Abstract
We present a coarse residue-based computational method to rapidly compute the solution scattering profile from a protein with dynamical fluctuations. The method is built upon a coarse-grained (CG) representation of the protein. This CG representation takes advantage of the intrinsic low-resolution and CG nature of solution scattering data. It allows rapid scattering determination from a large number of conformations that can be extracted from CG simulations to obtain scattering characterization of protein conformations. The method includes several important elements, effective residue structure factors derived from the Protein Data Bank, explicit treatment of water molecules in the hydration layer at the surface of the protein, and an ensemble average of scattering from a variety of appropriate conformations to account for macromolecular flexibility. This simplified method is calibrated and illustrated to accurately reproduce the experimental scattering curve of Hen egg white lysozyme. We then illustrated the applications of this CG method by computing the solution scattering patterns of several representative protein folds and multiple conformational states. The results suggest that solution scattering data, when combined with the reliable computational method that we developed, show great potential for a better structural description of multidomain complexes in different functional states, and for recognizing structural folds when sequence similarity to a protein of known structure is low.
Introduction
Small-angle x-ray solution scattering (SAXS) is an increasingly powerful technique to characterize structurally large macromolecular complexes (1–6). It takes less effort for sample preparation relative to crystallography, and avoids the challenge of growing crystals of good diffraction quality. It provides native structural data at physiological conditions such as in NMR, but without the inherent size limitations. It also allows rapid data collection with the current high flux synchrotron sources. The tradeoff is that only low-resolution information (in the range of 10–50 Å; see (1) for more discussion about SAXS resolution) about overall shape can be obtained because of the spherical averaging of protein scattering from multiple random orientations adopted in solution.
Low-resolution SAXS data can be combined with computational methods to reconstruct low-resolution structural models of large multidomain complexes. The scattering data can serve as independent constraints for computational modeling to ultimately characterize the structures/shapes of large protein complexes, especially when the structure of each individual domain in the complex is known at high resolution (1). This combination of solution scattering with computation and atomic resolution structures from crystallography provides an alternative approach to achieving structural characterization of multidomain proteins (1). Recent studies suggest that such a combination can be used to obtain the structural multiplicity of multidomain complexes in solution (7,8). However, this application is often limited by the efficiency of the scattering calculations for a large number of conformations generated from extensive sampling in the configurational space. Before this combination can be achieved, we need to develop a rapid scattering determination method to efficiently and accurately compute the scattering profiles so that it can be productively applied to an ensemble of protein states, e.g., tens of thousands of protein configurations.
Recently, several computational approaches have been introduced to address this question, at both the all-atom and coarse-grain levels of detail. In most current all-atom methods, the details of both the protein itself and the surrounding water molecules are approximately taken into account (9–12). One of the most widely used all-atom methods is provided by the CRYSOL program (9), which has been shown to be quite successful in many cases (13). The current treatment in CRYSOL makes assumptions about the structure of primary hydration shell around the protein using an implicit solvent model and about the electron density in the shell. Additionally, the treatment of the density contrast of bound waters in the hydration shell relative to the remaining bulk water remains uncertain (14–17). For large macromolecules, the conformational flexibility must be also taken into account. This flexibility is an important aspect of multidomain complexes and enzymes (18,19) and can be reflected in dynamic transitions among those states accessible to a protein in solution (20). Complete and accurate computation of SAXS patterns would require that it be included in any model.
Because atomic details are included, these all-atom calculations are often computationally expensive for large complexes, especially when a large number of configurations is involved. Alternative methods for computing scattering patterns involve coarse-graining molecular representations. These are essentially based on the nature of SAXS as a low-resolution technique, making it well suited for use with coarse-graining. Ideally, it should significantly reduce computer time without compromising accuracy. Along this direction, multiple levels of coarse-grained (CG) models have been introduced, including a simple Cα model (21), a side-chain CG model (primarily for diffraction) (22), and a dummy-residue (DR) model (23). In the Cα model, all amino acids are assigned with the same number of electrons at the Cα positions (21,24). This simple Cα model approach has reportedly improved the prediction ability of protein folding and structure prediction (for example, see (25,26)). In the side-chain coarse-graining model, a procedure of simplifying each residue into backbone and side-chain groups was aimed to obtain low-resolution interpretation for diffraction data (22,27,28). In the DR model, each residue is represented by its Cα atom and explicit water molecules are used in the hydration shell (23). In addition, the use of a correction function for the structure factor of each residue was enforced to reproduce the CRYSOL-calculated scattering curves. Furthermore, in the DR treatment of the hydration shell, the density of explicit water molecules used to represent the hydration shell density is much lower than that of the bulk solvent.
Inspired by the successes of CRYSOL and the DR model, we aim here at developing a CG model that can be used to efficiently and accurately compute scattering curves from a given protein conformation. In this CG model, three major elements are addressed as follows.
First, a knowledge-based structure factor for each residue is developed based on their atomic models as in the Protein Data Bank (PDB). This differs from the Cα model by including a CG structure factor for each residue. It also differs from the DR model by avoiding the introduction of a correction function since the effective structure factors are essentially derived from experimentally observed conformers. Clearly, this coarse-graining is based on the low-resolution nature of SAXS data.
Second, an explicit solvent layer of dummy water molecules is placed around the protein, similar to the DR model, but with a proper electron density set to account for the excess electron density of the hydration layer relative to bulk solvent. An effective structure factor of dummy waters is derived and a proper weight is assigned based on the experimental data of lysozyme.
Third, we use molecular dynamics (MD) simulations to account for the conformational flexibility that occurs in proteins in solution. The CG model is capable of reproducing the SAXS scattering data of lysozyme accurately. More importantly, it provides a rapid determination method for computing scattering profiles with an ensemble of states incorporated. We further apply the CG model to characterize a variety of protein folds and multiple conformational states in terms of their distinct scattering profiles. This rapid computational method, when combined with solution scattering data and atomic resolution structures of individual components, is well positioned to provide a powerful tool to shape-reconstruct large multidomain complexes and to determine the population fraction of different conformational states of protein under different physiological conditions.
Theoretical background and developments
The SAXS from protein in solution essentially measures the difference in the electron density between protein molecules and bulk solvent. Proteins have an average electron density of ρm ∼0.43 e/Å3 (29), whereas pure water has an electron density of ρs ∼ 0.334 e/Å3 at 20°C (30). The difference makes SAXS particularly attractive for resolving the electron density contrast and potentially determining protein structures. In practice, the scattering curve I(q) is determined from the total scattering of protein samples by subtracting the capillary scattering and background buffer scattering after correction for the volume of buffer displaced by the protein (1,31). Theoretically, the intensity from dilute samples is proportional to the spherically averaged scattering of a single molecule minus the excluded volume contributions but with the hydration shell excess density (3),
| (1) |
where the amplitude of the wavevector transfer q = |q| = 2π/d = 4π sin θ/λ (d is the Bragg spacing, 2θ is the scattering angle, and λ is the x-ray wavelength). Am(q) is the scattering amplitude from the protein molecule in vacuum, As(q) is from the solvent with an excluded volume displaced by the protein, and Ab(q) is from the shell of bound waters reflected in the density excess (Δρb) relative to the bulk phase (29). The quantity stands for an average over all orientations in reciprocal space to account for the nature of protein adopting random orientations in solution.
