exterior solutions, resulting from a mixture of 200 mM NaCl and 200 mM KCl. In addition, the interior solution contained counterions to the negatively charged POPS lipids, which were also Na+ and K+ cations at a 1:1 ratio. This choice of equal and relatively high NaCl and KCl concentrations led to a fast convergence of results in terms of ion segregation at the membrane/solution interface. The second system was intended to mimic in a more realistic fashion the sodium/potassium imbalance in cells, and was denoted the NaK system. This setup contained a 200 mM NaCl solution in the exterior region. The interior region had 20 mM of Cl− anions, together with K+ counterions, which also compensated the charge of anionic lipids.
Table 1.
Exact compositions of studied systems
| NaK-NaK system (equal ion compositions) | NaK system (∼physiological ion composition) | |
|---|---|---|
| Outer leaflet | 30 POPC, 30 PSM | 30 POPC, 30 PSM |
| Inner leaflet | 44 POPE, 20 POPS | 44 POPE, 20 POPS |
| Interior | 6430 H2O, 44 Na+, 44 K+, 48 Cl− | 6430 H2O, 42 K+, 2 Cl− |
| Inner leaflet | 44 POPE, 20 POPS | 44 POPE, 20 POPS |
| Outer leaflet | 30 POPC, 30 PSM | 30 POPC, 30 PSM |
| Exterior | 6430 H2O, 24 Na+, 24 K+, 48 Cl− | 6430 H2O, 24 Na+, 24 Cl− |
Computational details
All simulations were performed using the program GROMACS, version 3.3.1 (23,24). The systems were kept within the constant temperature and pressure (NpT) ensemble using a semi-isotropic pressure coupling at 1 bar by means of a Parrinello-Rahman barostat with a coupling constant of 2 ps. Temperature coupling was established by a Nose-Hoover thermostat separately for the lipids and the solution, with a coupling time constant of 1.0 ps. Temperature was set at a slightly elevated value of 323 K, to prevent the bilayers from undergoing a possible phase transition. Short-range van der Waals interactions were cut off at 1 nm, and the effect of long-range Coulomb interactions was accounted for by using the particle mesh Ewald method (25), with a grid spacing of 0.12 nm and cubic interpolation. Application of the SETTLE algorithm (26) for constraining water and the LINCS algorithm (27) for lipids allowed us to use a 2 fs time step. Initially, lipids were randomly generated on an 8 × 8 array, followed by a minimization step to remove close contacts of atoms. Subsequently, simulations were performed for 260 ns. The first 160 ns were considered as equilibration, and discarded from further analysis. The equilibrated size of the unit cell was ∼5.7 × 5.7 × 21.5 nm, where membranes of a thickness of ∼3.7 nm were separated with 7 nm of solution.
Potential parameters for the lipids were based on the united-atom force field of Berger et al. (18), as defined for POPC (17,19), POPE (19), POPS (20), and PSM (22). For water, we used the Simple Point Charge (SPC) model (28) with which the lipid force field was developed, and the ionic parameters were taken from the GROMACS force field (24), with the exception of potassium. The depth of the Lennard-Jones potential for K+ is unrealistically small. Therefore, in previous studies, other parameters were used (13,29). We adopted a potassium ion (30) that was carefully parameterized by Dang et al. (30). In a lipid force field, all Lennard-Jones parameters for interactions between different atom types are explicitly defined. These values are typically obtained by using the combination rules and . Different pairs of C6 and C12 parameters are thus associated with each atom, to describe interactions with various partners. Therefore, to be fully consistent with the force field for Na+, we generated the nonbonded parameters for K+ interacting with any atom i, using the following relationships:
Results
Equilibration and membrane area
The time development of the membrane area provides important information about the equilibration process. This time development yields the averaged distributions (i.e., number of density profiles) of phospholipids across the unit cell, as depicted in Fig. 2 B. Membrane area is defined as the lateral area of the simulation box in the xy-plane. The equilibration process, monitored by the convergence of the membrane area within the unit cell (Fig. 3), exhibited two timescales. The first equilibration interval, during which a rapid decrease of membrane area was evident, lasted ∼20 ns. The second time regime, connected with a much slower decrease of membrane area, lasted ∼140 ns. After 160 ns, the systems reached equilibrium values of the membrane areas, i.e., 32.5 ± 0.2 nm2 for the NaK-NaK system, and 32.8 ± 0.3 nm2 for the NaK system (the errors are standard deviations). In terms of an averaged area per lipid, these values correspond to 0.542 nm2 for the outer leaflet and 0.508 nm2 for the inner leaflet in the NaK-NaK system, and 0.546 nm2 for the outer leaflet and 0.512 nm2 for the inner leaflet in the NaK system. At the same time, the equilibrium membrane thickness, measured as the distance between density peaks of lipids, was ∼3.7 nm. For an additional indication that the systems reached equilibrium, after 160 ns, the total number of all adsorbed ions fluctuated around constant mean values, and no drift was evident (Fig. 4).
Figure 3.