Equation 1 provides the theoretical basis for solution scattering. For a given protein conformation with N spherical atoms (e.g., Fig. 1), the scattering is contributed from 1), the protein itself in vacuum; 2), the excluded volume of solvent displaced by the protein; 3), the relative electron density in the hydration shell; and 4), in addition, protein conformational flexibility by the ensemble average over all conformations that are accessible to protein motions in solution. The ensemble average differs from the orientational average in Eq. 1. We briefly describe these four aspects as follows. First, the scattering I(q) from the protein itself is calculated by the Debye formula
| (2) |
where fi values are atomic form factors (i = 1, ···, N) and rij values are the interparticle distances. At the limit of q → 0, fi values are the electron numbers of each atom. Fig. 2 shows the scattering factor curves fi for C, N, O, H, and S using the Cromer-Mann scattering-factor coefficients (32).
Figure 1.
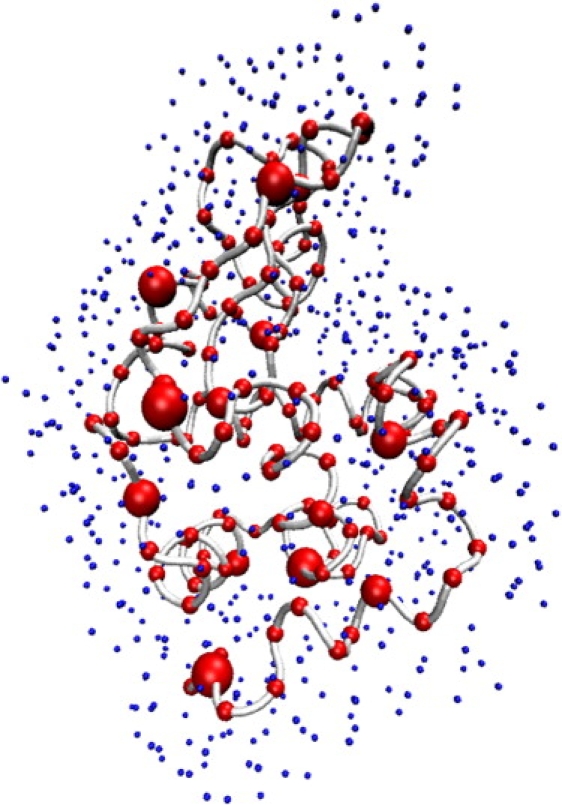
Schematic representation of lysozyme surrounded by a layer of “dummy” waters to model the local difference in relative solvent density at the surface of the protein. The scattering from a given protein conformation is conveniently and accurately represented by its N residues (red) via the Cα position, with M explicit water molecules (blue) inserted 3.5–6.5 Å away from Cα atoms to represent the hydration shell. Conformational flexibility also contributes to the scattering because of the intrinsic motions that are accessible to protein dynamics in solution.
Figure 2.
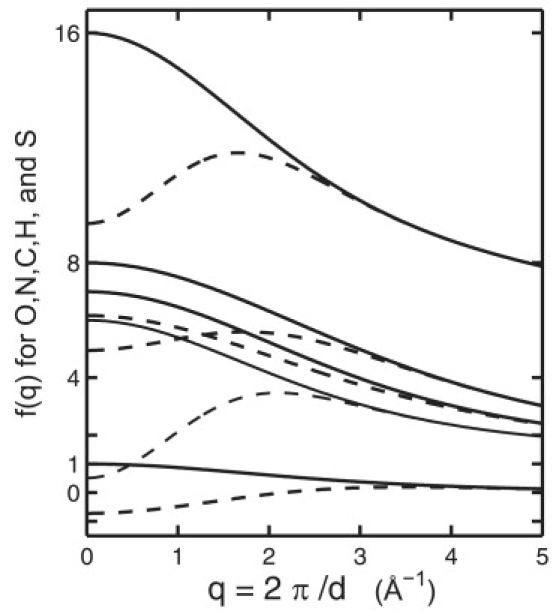
Atomic scattering form factors for atoms C, N, O, H, and S, before (solid line) and after (dashed line) the excluded volume correction. The effect of the excluded volume can be simulated by supposing that the volume displayed by the protein is filled with an electron gas with a density equal to the average electron density of pure water. This has been formulated by assigning a Gaussian sphere for each atom (30), according to Eq. 3.
Second, the effect of the excluded solvent can be incorporated into a correction for atomic scattering factors by assigning a Gaussian sphere for all the atoms (30),
| (3) |
where vi values are the observed volumes of each atom from experiments (30). This treatment has been implemented in the CRYSOL package (9). Therefore, the scattering from the protein taking into account the excluded volume is given by
| (4) |
where f′i(q) values are the corrected scattering factors in Eq. 3 and plotted in Fig. 2.
Third, the scattering is also contributed from the relative electron density of the primary solvation layer surrounding the protein to the extent that it differs from bulk solvent. It has been documented that the density of bound waters in the hydration shell is slightly higher relative to bulk water (15–17,29). The difference in density gives rise to the third term in the scattering I(q) (Eq. 1). In the CRYSOL calculations, the hydration shell is accounted for by using a water density 10% greater than bulk solvent by default. The level of contrast in the hydration shell can be adjusted to improve the fit to data. The validity of such a representation of the structure of the implicit solvent shell is uncertain (17,33), and therefore an explicit solvent representation will be implemented here.
Finally, the scattering is also affected by the protein conformational flexibility that reflects the nature of protein motions in solution. Modeling such flexibility is fundamentally important to describe the motions that are accessible to molecules outside of a crystal lattice (1). From a theoretical standpoint, we shall address this issue by adopting MD simulations.
Based on these four theoretical aspects of solution scattering, we have developed a new set of programs for computing solution scattering from atomic coordinates. In these new programs, the scattering intensity is computed by coarse-graining the protein representation with effective residue-based scattering factors, and coarse-graining the protein motions for conformational flexibility using MD simulations. The coarse-graining methods allow us to achieve a significant reduction of computer time, and the large timescale protein motions are required to adequately reflect the nature of protein dynamics in solution. These programs perform just as well on the test of lysozyme as CRYSOL, and make better assumptions about the solvent density in the primary hydration shell by including the solvent explicitly in the calculations.
Derivation of coarse residue structure factors
Solution scattering has been traditionally used to characterize the protein shape at low resolution (1). Calculations of such low resolution scattering profiles can be accommodated by simplifying the protein representation. We represent the protein as a chain of effective residues specified by the Cα position. Coarse-graining the protein representation is computationally advantageous, though one has to be careful in replacing the atomic scattering factors by effective residue-based structure factors to account for the internal detail of each amino acid (22,28). For each amino acid, an effective structure factor is derived from atomic coordinates of its n spherical atoms using the Debye formula (34)
| (5) |
where f′i(q) values are the scattering factors, corrected for excluded volume (Eq. 3 and Fig. 2). The procedure of simplifying a group of spherical atoms into a “glob” for each residue is illustrated in the Appendix. This idea of residue-based coarse-graining bears some similarity with the side-chain “globbicity,” introduced by Harker (27) and extended by Guo et al. (22,28). The brackets indicates the scattering factor was averaged over backbone conformers and side-chain rotamers of each residue in a set of high-resolution crystal structures of 434 protein chains selected from the PDB (as of July 2008) using the PISCES program (35).
A layer of explicit “dummy” waters
To represent the bound waters in the hydration shell, a layer of explicit water molecules are placed around the surface of protein. For the water molecule, similar to the procedure for amino acids, an effective scattering factor is derived and plotted in Fig. 3 according to
| (6) |
where rij values are the internal distances taken from a TIP3P model water (36). In practice, such a layer of “dummy” waters were placed at their Oxygen positions in a density of bulk solvent (ρs) with positions 3.5–6.5 Å away from the protein Cα atoms (Fig. 1), generated using a large equilibrated TIP3P waterbox.
Figure 3.