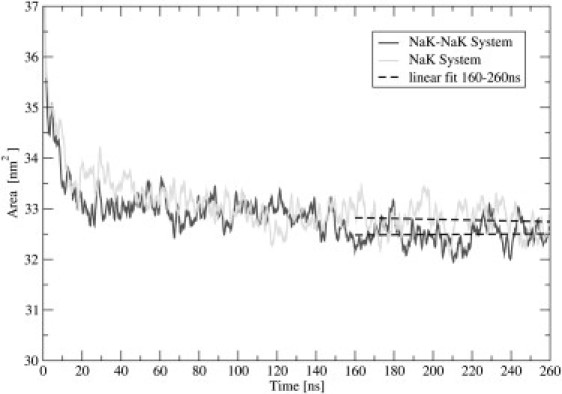
Time dependence of total membrane area within unit cell for NaK-NaK and Na-K systems. Dashed lines depict linear fits to last 100 ns.
Figure 4.
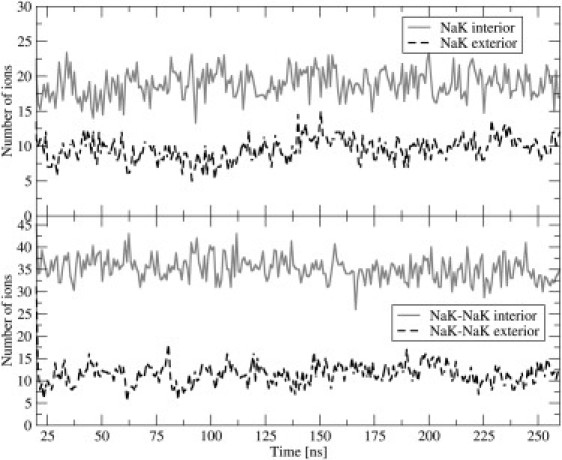
Total amounts of adsorbed ions at membrane, determined as number of ions within 0.7 nm from phosphate groups.
Adsorption of ions
The number density profiles of ions are plotted in Fig. 5. In the NaK system, Na+ was enhanced at the carbonyl region of the outer leaflet, whereas the density profile of K+ ions on the inner leaflet displayed two peaks: one at the carbonyl region, and one close to the carboxyl oxygens. The ion adsorption was stronger at the carbonyl compared with the carboxyl oxygens. In the NaK-NaK system, both cations were competing for adsorption on both sides of the membrane. The ion-adsorption pattern was the same as in the NaK system. The sodium cation was preferred over potassium, with the exception of the carboxyl region. The Cl− anion exhibits a weak enhancement at the outer leaflets, but no enhancement at the inner leaflets, in both systems under study.
Figure 5.
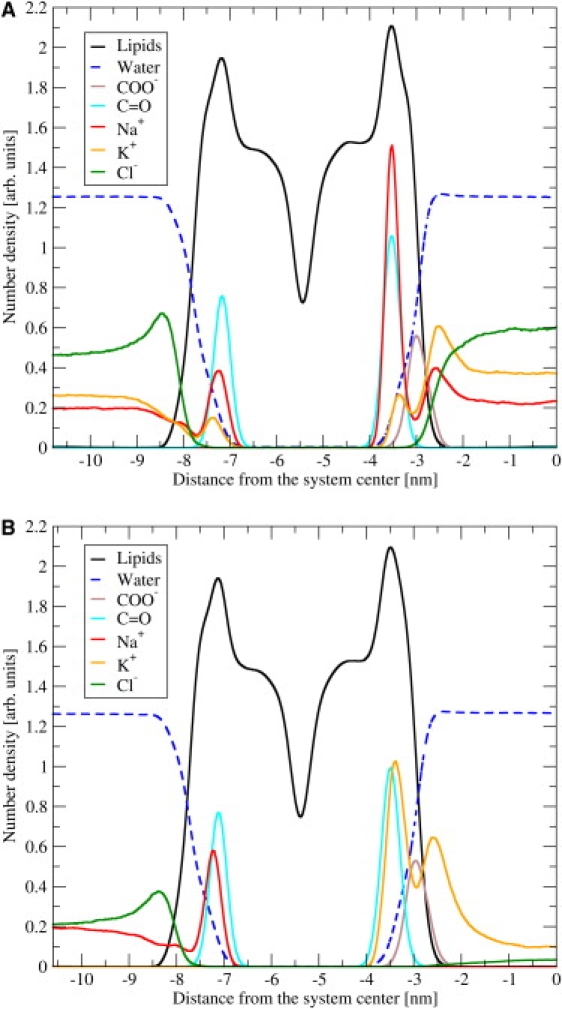
Number density profiles of ions across asymmetric membrane, with inner leaflet at right. For easier localization of ion peaks, density profiles of carbonyl and carboxyl oxygens are also depicted. Only half of the unit cell is shown. The results were averaged over two equivalent halves. (A) NaK-NaK system. (B) NaK system.