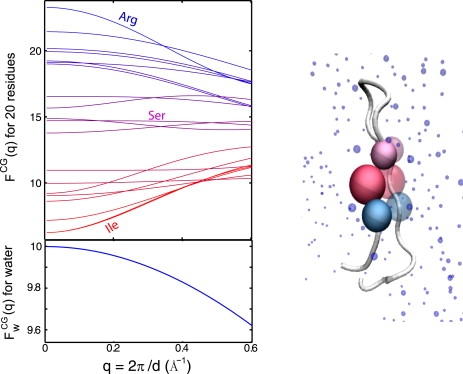
(Top) Effective residue-based scattering structure factors for 20 residues derived from a set of high-resolution crystal structures from the Protein Data Bank (PDB) (Eq. 5). (Bottom) The blue curve is the theoretical scattering factor of a TIP3P model water before the weighting (Eq. 6). An average scattering factor was calculated to account for backbone conformers and side-chain rotamers of each residue. For simplicity, we used an average over all residues in a set of high-resolution crystal structures. The data set consists of 434 protein chains derived from the PDB (as of July 2008). A large number of resulting atomic conformations were used for the averaging of each residue, ranging from 1308 for Cysteine, to 4400 for Proline, and to 8379 for Alanine. The ordering from large to small for 20 residues, according to the values of the intensity at q → 0, is: Arg, His, Asp, Asn, Glu, Cys, Gln, Met, Trp, Tyr, Ser, Thr, Lys, Gly, Phe, Ala, Pro, Val, Leu, and Ile.
To model the electron density contrast in the primary hydration shell relative to the bulk phase, a weight is assigned for the scattering factor of dummy waters:
| (7) |
As we shall see, the value of the weighting factor w is empirically calibrated using the experimental scattering data of lysozyme. With this strategy, the scattering from a given protein conformation is conveniently and accurately represented by its N residues via the Cα position and the surrounding M explicit water molecules via the Oxygen position. For this CG model of a protein with the accompanying hydration shell, the solution scattering can be calculated using the Debye formula
| (8) |
where FiCG(q) values are the effective CG scattering factors for both amino acids and water molecules (Eqs. 5 and 7). Thus, for a given protein conformation, a CG model of protein for scattering is achieved as a chain of N residues at their Cα positions and a layer of M dummy waters in the primary solvation shell.
Modeling protein flexibility in solution
The randomness of protein orientations in SAXS measurements requires spherical averaging in the theoretical framework. An additional level of disorder arises from the conformational flexibility of the protein. Proteins in solution fluctuate among accessible conformations, and the observed scattering reflects this ensemble. This is accomplished by the ensemble average of scattering , where stands for an MD average over the ensemble of structures around the local free energy minimum of folded proteins. Alternatively, they could be sampled by computational techniques such as Monte Carlo simulations, or normal mode analysis.
All aspects of the computation can be easily incorporated by sampling the local configurational space with CG simulations as a method of choice (37–42). In such a model, a protein is treated as a chain of Cα atoms with Lennard-Jones potentials to stabilize the native folded conformation. More can be found in Computational details, below.
Finally, an average scattering pattern of I(q) is computed by taking into account 1), the effective scattering factors for amino acids and water molecules (with a proper weight); and 2), an ensemble of folded structures generated from the CG simulations that allow the protein to fluctuate around the native conformations.
Computational details
The protein conformational flexibility is modeled by using an ensemble of structures extracted from MD simulations with a simplified model built from the native conformations. The energy function for a Cα-based CG model is similar to the one used in Yang et al. (42), i.e.,
| (9) |
where Kr, Kθ, and Kϕ are the force constants of the bond, angle, and dihedral angle for adjacent Cα atoms, respectively, and we chose Kr = 100 kcal/mol, Kθ = 20 kcal/mol, Kϕ(1) = 1 kcal/mol, and Kϕ(3) = 0.5 kcal/mol. All native folded structures with r0, θ0, ϕ0, and σij are taken from the PDB. The value σij is the distance of a pair of residues that are in contact in the native state. We chose ɛ1 = 1 kcal/mol for native contacts and ɛ2 = 0.001 kcal/mol for repulsive interactions for all pairs of residues that are not in contact in the native state (σo = 3.8 Å).
This CG model is simulated by a Langevin dynamics with a leveraged friction coefficient and an increased time step. The friction coefficient was set to 50 ps−1 and a time step to 0.01 ps. The value of friction coefficient for Cα atoms was chosen to mimic the friction for the all-atomic-detailed residues (43). All simulations were carried out at a temperature of 300 K for a period of 1–5 ns.
For each configuration generated from CG simulations, a layer of water molecules is placed at the surface of the protein. This placement is done by removing water molecules overlapping with protein atoms from a large equilibrated TIP3P waterbox. The water molecules are inserted in a density of bulk solvent, ρs = 0.334 e/Å3. Finally, only those water molecules with positions 3.5–6.5 Å away from the protein Cα atoms are kept, to represent the hydration shell (Fig. 1 and Fig. 4).
Figure 4.

The choice of the thickness of the hydration layer. A combination of varying the thickness (d) of the layer and the weighting factor (w) of water molecules in the layer to maintain the overall net density yields very similar scattering results for lysozyme. Shown are two examples with w = 3%, d = 3 Å (red) and w = 4.5%, d = 2 Å (blue), respectively.
Results and Discussion
Here, we first derive knowledge-based coarse residue structure factors for all 20 residues and then calibrate the scattering of water molecules in the hydration shell using the well-studied protein lysozyme. We finally apply the CG method to several representative protein folds and to proteins with multiple biological conformational states.
Coarse-grained residue structure factors
Equation 5 was used to compute the effective residue-based scattering structure factors for 20 residues. The coarse-graining procedure was based on a set of high-resolution crystal structures. A set of 434 protein structures was derived from the PDB (as of July 2008) by using the PISCES program (35), based on the criteria: 1), sequence identity <10%; 2), protein chain length from 40 to 10,000; 3), resolution <1.8 Å; and 4), R-factor value <0.15. This results in a large number of atomic conformations for each residue, ranging from 1308 for Cysteine, to 4400 for Proline, and to 8379 for Alanine. The scattering factor was derived from an average over all these conformers to account for different backbone and side-chain orientations of each residue (see Eq. 5).
Fig. 3 shows the CG residue scattering factors. The calculations were based on all appropriate conformers that are available in a subset of structure deposited in PDB using Eq. 5. The ordering of scattering intensity from large to small for 20 residues, according to the values at q → 0, is: Arg, His, Asp, Asn, Glu, Cys, Gln, Met, Trp, Tyr, Ser, Thr, Lys, Gly, Phe, Ala, Pro, Val, Leu, and Ile. For instance, the residue of Arginine has a positive electron density relative to the bulk solvent, whereas Isoleucine has a relative negative density. Therefore, a CG scattering method can be constructed as a chain of effective residues at their Cα positions and having the effective structure factors derived.
Lysozyme: a model system to calibrate the hydration shell
We used the HEW lysozyme (PDB code: 6LYZ) as our test case, since its SAXS data are publicly available through the CRYSOL package (9). We first simulated the lysozyme by a CG MD following Eq. 9 and then solvated the protein for each snapshot extracted from simulations by placing a layer of water molecules with an initial density of ρs ∼0.334 e/Å3 (see details above). Finally, the average scattering was calculated with CG residue scattering factors and a proper weight for dummy waters (to be determined) using Eq. 8.
Fig. 5 shows that the theoretical scattering intensity for the lysozyme with different lengths of simulation time from 1 ns to 5 ns. Clearly, the protein conformational flexibility is reflected by the standard deviations (and the averages as well) of I(q), which are represented by the shades of each curve (and the inset). In the low q region, where the overall protein shape is not sensitive to the conformation flexibility, the standard deviation of I(q) is quite small. It gets larger in the higher q region, where the contributions of internal detailed fluctuations begin to dominate. For proteins with large conformational flexibility in solution, this larger uncertainty of I(q), together with an intrinsic low signal/noise ratio in the high-q regions, may contribute to very noisy experimental observations as q increases.
Figure 5.

An ensemble average of configurations for scattering of the HEW lysozyme. The curves were calculated from an ensemble of snapshots taken from a simulation period of time of 1 ns, 2 ns, and 5 ns, respectively. The shades of each curve represent the standard deviations at each q position (q = 2π/d), which are enlarged by the plot in the inset. Comparison of these curves suggests that it is representative to take the scattering with an average over a period of 2-ns MD simulations. The weighting factor for dummy waters, w = 3% (Eq. 7), was used for the calculations. The intensity was plotted in a log-scale. For clarity, the curves with difference were shifted along the vertical direction.
Fig. 5 also shows that the standard deviations of I(q) start to converge with a length of 2-ns simulations in the case of lysozyme. Although the convergence of the length of simulations has to be examined by a case-by-case basis for different proteins, we take as representative an ensemble average over a period of 2-ns simulations for the following discussion. In these calculations, the weighting factor for dummy waters w = 3% (Eq. 7) was used for the calculations, where the value of the factor is calibrated as follows.
To account for the local electron density difference in the hydration shell relative to the rest of bulk solvent, we modeled such a density contrast by assigning a proper weighting factor for the dummy water scattering according to Eq. 7. In this equation, there is one free parameter, w, that remains to be determined, to calculate the scattering curve. Fig. 6 shows the theoretical scattering intensities with several different weighting factors for waters of w = 0%, 3%, and 10%. The weighting factor w for dummy waters reflects the excess electron density of the hydration shell relative to bulk solvent. In other words, the density of the shell is effectively w greater than that of the rest of bulk solvent. Fitted to the scattering data of lysozyme, a proper weight of w = 3% was chosen to reflect the relative difference in the primary solvent shell. Remarkably, the CG approach with a single fitting parameter for dummy waters can well reproduce the lysozyme data I(q) up to q = 0.5 Å−1. The high accuracy of reproducing the experimental scattering appears to be due to a combination of both the density difference and conformation flexibility.
Figure 6.
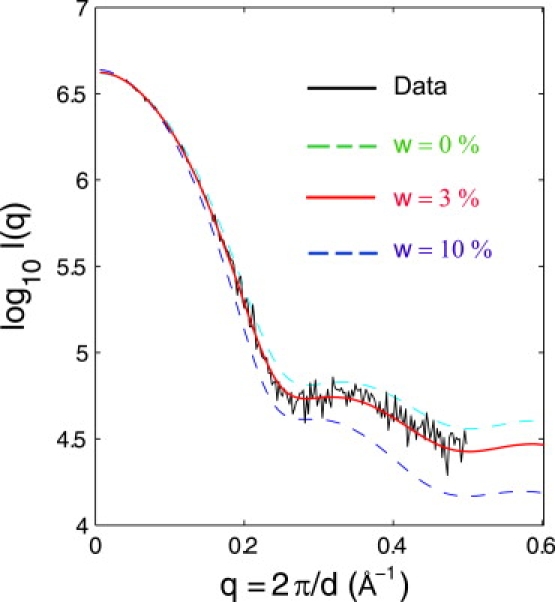
The weighting factor (w) for water molecules in the hydration layer with w of 0%, 3%, and 10%, respectively. The calculations were from 2-ns MD simulations. The theoretical curves were compared with the experimental data taken from the CRYSOL package. From the plot, a weight of w = 3% is chosen to fit the experimental curve. The data curve was shifted along the vertical direction to achieve an optimal overlap at the low q region.
We also note that different choices of the thickness of the hydration give rise to very similar scattering curves. Fig. 4 shows that a combination of varying the thickness (d) of the layer and the weighting factor (w) of water molecules in the layer to maintain the overall net density yields very similar scattering results for lysozyme, by two examples: with w = 3%, d = 3 Å (red) and w = 4.5%, d = 2 Å (blue), respectively. This would potentially further reduce the computational cost by using a thinner layer but with higher weighting factor for water molecules. From a physical consideration, we set the thickness of the hydration shell equal to 3 Å for the rest of discussion.
The CG residue scattering computational method can be advantageous. This CG calculation is much faster than the all-atom scattering calculations (9,11,12). For example, in the framework of the Debye formula, it is ∼N2 times faster to compute the scattering of a protein without surrounding water molecules (N is the average number of atoms per residue). It should be noted that this advantage is less pronounced when explicit dummy water molecules are included in the calculation. Here, we make a very brief comparison of the CRYSOL calculations with our CG results. We computed the intensity of lysozyme from the widely used all-atom CRYSOL calculation, in which the atomic details for scattering factors were included. The CRYSOL calculation was carried out with default parameters. Fig. 7 shows the difference between the CG model and CRYSOL for lysozyme. Comparison shows that the CRYSOL calculation with the default parameters (solvation shell electron density 0.030 e/Å3, i.e., 10% of bulk solvent electron density ρs) gives a very good intensity fit at the low q region, but shows a systematic shift from experiments at high q, whereas the CG model with w = 3% accurately reproduces the scattering curve. Putnam et al. also pointed out that this adjusted parameter with less density contrast (solvation shell electron density 0.010 e/Å3, i.e., 3% of ρs) in CRYSOL gives a better scattering curve compared with data (1). This notion is also supported by our CG residue-based scattering calculations. Although there is room for refinement of the weight factor w when more reliable SAXS data from additional protein samples become available, we use w = 3% for the following discussion.
Figure 7.
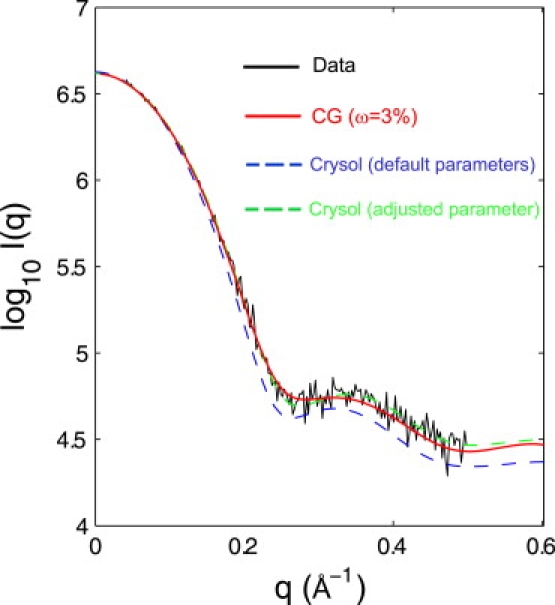
Theoretical scattering profiles of lysozyme from the CG model with a weight of w = 3% (red) and from CRYSOL (blue and green). The calculations from CRYSOL were performed with the default parameters (solvation shell contrast = 0.030 e/Å3, 10% of bulk solvent electron density ρs) and an adjusted parameter (solvation shell contrast = 0.01 e/Å3, 3% of ρs). The scattering curves were shifted along the vertical direction to achieve an optimal overlap at the low q region.
Comparison of our residue-based method with other simplified models is quite clear. Our method requires no additional computational cost to the simple Cα model (21,25,26), but includes effective residue structure factors and thus has a better accuracy (data not shown). In contrast to the DR model (23), our CG method requires no use of a correction function of q for scattering factors in order to reproduce the scattering patterns derived from the all-atom CRYSOL calculations. Instead, a knowledge-based, coarse-residue structure factor for each amino acid was calculated, and a single parameter for the density contrast was fitted for the solvent layer. Nonetheless, our CG method can well reproduce the experimental scattering of lysozyme, which provides us with a reasonable start point for further investigation.