Ion adsorption was further quantified by evaluating the numbers of adsorbed ions. These were determined as the numbers of ions observed within a distance of 0.7 nm from the phosphate groups, averaged over the last 100 ns of the simulation run. This distance was chosen to include the highest peak of the radial distribution function between ions and phosphates (data not shown). The results are presented in Table 2. These data are consistent with the density profiles, and show stronger adsorption at the membrane of sodium over potassium. The residence times of adsorbed ions were calculated as the mean time that cations spend without interruption within a distance of 0.7 nm from the phosphate atoms. For potassium, the mean residence time was ∼50 ps, whereas for sodium it was about three times longer.
Table 2.
Number of ions adsorbed at membrane (with values per lipid in leaflet given in parentheses) and their mean residence times
| Adsorption |
Residence times (ps) |
|||
|---|---|---|---|---|
| Outer leaflet | Inner leaflet | Outer leaflet | Inner leaflet | |
| NaK-NaK system: Na+ | 8.0 (0.07) | 23.7 (0.18) | 316 | 294 |
| NaK-NaK system: K+ | 3.8 (0.03) | 10.7 (0.09) | 114 | 69 |
| NaK system: Na+ | 10.0 (0.08) | 465 | ||
| NaK system: K+ | 18.6 (0.15) | 133 | ||
Ion-lipid complexes
The weakly bound complexes between ions and lipids were analyzed in more detail, and the populations of individual complexes are shown in Fig. 6. An ion was considered to be in a complex with a lipid when any of the lipid atoms were within the ion's first solvation shell. The size of the first solvation shell was determined from the radial distribution function between the ion and lipids to be 0.275 nm for Na+, and 0.311 nm for K+. The ion-lipid separation was defined as the minimal distance between the ion and lipid atoms. The populations of complexes were normalized to the number of contacts.
Figure 6.
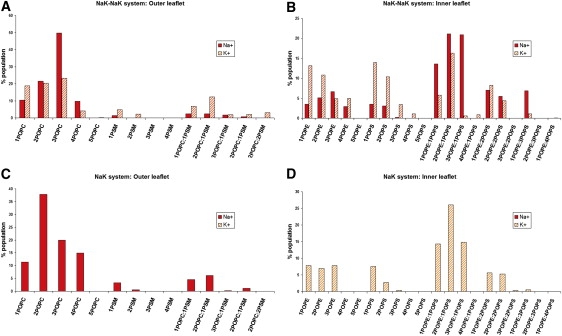
Populations (in percentages) of weakly bound complexes of lipids with sodium (solid) and potassium (dashed) normalized to total number of complexes for each ion. (A, B) NaK-NaK system. (C, D) NaK system.
First, we analyzed the NaK-NaK system (Fig. 6, A and B). At the outer leaflet, the ions create complexes mostly with POPC for both cations. More specifically, the most abundant complex is with three POPC lipids, followed by those with a pair of POPCs. Complexes with PSM are much less frequent, and are preferred by potassium slightly more than by sodium. At the inner leaflets, the complexes with three lipids were also the most populated. Sodium prefers mostly mixed complexes with POPE and POPS lipids (∼70%). The most abundant of these are complexes with 2 POPEs and 1 POPS, and with 3 POPEs and 1 POPS, which together represent almost half of all sodium complexes at the inner leaflet. For potassium, the complex with 2 POPEs and 1 POPS is also the most populated. However, compared with sodium, complexes containing potassium with a single lipid type are more abundant (30% with POPE, 30% with POPS, and 40% mixed).
The NaK system is simpler (Fig. 6, C and D). Sodium at the outer leaflet mainly forms complexes with POPC, so that complexes containing sodium and two POPCs are the most populated. Potassium in contact with the inner leaflet forms mostly mixed complexes, and the most common one contains 2 POPEs and 1 POPS molecule.
Electrostatic potentials
The electrostatic potentials depicted in Fig. 7 were calculated from charge densities, using Poisson's equation with periodic boundary conditions according to Sachs et al. (31). The absolute value of the potential was set at zero in the middle of the exterior solution, i.e., at the edge of the unit cell. It follows from the simulations presented here that the electrostatic potential across the membrane is zero for both systems containing salts. For comparison, we also evaluated the total electrostatic potential for the system without added salt (Fig. 7 A). For this system, a total electrostatic potential of 85 mV across the membrane was found. The error in calculations is estimated to be on the order of 10 mV, judging by the asymmetric character of the potential profile. The shape of the potential profile in the interior is system-dependent: it decreases from the membrane surface toward the center of the interior solution in the NaK-NaK system, but it increases in the NaK system. Fig. 7 B shows a decomposition of the electrostatic potential into contributions from two overall neutral subsystems: lipids and ions as one subsystem, and water as the other. As is usually the case in hydrated bilayers, water efficiently compensates the electric fields attributable to the lipid headgroups and ions. Consistent with the screening effect of salt ions, at high salt concentrations, the electrostatic profile is flat in the interior solution, whereas the effect of the headgroups propagates deeper into water at low salt.
Figure 7.