It is worth noting that at high concentrations, interparticle correlations may lead to an interparticle form factor that is different from unity and will modulate the observed scattering at very small angles (44–46). In this article, since we assume that the scattering is from a dilute solution of protein particles, these interparticle effects are negligible.
Scattering characterization of protein folds
We now have a working model for calculating the scattering intensity from atomic models. Such a calculation can be very useful in many scenarios. For example, combined with SAXS data, the CG model can be used to model the biological assembled structures of multidomain complexes in cases where high-resolution crystal structures of each individual domain are known. Such an example will be presented in future communication. Currently, calculations from known atomic models can provide a scattering-signature of each protein fold. In fact, such an effort has been put forward by building a database of the CRYSOL-calculated scattering curves for a portion of structures deposited in the PDB (47,48). Similar efforts have been performed to characterize protein folds using wide-angle scattering calculations (49). In general, such a theoretical effort could be potentially useful for providing the ranking scores for experimental scattering data and further identifying top candidates from this kind of database for refinement. From this point of view, despite the possibility that multiple protein folds could share a similar scattering curve, we envision that the CG model can serve for a similar purpose to achieve scattering characterization of different protein folds.
Fig. 8 shows the scattering for several representative protein folds/structures, including α-helical, β-strand, and multidomain proteins. For a quantitative assessment of conformation flexibility, three scattering curves for each protein are computed and reported from 2-ns CG MD simulations. Conformational flexibility from simulations is reflected in the average (lines) and standard deviations (shades) in each curve. Three proteins are used to illustrate the scattering patterns of α-helical proteins.
Figure 8.
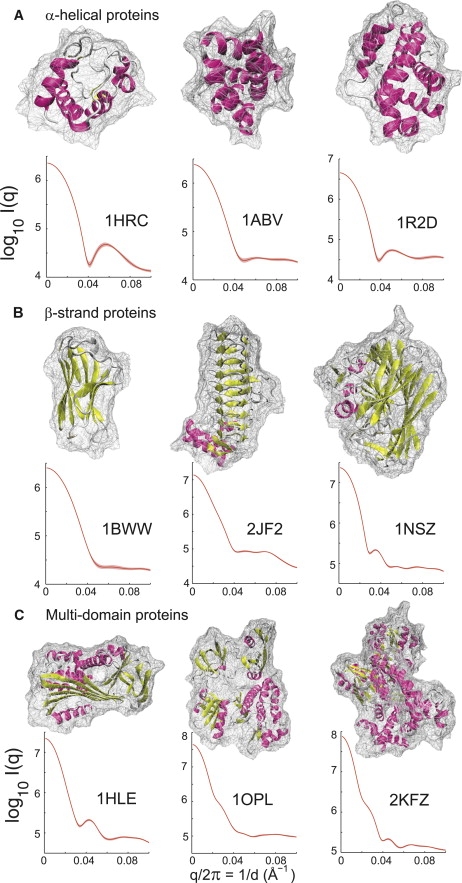
Scattering characterization of protein folds: α-helical, β-strand, and multidomain proteins. The low-angle scattering contains information about protein size and overall shape; the wider-angle scattering provides information about secondary-structure packing and domain motions. (A) The α-helical proteins: cytochrome c (PDB code: 1HRC (70)), ATPsynthase (PDB code: 1ABV (71)), and Bcl-X (PDB code: 1R2D (72)). (B) The β-strand proteins: immunoglobulin (PDB code: 1BWW (73)), acyltransferase (PDB code: 2JF2 (74)), and galactose mutarotase (PDB code: 1NSZ (75)). (C) Multidomain proteins: serpin (PDB code: 1HLE (76)), c-Abl (PDB code: 1OPL (59)), and DNA polymerase (PDB code: 2KFZ (60)). Each curve was calculated from an ensemble of snapshots taken from a 2-ns MD simulation trajectory. A log-scale of the scattering intensity was used for all the proteins.
In general, the SAXS scattering pattern contains less information than the pattern in crystallography, although it is still rich in details about the overall shape and internal structure of a macromolecule. In the low q region, the scattering can be used to measure the protein radius of gyration (RG) by the Guinier approximation (50), . In the q region beyond RG where the intensity starts to fall off, I(q) shows a systematic trend for folded proteins, I(q) ∝ q−d, referred to as Porod's law (51,52). A value of d = 4 was found for many folded single-domain proteins. For multidomain complexes, our experience demonstrates that scattering from large domain-domain separations causes a modulation of the curve in this region. As q further increases, the power-law pattern breaks down where more detailed substructures start to contribute to the total scattering profile. These peaks are the collective contributions of scattering due to a spatial separation between large groups of atoms, such as the domain-domain separation and the secondary-structure packing.
Several interesting features might be noted from the theoretical patterns. First, the low-angle scattering contains information about protein size and overall shape/envelope: the larger the protein, the greater the scattering intensity at q → 0 (in theory, I(q = 0) is proportional to protein size (1–3)). For example, Bcl-X with a total of 196 residues (PDB code: 1R2D) has a higher scattering intensity than cytochrome c (104 residues).
Second, the higher angle scattering provides information about secondary-structure packing, e.g., the helix-helix organization in the case of all-α proteins as observed as in cytochrome c and Bcl-X. However, such a peak is not generic for the proteins in the family of α-helical proteins. For example, the curve is relatively flat in ATPsynthase (PDB code: 1ABV) or a peak is found at a lower angle at ∼q/2π = 1/d ∼ 0.05 Å−1 in Bcl-X. Further wider-angle scattering (q > 0.6 Å−1) was not investigated here, but has been studied by several experimental groups (53,54).
Similar features are observed in β-strand proteins including the all-β immunoglobulin (PDB code: 1BWW), the β-helix acyltransferase (PDB code: 2JF2), and the supersandwich fold of galactose mutarotase (PDB code: 1NSZ). In particular, the β-helix fold has recently become a very interesting research focus, in part because it has been proposed as a structural candidate for amyloid proteins such as the prion protein (55–57) and the Aβ protein (58). It suggests that SAXS combined with computation has a significant potential of elucidating the basic structural details of cross-β fingerprints implicated in many amyloid diseases such as Alzheimer's. In the case of acyltransferase, the scattering curve displays a plateaulike flat pattern in a quite wide q-range (1/d ∼ 0.04–0.07 Å−1), before falling off at higher q (1/d ∼ 0.08 Å−1). It differs from the flat pattern in the all-β immunoglobulin, where the scattering intensity does not start to fall off even at 1/d ∼ 0.09 Å−1.
SAXS scattering appears to offer significant potential advantage for examining the structures of multidomain proteins. Shown in Fig. 8 are the theoretical scattering patterns from serpin (PDB code: 1HLE), c-Abl (PDB code: 1OPL), and DNA polymerase (PDB code: 2KFZ). As mentioned earlier, rich scattering information such as one or multiple peaks at the power-law or high q regions can be observed because of the collective separation of two large groups. This is clearly represented in all three cases because of the domain-domain organization. For example, there is a peak at ∼1/d ∼ 0.04 Å−1 in serpin, which represents a major separation of d ∼ 25 Å between two domains. We note that the c-Abl tyrosine kinase has been the target of drug design for cancer treatment (59). The scattering of c-Abl shows a very detailed pattern in a wide q-range (e.g., 1/d ∼ 0.03 Å−1 and 1/d ∼ 0.07 Å−1), reflecting the complex assembly among two regulatory domains and one catalytic domain. Similarly, peaks are superimposed on the power-law region and the high q region for a larger complex of the DNA polymerase (60). Thus, combined with computation including MD simulations, the solution scattering can have a great potential in understanding how multiple domains assemble in physiological conditions.
Use of SAXS for fold recognition
As described above, different proteins display distinct scattering patterns by protein size at q → 0, RG at low q, structural packing at higher q, etc. Such distinctions suggest that a SAXS scattering curve serve as a characteristic or semisignature of each protein fold/structure. As mentioned earlier, the knowledge about the theoretical scattering could be potentially useful by creating a database of theoretical scattering curves, similar to the CRYSOL-based DARA (47). Ideally, experimental scattering from an unknown fold is fitted to the precalculated theoretical scattering curves to obtain a list of top hits by ranking. A ranking score for such a measure could be developed based on a χ2 parameter
| (10) |
where Nq is the number of data points in the scattering curve and Nq = 100 was used for theoretical calculations throughout the rest of the article. The value Δ is a normalization factor, which is used to offset the difference of scattering intensity at q → 0. The value σ(q) is experimental uncertainty of log Iexp(q) or simulated standard deviation of log ICG(q), in cases where the experimental errors are not available.
To illustrate the concept of fold recognition by the use of SAXS, we first computed the pairwise χ2 derivations between the scattering signature of the nine protein folds above-described. Table 1 shows that the χ2 parameter ranges from 103 to 106, suggesting that there is a large separation between different protein folds. Such a large separation makes possible the construction of a theoretical database for fold recognition. To illustrate this concept, we used two representative proteins, the Bcl-2 homolog from myxoma virus (PDB code: 2O42) and the YDCK from Salmonella cholerae (PDB code: 2F9C), which have similar structures to Bcl-X (PDB code: 1R2D) and acyltransferase (PDB code: 2JF2), respectively. The computed scattering from these two proteins were ranked against the database of the precomputed nine curves, according to χ2 (Eq. 10). Fig. 9 shows that the best hit for both cases is indeed the one which is the best match (marked by arrows), according to the CE alignment calculations (61). We note that in both cases proteins are similar in fold but very different in sequence, e.g., the sequence identities are 10.7% and 16.9%, respectively. These encouraging results suggest that SAXS might provide an alternative approach for protein structure prediction by taking advantage of the ease of use of solution scattering to support and complement current homology modeling or ab initio protein structure predictions (62). We envision that the concept of best-fitting to a theoretical scattering database of all known folds could play an important role in fold recognition. For this purpose, the rapid determination of scattering profiles from our CG method can provide a fast and efficient way to create a database of SAXS profiles for all appropriate protein folds as deposited in the PDB.
Table 1.
The pairwise χ2 distances in the scattering space between the nine proteins as shown in Fig. 8
|
α-Helical proteins |
β-Strand proteins |
Multidomain proteins |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| χ2(× 106) | 1HRC | 1ABV | 1R2D | 1BWW | 2JF2 | 1NSZ | 1HLE | 1OPL | 2KFZ |
| 1HRC | 0 | 0.0031 | 0.0110 | 0.0038 | 0.1037 | 0.1574 | 0.2038 | 0.4043 | 0.6624 |
| 1ABV | 0 | 0.0098 | 0.0016 | 0.0752 | 0.1275 | 0.1510 | 0.2882 | 0.4609 | |
| 1R2D | 0 | 0.0051 | 0.0382 | 0.0537 | 0.0770 | 0.1850 | 0.3291 | ||
| 1BWW | 0 | 0.1759 | 0.2877 | 0.3464 | 0.6307 | 1.0118 | |||
| 2JF2 | 0 | 0.0581 | 0.0452 | 0.1075 | 0.2069 | ||||
| 1NSZ | 0 | 0.0149 | 0.1217 | 0.3385 | |||||
| 1HLE | 0 | 0.0273 | 0.0811 | ||||||
| 1OPL | 0 | 0.0119 | |||||||
| 2KFZ | 0 | ||||||||
The large separation between them suggests that a database of scattering would be useful for fold recognition. The calculations were based on Eq. 10, where the standard deviation of theoretical curves was used for σ(q).
Figure 9.
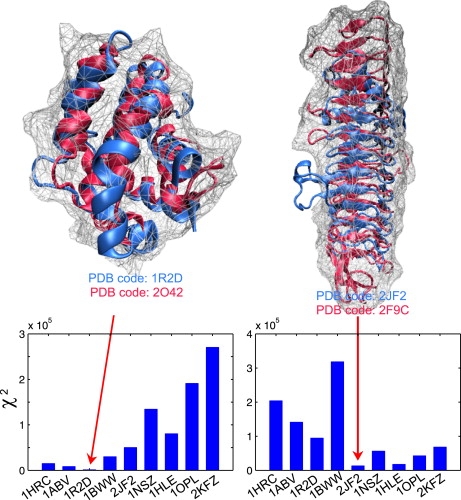
Two illustrative examples for fold recognition by χ2 parameters. (Left) The computed scattering curve from the Bcl-2 homolog from myxoma virus (red; PDB code: 2O42 (77)) best-matches that of Bcl-X (blue; PDB code: 1R2D). (Right) The computed scattering curve from the YDCK from Salmonella cholerae (red; PDB code: 2F9C) best-fits with that of acyltransferase (blue; PDB code: 2JF2); both have a similar β-helical fold. In both cases, the sequence identities are quite low, 10.7% and 16.9%, respectively, according to the CE alignment calculations (61). The calculations for χ2 were based on Eq. 10 and the standard deviation of scattering curves was used for σ(q).
Further application of SAXS data to the so-called natively disordered proteins has been shown to elucidate their structural features (e.g., (63,64).). In this case, because a large ensemble of unfolded structures must be sampled, our rapid CG computational method might provide an efficient tool for characterizing structural features of disordered states. Incorporation of SAXS data with NMR data for structure refinements (65,66) is beyond the scope of this article, but our CG method can provide an alternative approach for scattering calculations, especially for low- and medium-angle scattering.
Scattering characterization for multiple conformational states
Protein can adopt multiple conformational states in equilibrium under specific physiological conditions. Specific binding to ligands, substrates, or target proteins can shift the population from one state to another (19). In cases where each individual state is well defined, theoretical scattering can be used to characterize the conformational states from atomic protein models. Such a theoretical scattering calculation can be an important tool to deconvolute the relative population of each state in the SAXS sample with mixing states in solution. Here, several examples are given for theoretical scattering patterns of distinct conformational states from given atomic models.
Fig. 10 shows the theoretical scattering curves of two distinct conformational states from three proteins: transferrin, calmodulin, and ParM. In transferrin, ligand-induced conformational change occurs between two lobes when iron binds. The structures of the apo- and holoforms are shown in blue and red, respectively. The differences in theoretical curves are at 1/d ∼ 0.03 Å−1 and 1/d ∼ 0.06–0.08 Å−1, which suggests a large-scale conformational change upon the binding.
Figure 10.