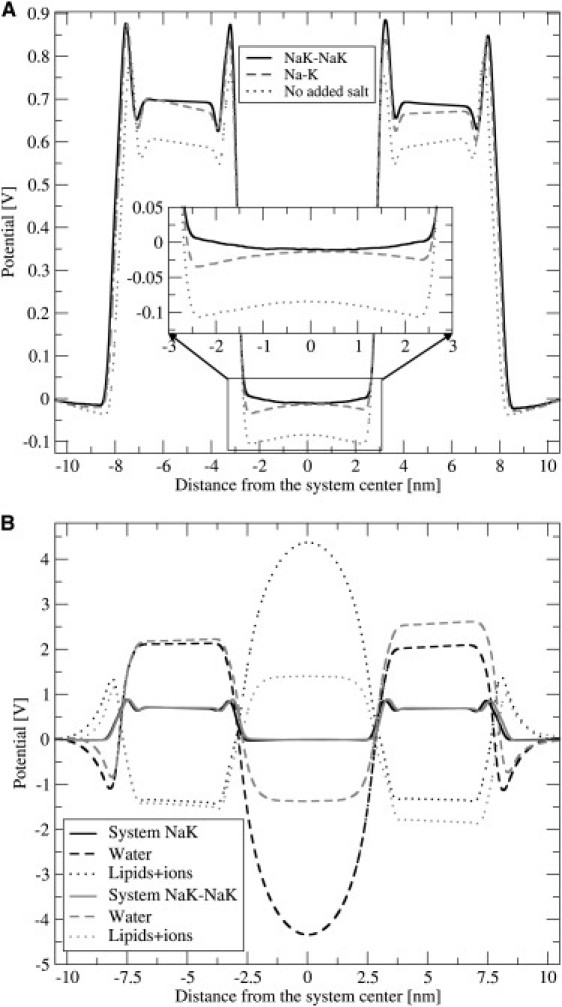
(A) Total electrostatic potentials of both studied systems, plus system with no added salt (where only counterions for POPS were present). The inlet shows the magnified electrostatic potential of inner solution. (B) Partial electrostatic potentials for NaK-NaK and Na-K systems.
Water permeation
During the simulations presented here, we observed water permeation through the membrane. In total, three water molecules penetrated the membrane from the exterior to the interior in the NaK-NaK system, whereas eight waters moved in the opposite direction during the last 100 ns of the simulation. Similar permeation was also evident for the NaK system, where 10 water molecules penetrated from the exterior to the interior, and three waters moved in the opposite way. There were also events where water penetrated inside the bilayer reemerging later at the original solvent side. The density of water inside the membrane can be roughly estimated from water-density profiles, based on the observed 24 permeating events, to be three orders of magnitude lower than in bulk water. There was no penetration of ions within the whole period of 260 ns of the simulation run.
The process of water permeation through the membrane is depicted as snapshots in Fig. 8. Fig. 9 depicts four representative trajectories of water molecules, projected in the direction perpendicular to the membrane. Observations based on Figs. 8 and 9 allow us to divide the permeation process into motion within distinct regions:
-
1.
Diffusion in aqueous solution;
-
2.
Motion across the membrane interface, particularly across the carbonyl region, where water was observed to reside for up to several nanoseconds, and to diffuse slowly;
-
3.
Motion across the hydrophobic carbon chain region, where water diffuses very fast. The maximum observed duration was tens of picoseconds;
-
4.
Motion in the middle of the membrane, where the density of chains is low, and the water molecule remains for up to nanoseconds and is very mobile.
Finally, we found that the permeating water molecule forms a hydrogen bond with the carbonyl oxygen while moving from the headgroup region into the tail region. This may help stabilize the water molecule while it is loosing hydrogen bonds with other waters.
Figure 8.
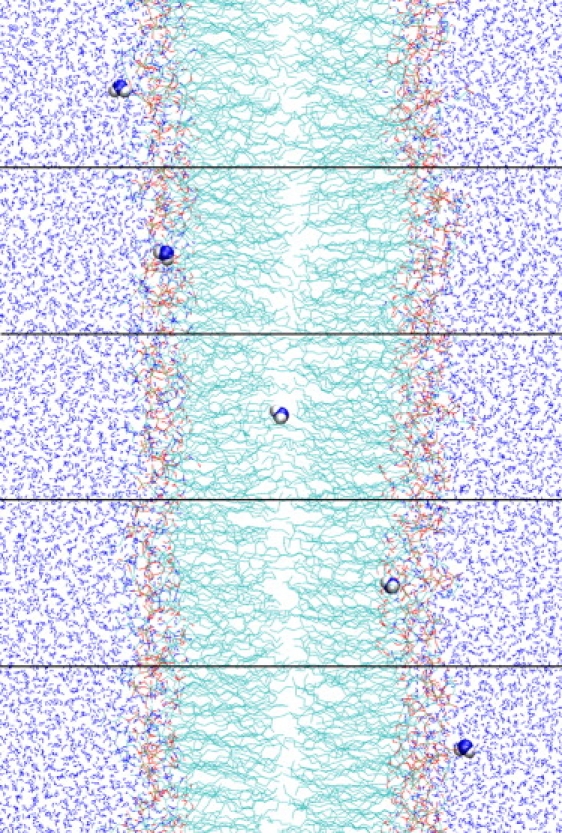
Five snapshots depict process of water permeation.
Figure 9.
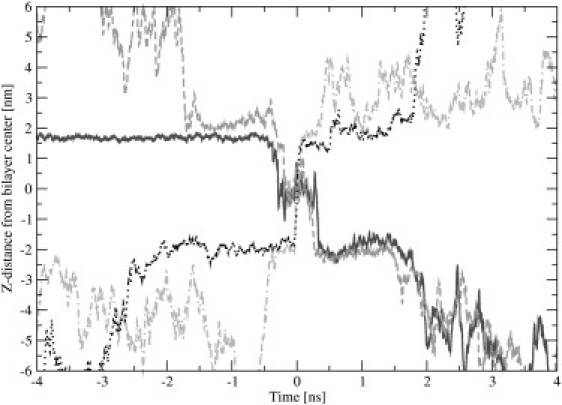
Four representative trajectories of water molecules penetrating through bilayer. Only the coordinate perpendicular to the bilayer is plotted, and time is synchronized to moment when water molecule is in center of membrane for each trajectory.
Lipid clustering
Fig. 10 contains snapshots of the membrane at time points of 40 ns and 200 ns (snapshots at the end of the simulation at 260 ns are very similar). Based on top views of both the outer and inner leaflets, we conclude that during the several hundreds of nanoseconds of the simulation, no domains were created, and the distribution of lipids remained random, as generated at the beginning.
Figure 10.
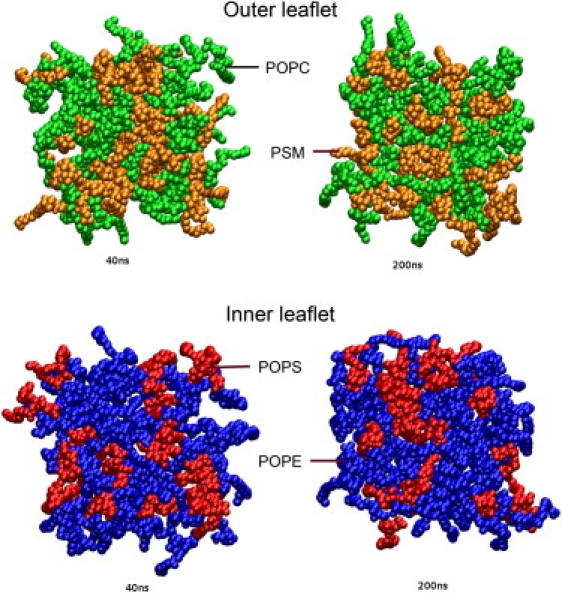
Snapshots of system at 40 ns and 200 ns. Top views of both inner and outer leaflets.
Discussion
Equilibration and membrane area
We observed in our simulations that the equilibration time was relatively long, at ∼160 ns, and that it contained two time regimes: a fast one of ∼20 ns, followed by a period of more than 100 ns within which the system slowly reached equilibrium (Fig. 3 depicts the equilibration process, monitored in terms of the total membrane area in the unit cell). We also observed that the equilibrated area per lipid was a bit smaller in the NaK-NaK system compared with the NaK system, consistent with a previously observed decrease of the area per POPC lipid for higher NaCl concentrations (21).
Adsorption of ions
Our simulations show that sodium ions adsorb more strongly than potassium on both sides of an asymmetric membrane. This difference is more pronounced at the inner leaflet, which contains negatively charged lipids. The strength and spatial localization of the adsorption of alkali cations are consistent with the longer residence times for smaller cations (32), and with previous studies of single-lipid membranes using the same force field (11,13,14,20,21,33,34). The observed double peak of cationic density profiles next to the inner leaflet is in agreement with results for a pure POPS bilayer (20). Therefore, we conclude that the adsorption of cations at the inner leaflet is mainly determined by anionic POPS, even though POPS represents only one third of the lipids in the inner leaflet. Not surprisingly, and consistent with previous observations for pure POPS (20), there was no enhancement of Cl− at the inner leaflet.
In general, sodium is adsorbed at the membrane more than potassium. The only exception is the carboxyl-group region of the inner leaflet in the NaK-NaK system, which constitutes a finite concentration effect. Because of the strong sodium adsorption to the carbonyl region, there are not as many sodium cations left in the solution as there are potassium cations. Subsequently, we find the preferred adsorption of potassium on carboxyl, despite the different affinities. Indeed, if one constructs normalized radial distribution functions between the carboxyl group and cations (data not shown), sodium is the preferred ion. Moreover, the most preferred configuration between the carboxyl group and alkali cations is a water-separated pair, with an oxygen-cation distance of ∼0.4 nm. This is likely due to the vicinity of the positively charged ammonium group, which leads to a destabilization of the contact ion pair that is otherwise formed (e.g., for acetate anion in solution) (35).