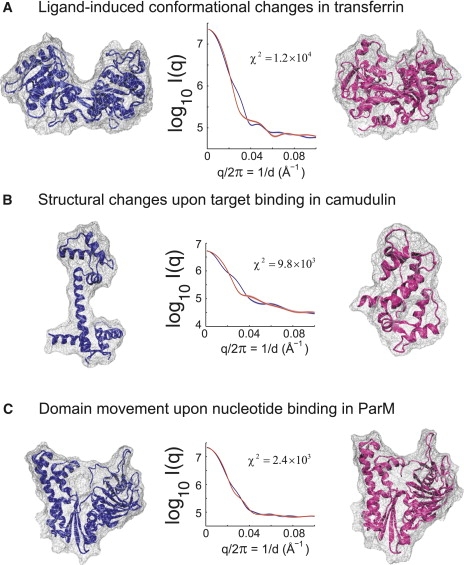
Scattering difference of multiple conformation states. The differences in scattering curves are found in a wide q-range from low to high, suggesting that SAXS can be used to investigate the large-scale conformational motions in solution. The differences are also reflected by a χ2 parameter between two curves. (A) Ligand-induced conformational change in transferrin before (blue) and after (red) the ion binding (PDB codes: 1BP5 (78) and 1A8E (79), respectively). (B) The dumbbell shape (blue) and the compact shape (red) of calmodulin (PDB codes: 1CLL (67) and 1CDL (80), respectively). (C) Domain-domain conformations before and after nucleotide binding in ParM (PDB codes: 1MWK (81) and 1MWM (81), respectively). All scattering curves in a log-scale were calculated from an ensemble of snapshots taken from an MD simulation period of time of 2 ns. The χ2 parameter was calculated using Eq. 10, where the standard deviation of the theoretical scattering of the left state was used for σ(q).
Another well-studied example is the Ca2+-bonded calmodulin where a major conformational change occurs when it binds to a target protein (67). The difference in theoretical curves at low q (1/d ∼ 0.01–0.02 Å−1 and 1/d ∼ 0.03 Å−1) in Fig. 10 clearly represents the distinct protein shapes (e.g., RG) and helix-helix arrangements. This arises from the range of forms calmodulin displays in solution, from extended to more compact. The equilibrium can be shifted from one to another, dependent on whether it is in solution, a crystal lattice, or binding to specific agents. Thus, SAXS has been a method of choice for studying such a multistate calmodulin in both experimental and computational aspects (68,69).
A similar feature is also found in theoretical scattering curves in the apo- and holoforms of ParM, a member of the actin filament protein family, where domains move upon nucleotide binding. Scattering differences can be found in several places such as 1/d ∼ 0.03 Å−1 and 1/d ∼ 0.06 Å−1 (Fig. 10). Again, this demonstrates the usefulness of SAXS for characterizing distinct functional states in solution. The χ2 calculations also indicate scattering differences in distinct states (Fig. 10).
To summarize, several multiconformational-state proteins examined here show that a major difference in scattering of distinct states is observed in or near the power-law region of scattering curves. Such distinct patterns can be detected by SAXS experiments. This suggests that the solution scattering at relative small angles has the capability of identifying the assembly mechanisms of multiple-state proteins, often with multiple domains. These kinds of SAXS experiments are obviously attractive because of the difficulties of growing crystals for large protein complexes.
Conclusion
SAXS is an increasingly important technique for characterizing macromolecular folds, conformations and assembly states in physiological conditions. It can provide low-resolution structural information without the challenges and limitations of crystallography or solution NMR. In principle, the scattering data can be used as an input for computational modeling to reconstruct the structures for multiprotein complexes and to deconvolute the equilibrium population of each conformational state of proteins in solution. However, a reliable and efficient computational approach is needed to achieve this goal.
A theoretical CG model was developed to compute the scattering pattern from a protein in a given conformation. The model is residue-based, but the scattering for each amino acid was built from atomically detailed conformers. Such a CG representation for calculating scattering is advantageous, because it significantly reduces the computational cost and it can be combined with CG simulations that can be used to sample broad configurational spaces exhibited by many large complexes. The CG representation of the protein takes advantage of the low-resolution character of SAXS. The computational methods were further illustrated by characterizing a variety of protein folds and multiple conformational states. Preliminary tests show that a given fold can be detected via the scattering signature of the protein. This suggests that the structural information from SAXS, when combined with computations, could provide a powerful route for rapid fold recognition and shape reconstruction of large macromolecular complexes.
The program for this rapid coarse residue-based computational method for proteins will be released under the GNU General Public License with the code name of Fast-SAXS.
Acknowledgments
We thank Jan Lipfert for very helpful comments and suggestions on the article; we also thank Albert Lau, Nilesh Banavali, Jaydeep Bardhan, and Franci Merzel for valuable discussions.
This work was supported by the National Institute of Health through grant No. CA-093577 and by a joint grant from the University of Chicago Cancer Center and Argonne National Laboratory (grant No. UCCC/ANL).
Appendix
In this Appendix, the procedure of simplifying each amino acid into a “glob” in Eq. 5 is demonstrated in the case of a group of spherical atoms. The scattering from these atoms within a given protein conformation is given by Eq. 4, which can be rewritten as
| (11) |
where NG is the number of amino acids and F(q) is the scattering factor of the gth residue in the protein. The notations j and j′ refer to atoms in different residues, and rjj′ are the distances between atoms j and j′.
In the case of spherical atoms where scattering factors are independent of direction, Harker pointed out that Eq. 11 can be essentially given by (27)
| (12) |
where rgg′ values are the distances between amino acids g and g′.
References
- 1.Putnam C.D., Hammel M., Hura G.L., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 2.Lipfert J., Doniach S. Small-angle x-ray scattering from RNA, proteins, and protein complexes. Annu. Rev. Biophys. Biomol. Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 3.Koch M.H.J., Vachette P., Svergun D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 4.Chu B., Hsiao B. Small-angle x-ray scattering of polymers. Chem. Rev. 2001;101:1727–1762. doi: 10.1021/cr9900376. [DOI] [PubMed] [Google Scholar]
- 5.Doniach S. Changes in biomolecular conformation seen by small angle x-ray scattering. Chem. Rev. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 6.Perkins S.J. Structural studies of proteins by high-flux x-ray and neutron solution scattering. Biochem. J. 1988;254:313–327. doi: 10.1042/bj2540313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forster F., Webb B., Krukenberg K.A., Tsuruta H., Agard D.A. Integration of small-angle x-ray scattering data into structural modeling of proteins and their assemblies. j. mol. biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernado P., Pérez Y., Svergun D.I., Pons M. Structural characterization of the active and inactive states of Src kinase in solution by small-angle x-ray scattering. J. Mol. Biol. 2008;376:492–505. doi: 10.1016/j.jmb.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 9.Svergun D., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystal. 1995;28:768–773. [Google Scholar]
- 10.Merzel F., Smith J.C. SASSIM: a method for calculating small-angle x-ray and neutron scattering and the associated molecular envelope from explicit-atom models of solvated proteins. Acta Crystallogr. D Biol. Crystallogr. 2002;58:242–249. doi: 10.1107/s0907444901019576. [DOI] [PubMed] [Google Scholar]
- 11.Tiede D., Zhang R., Seifert S. Protein conformations explored by difference high-angle solution x-ray scattering: oxidation state and temperature dependent changes in cytochrome c. Biochemistry. 2002;41:6605–6614. doi: 10.1021/bi015931h. [DOI] [PubMed] [Google Scholar]
- 12.Tjioe E., Heller W.T. ORNL_SAS: software for calculation of small-angle scattering intensities of proteins and protein complexes. J. Appl. Cryst. 2007;40:782–785. [Google Scholar]