Ion-lipid complex
At the outer leaflets, the most important complexes with sodium and potassium ions were those with POPC lipids for both systems under investigation. This behavior likely occurs because the PSM lipid has only one carbonyl oxygen, and its partial charge is smaller than the one on POPC. The most common ion-lipid complexes had 2 POPCs in the NaK system, but 3 POPCs in the NaK-NaK system, which points to an effect of the concentration.
The inner leaflets contained mostly mixed complexes, with sodium preferred over potassium. Thus, none of the lipid-type molecules are strongly preferred by the ions, even though the POPS lipids are negatively charged. The most populated are complexes with three lipids, and the most abundant contain 2 POPE and 1 POPS molecules. Incidentally, this complex has the same ratio of lipids as the ratio in the leaflet. Although sodium prefers mixed lipid complexes, the more weakly bound potassium is left to form complexes mainly with a single type of lipid.
Electrostatic potentials
The observed vanishing electrostatic potential across the membrane in salt solutions can be attributed to the effect of ion screening of the membrane surface. The negative membrane potential is observed for systems containing negatively charged lipids without added salt, consistent with the results of a recent study of pure PS/PC and PS/PE membranes (36), and is caused by incomplete screening of negatively charged lipids. In salt solution (the NaK system), cations are adsorbed to the membrane, leading to the screening of electrostatic potential at shorter distances. At higher salt concentrations (the NaK-NaK system), the ion adsorption becomes strong enough to overcompensate the POPS negative charge. This explains why the electrostatic potential monotonously approaches zero without acquiring negative values (as in previous cases).
The observed slightly negative potential on the outer leaflets is in accordance with previous studies of zwitterionic lipids (21,32,33), and was shown to be caused by water orientation at the headgroups (21).
Water permeation
Water permeation through membrane has been found to be a relatively fast (in nanoseconds) process for all 24 observed cases. Diffusion of a water molecule through the hydrophobic tails was faster than in bulk water (of the order of tens of ps) peaking in the middle of the membrane. The rate limiting step was the initial water permeation at the carbonyl region which took up to several ns. This is in agreement with recent experiments (37,38), where it was shown that water permeation is strongly dependent on the area per lipid rather than on the membrane thickness. Our results are also consistent with previous simulations (39–41), where the free-energy profile and local diffusion of water through a PC membrane was calculated. In these studies, a free-energy barrier at the dense part of the lipid tails, located below the carbonyl region, was found to be a limiting step for water to enter the hydrophobic part of the membrane. Furthermore, there is a small free energy minimum in the middle of the membrane that codetermines water transport through lipids. Marrink and Berendsen (41) also calculated the water lateral diffusion rate, which was found to be highest in the middle of the membrane, decreasing toward the carbonyl region, and increasing again upon moving toward the aqueous bulk, in agreement with our findings.
Conclusions
In our simulation, we increased the complexity level of previously simulated single-component membranes or leaflets in water or salt solutions by means of a multicomponent asymmetric bilayer in aqueous solutions containing mixtures of salts. The choice of the four particular lipids, and of sodium and potassium chloride salts, was dictated by biological considerations. The study of such systems brings us closer to a description of real biological membranes. Moreover, thanks to the geometry of our unit cell, which contained two bilayers, the aqueous solutions in our systems were represented by two pools (exterior and interior). This also represents a step toward biological reality, because membranes separate different intracellular and extracellular solutions.
We observed in our simulations that specific effects play a role in the distribution of ions at the membrane/aqueous solution interface. Thus, at equal concentrations, Na+ ions are more strongly adsorbed to the carbonyl groups of lipid molecules than K+ ions, because of specific interactions (42). Only in cases of concentration excess can K+ win over Na+ in terms of abundance at the membrane. The preference for sodium over potassium in the headgroup region is consistent with our previous simulations of a single-component DOPC membrane, using an all-atom force field (43). Furthermore, detailed results on the specific character of ion-lipid headgroup interactions somewhat depend on the force field in use. In our previous simulation with the all-atom force field, the preferred adsorption site for cations was near the phosphate group (43). A simulation using the CHARMM force field, which has a larger sodium radius, showed a weaker surface preference (31). This is consistent with the weaker surface affinity of potassium over sodium observed here. In addition, we observed that cations can form various weak complexes with more than a single headgroup. The most common complexes are those with three lipids. In the outer leaflet, the most abundant complex is with 3 POPCs, whereas in the inner leaflet, the most abundant complex is with 2 POPE and 1 POPS molecules. An interesting question arises whether the observed preference for Na+ over K+ by the membrane interior surface has anything to do with the strong asymmetry in concentration of these ions in the interior and exterior of the cell, as maintained by ion pumps and channels.
Concentration-dependent specific ion-membrane interactions are also evident in the behavior of the electrostatic potential in the inner pool of the aqueous solution. The charge on the negative inner membrane can be partially screened by salt and counterions (in the case of NaK) or even slightly overcompensated (in the NaK-NaK system) by the adsorption of ions from the solution. Moreover, the concentration of ions, even in the case of the NaK system, which is of physiological relevance, is large enough to produce effective screening, which reduces the potential to zero in the middle of the relatively small aqueous interior pool.
We also observed that water can permeate across the membrane in our simulations, and the qualitative features describing this permeation are consistent with those observed for systems simpler than ours (41). The presence of ions produces a slight condensation of the lipid bilayer, but we did not observe the creation of domains that are speculated to exist in membranes because of the salt effect (44). This can be attributed to the limited duration (260 ns) of our simulation, which also prevented us from observing passive ion transport across membranes possibly facilitated by the creation of water channels. To model faithfully the transport of ions across channels and active transport across pumps, one needs to include proteins in bilayers that contain an asymmetry in their leaflet compositions, to regulate both the mechanical and electrical properties of the protein. These electrical properties can be accomplished by adjusting, via salt-screening and adsorption, the cross-membrane potential. Our simulation thus represents a step toward detailed studies of the complicated biological membrane/aqueous solution interface.
Acknowledgments
R.V. acknowledges support from the Grant Agency of Charles University in Prague (grant 99408), the Czech Science Foundation (grant 203/05/H001), and the International Max-Planck Research School. Support to P.J. from the Czech Ministry of Education (grant LC512) and the Czech Science Foundation (grant 203/08/0114) is gratefully acknowledged. Part of the work in Prague was supported by the governmental Project Z40550506. M.L.B. acknowledges support from National Science Foundation grant MCB-0615469.
Supporting Material
References
- 1.Alberts B., Johnson A., Lewis J., Raff M., Roberts K. Garland Science; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- 2.Parthasarathy R., Groves J.T. Curvature and spatial organization in biological membranes. Soft Matter. 2007;3:24–33. doi: 10.1039/b608631d. [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 4.Pike L.J. Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Epand R.M. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Devaux P.F., Morris R. Transmembrane asymmetry and lateral domains in biological membranes. Traffic. 2004;5:241–246. doi: 10.1111/j.1600-0854.2004.0170.x. [DOI] [PubMed] [Google Scholar]
- 7.Aroti A., Leontidis E., Dubois M., Zemb T. Effects of monovalent anions of the Hofmeister series on DPPC lipid bilayers part I: swelling and in-plane equations of state. Biophys. J. 2007;93:1580–1590. doi: 10.1529/biophysj.106.094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leontidis E., Aroti A., Belloni L., Dubois M., Zemb T. Effects of monovalent anions of the Hofmeister series on DPPC lipid bilayers part II: modeling the perpendicular and lateral equation-of-state. Biophys. J. 2007;93:1591–1607. doi: 10.1529/biophysj.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Celma J.J., Hatahet L., Kunz W., Fendler K. Specific anion and cation binding to lipid membranes investigated on a solid supported membrane. Langmuir. 2007;23:10074–10080. doi: 10.1021/la701188f. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Manyes S., Oncins G., Sanz F. Effect of temperature on the nanomechanics of lipid bilayers studied by force spectroscopy. Biophys. J. 2005;89:4261–4274. doi: 10.1529/biophysj.105.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.J., Song Y., Baker N.A. Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+-lipid interactions and finite size effects. Biophys. J. 2008;94:3565–3576. doi: 10.1529/biophysj.107.116335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurtovenko A.A., Vattulainen I. Lipid transmembrane asymmetry and intrinsic membrane potential: two sides of the same coin. J. Am. Chem. Soc. 2007;129:5358–5359. doi: 10.1021/ja070949m. [DOI] [PubMed] [Google Scholar]
- 13.Gurtovenko A.A., Vattulainen I. Effect of NaCl and KCl on phosphatidylcholine and phosphatidylethanolamine lipid membranes: insight from atomic-scale simulations for understanding salt-induced effects in the plasma membrane. J. Phys. Chem. B. 2008;112:1953–1962. doi: 10.1021/jp0750708. [DOI] [PubMed] [Google Scholar]
- 14.Berkowitz M.L., Bostick D.L., Pandit S. Aqueous solutions next to phospholipid membrane surfaces: Insights from simulations. Chem. Rev. 2006;106:1527–1539. doi: 10.1021/cr0403638. [DOI] [PubMed] [Google Scholar]
- 15.Virtanen J.A., Cheng K.H., Somerharju P. Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model. Proc. Natl. Acad. Sci. USA. 1998;95:4964–4969. doi: 10.1073/pnas.95.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zachowski A. Phospholipids in animal eukaryotic membranes—transverse asymmetry and movement. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tieleman D.P., Sansom M.S.P., Berendsen H.J.C. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys. J. 1999;76:40–49. doi: 10.1016/S0006-3495(99)77176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger O., Edholm O., Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tieleman D.P., Berendsen H.J.C. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 1998;74:2786–2801. doi: 10.1016/S0006-3495(98)77986-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay P., Monticelli L., Tieleman D.P. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys. J. 2004;86:1601–1609. doi: 10.1016/S0006-3495(04)74227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bockmann R.A., Hac A., Heimburg T., Grubmuller H. Effect of sodium chloride on a lipid bilayer. Biophys. J. 2003;85:1647–1655. doi: 10.1016/S0006-3495(03)74594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemela P., Hyvonen M.T., Vattulainen I. Structure and dynamics of sphingomyelin bilayer: insight gained through systematic comparison to phosphatidylcholine. Biophys. J. 2004;87:2976–2989. doi: 10.1529/biophysj.104.048702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berendsen H.J.C., Vanderspoel D., Vandrunen R. GROMACS—a message-passing parallel molecular-dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 24.Lindahl E., Hess B., van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 25.Darden T., York D., Pedersen L. Particle mesh Ewald—an N.LOG(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 26.Miyamoto S., Kollman P.A. SETTLE—an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 27.Hess B., Bekker H., Berendsen H.J.C., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 28.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 29.Gurtovenko A.A., Vattulainen I. Ion leakage through transient water pores in protein-free lipid membranes driven by transmembrane ionic charge imbalance. Biophys. J. 2007;92:1878–1890. doi: 10.1529/biophysj.106.094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang L.X., Schenter G.K., Glezakou V.A., Fulton J.L. Molecular simulation analysis and x-ray absorption measurement of Ca2+, K+ and Cl− ions in solution. J. Phys. Chem. B. 2006;110:23644–23654. doi: 10.1021/jp064661f. [DOI] [PubMed] [Google Scholar]
- 31.Sachs J.N., Crozier P.S., Woolf T.B. Atomistic simulations of biologically realistic transmembrane potential gradients. J. Chem. Phys. 2004;121:10847–10851. doi: 10.1063/1.1826056. [DOI] [PubMed] [Google Scholar]
- 32.Siu S.W.I., Vácha R., Jungwirth P., Bockmann R.A. Biomolecular simulations of membranes: physical properties from different force fields. J. Chem. Phys. 2008;128:125103. doi: 10.1063/1.2897760. [DOI] [PubMed] [Google Scholar]
- 33.Pandit S.A., Bostick D., Berkowitz M.L. Mixed bilayer containing dipalmitoylphosphatidylcholine and dipalmitoylphosphatidylserine: lipid complexation, ion binding, and electrostatics. Biophys. J. 2003;85:3120–3131. doi: 10.1016/S0006-3495(03)74730-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordomi A., Edholm O., Perez J.J. Effect of ions on a dipalmitoyl phosphatidylcholine bilayer. A molecular dynamics simulation study. J. Phys. Chem. B. 2008;112:1397–1408. doi: 10.1021/jp073897w. [DOI] [PubMed] [Google Scholar]
- 35.Jagoda-Cwiklik B., Vácha R., Lund M., Srebro M., Jungwirth P. Ion pairing as a possible clue for discriminating between sodium and potassium in biological and other complex environments. J. Phys. Chem. B. 2007;111:14077–14079. doi: 10.1021/jp709634t. [DOI] [PubMed] [Google Scholar]
- 36.Gurtovenko A.A., Vattulainen I. Membrane potential and electrostatics of phospholipid bilayers with asymmetric transmembrane distribution of anionic lipids. J. Phys. Chem. B. 2008;112:4629–4634. doi: 10.1021/jp8001993. [DOI] [PubMed] [Google Scholar]
- 37.Mathai J.C., Tristram-Nagle S., Nagle J.F., Zeidel M.L. Structural determinants of water permeability through the lipid membrane. J. Gen. Physiol. 2008;131:69–76. doi: 10.1085/jgp.200709848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagle J.F., Mathai J.C., Zeidel M.L., Tristram-Nagle S. Theory of passive permeability through lipid bilayers. J. Gen. Physiol. 2008;131:77–85. doi: 10.1085/jgp.200709849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinoda K., Shinoda W., Mikami M. Efficient free energy calculation of water across lipid membranes. J. Comput. Chem. 2008;29:1912–1918. doi: 10.1002/jcc.20956. [DOI] [PubMed] [Google Scholar]
- 40.Bemporad D., Luttmann C., Essex J.W. Behaviour of small solutes and large drugs in a lipid bilayer from computer simulations. Biochim. Biophys. Acta. 2005;1718:1–21. doi: 10.1016/j.bbamem.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Marrink S.J., Berendsen H.J.C. Simulation of water transport through a lipid-membrane. J. Phys. Chem. 1994;98:4155–4168. [Google Scholar]
- 42.Vrbka L., Vondrasek J., Jagoda-Cwiklik B., Vacha R., Jungwirth P. Quantification and rationalization of the higher affinity of sodium over potassium to protein surfaces. Proc. Natl. Acad. Sci. USA. 2006;103:15440–15444. doi: 10.1073/pnas.0606959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vácha R., Siu S.W.I., Petrov M., Böckmann R.A., Jurkiewicz P. Effects of alkali cations and halide anions on the DOPC lipid membrane. J. Phys. Chem. B. 2009 doi: 10.1021/jp809974e. In press. [DOI] [PubMed] [Google Scholar]
- 44.Reviakine I., Simon A., Brisson A. Effect of Ca2+ on the morphology of mixed DPPC-DOPS supported phospholipid bilayers. Langmuir. 2000;16:1473–1477. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.