- 13.Petoukhov M., Svergun D. Analysis of x-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Hubbard S., Hodgson K., Doniach S. Small-angle x-ray scattering investigation of the solution structure of troponin C. J. Biol. Chem. 1988;263:4151–4158. [PubMed] [Google Scholar]
- 15.Svergun D.I., Richard S., Koch M.H.J., Sayers Z., Kuprin S. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merzel F., Smith J.C. Is the first hydration shell of lysozyme of higher density than bulk water? Proc. Natl. Acad. Sci. USA. 2002;99:5378–5383. doi: 10.1073/pnas.082335099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koizumi M., Hirai H., Onai T., Inoue K., Hirai M. Collapse of the hydration shell of a protein prior to thermal unfolding. J. Appl. Cryst. 2007;40:s175–s178. [Google Scholar]
- 18.Boehr D.D., McElheny D., Dyson H.J., Wright P.E. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 19.Vendruscolo M., Dobson C.M. Dynamic visions of enzymatic reactions. Science. 2006;313:1586–1587. doi: 10.1126/science.1132851. [DOI] [PubMed] [Google Scholar]
- 20.Makowski L., Rodi D.J., Mandava S., Minh D.D., Gore D.B. Molecular crowding inhibits intramolecular breathing motions in proteins. J. Mol. Biol. 2008;375:529–546. doi: 10.1016/j.jmb.2007.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther D., Cohen F.E., Doniach S. Reconstruction of low-resolution three-dimensional density maps from one-dimensional small-angle x-ray solution scattering data for biomolecules. J. Appl. Cryst. 2000;33:350–363. [Google Scholar]
- 22.Guo D.Y., Blessing R.H., Langs D.A., Smith G.D. On “globbicity” of low-resolution protein structures. Acta Crystallogr. D Biol. Crystallogr. 1999;55:230–237. doi: 10.1107/S0907444998008208. [DOI] [PubMed] [Google Scholar]
- 23.Svergun D.I., Petoukhov M.V., Koch M.H.J. Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chacón P., Morán F., Díaz J., Pantos E., Andreu J. Low-resolution structures of proteins in solution retrieved from x-ray scattering with a genetic algorithm. Biophys. J. 1998;74:2760–2775. doi: 10.1016/S0006-3495(98)77984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng W., Doniach S. Protein structure prediction constrained by solution x-ray scattering data and structural homology identification. J. Mol. Biol. 2002;316:173–187. doi: 10.1006/jmbi.2001.5324. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Tian X., Lu M., Chen M., Wang Q. Folding of small helical proteins assisted by small-angle x-ray scattering profiles. Structure. 2005;13:1587–1597. doi: 10.1016/j.str.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Harker D. The meaning of the average of |F|2 for large values of the interplanar spacing. Acta Crystallogr. 1953;6:731–736. [Google Scholar]
- 28.Guo D.Y., Smith G.D., Griffin J.F., Langs D.A. Use of globic scattering factors for protein structures at low resolution. Acta Crystallogr. A. 1995;51:945–947. doi: 10.1107/s0108767395010038. [DOI] [PubMed] [Google Scholar]
- 29.Bragg L., Perutz M.F. The structure of hemoglobin. Proc. Roy. Soc. A (Lond.) 1952;213:425–435. [Google Scholar]
- 30.Fraser R.D.B., MacRae T.P., Suzuki E. An improved method for calculating the contribution of solvent to the x-ray diffraction pattern of biological molecules. J. Appl. Cryst. 1978;11:693–694. [Google Scholar]
- 31.Lee S., Eisenberg D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat. Struct. Biol. 2003;10:725–730. doi: 10.1038/nsb961. [DOI] [PubMed] [Google Scholar]
- 32.Cromer D.T., Mann J.B. X-ray scattering factors computed from numerical Hartree-Fock wave functions. Acta Cryst. A. 1968;24:0567–7394. [Google Scholar]
- 33.Lau A.Y., Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debye P. Dispersion of Roentgen rays. Ann. Phys. (Leipzig) 1915;46:809–823. [Google Scholar]
- 35.Wang G., Dunbrack J., Roland L. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Nymeyer H., García A.E., Onuchic J.N. Folding funnels and frustration in off-lattice minimalist models. Proc. Natl. Acad. Sci. USA. 1998;95:5921–5928. doi: 10.1073/pnas.95.11.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clementi C., Nymeyer H., Onuchic J.N. Topological and energetic factors: what determines the structural details of the transition state ensemble and “on-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 39.Koga N., Takada S. Roles of native topology and chain-length scaling in protein folding: a simulation study with a Gō-like model. J. Mol. Biol. 2001;313:171–180. doi: 10.1006/jmbi.2001.5037. [DOI] [PubMed] [Google Scholar]
- 40.Cheung M.S., Garcia A.E., Onuchic J.N. Protein folding mediated by solvation: water expulsion and formation of the hydrophobic core occur after the structural collapse. Proc. Natl. Acad. Sci. USA. 2002;99:685–690. doi: 10.1073/pnas.022387699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karanicolas J., Brooks C.L. The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S., Onuchic J.N., Levine H. Effective stochastic dynamics on a protein folding energy landscape. J. Chem. Phys. 2006;125:054910. doi: 10.1063/1.2229206. [DOI] [PubMed] [Google Scholar]
- 43.Yang S., Roux B. Src kinase conformational activation: thermodynamics, pathways, and mechanisms. PLoS Comput. Biol. 2008;4:e1000047. doi: 10.1371/journal.pcbi.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stradner A., Sedgwick H., Cardinaux F., Poon W.C.K., Egelhaaf S.U. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature. 2004;432:492–495. doi: 10.1038/nature03109. [DOI] [PubMed] [Google Scholar]
- 45.Shukla A., Mylonas E., Di Cola E., Finet S., Timmins P. Absence of equilibrium cluster phase in concentrated lysozyme solutions. Proc. Natl. Acad. Sci. USA. 2008;105:5075–5080. doi: 10.1073/pnas.0711928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trewhella J. The different views from small angles. Proc. Natl. Acad. Sci. USA. 2008;105:4967–4968. doi: 10.1073/pnas.0801324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sokolova A., Volkov V., Svergun D. Database for rapid protein classification based on small-angle x-ray scattering data. Crystallogr. Rep. 2003;48:959–965. [Google Scholar]
- 48.Sokolova A.V., Volkov V.V., Svergun D.I. Prototype of a database for rapid protein classification based on solution scattering data. J. Appl. Cryst. 2003;36:865–868. [Google Scholar]
- 49.Makowski L., Rodi D.J., Mandava S., Devarapalli S., Fischetti R.F. Characterization of protein fold using wide-angle x-ray solution scattering. J. Mol. Biol. 2008;383:731–744. doi: 10.1016/j.jmb.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 50.Guinier A., Fournet G. Wiley; New York: 1955. Small-Angle Scattering of X-Rays. [Google Scholar]
- 51.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Cryst. 1977;10:415–421. [Google Scholar]
- 52.Roe R.-J. Oxford University Press; New York: 2000. Methods of X-Ray and Neutron Scattering in Polymer Science. [Google Scholar]
- 53.Hirai M., Iwase H., Hayakawa T., Miura K., Inoue K. Structural hierarchy of several proteins observed by wide-angle solution scattering. J. Synchrotron Radiat. 2002;9:202–205. doi: 10.1107/s0909049502006593. [DOI] [PubMed] [Google Scholar]
- 54.Fischetti R.F., Rodi D.J., Gore D.B., Makowski L. Wide-angle x-ray solution scattering as a probe of ligand-induced conformational changes in proteins. Chem. Biol. 2004;11:1431–1443. doi: 10.1016/j.chembiol.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Govaerts C., Wille H., Prusiner S.B., Cohen F.E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S., Levine H., Onuchic J.N., Cox D.L. Structure of infectious prions: stabilization by domain swapping. FASEB J. 2005;19:1778–1782. doi: 10.1096/fj.05-4067hyp. [DOI] [PubMed] [Google Scholar]
- 57.Kunes K.C., Clark S.C., Cox D.L., Singh R.R. Left-handed β-helix models for mammalian prion fibrils. Prion. 2008;2:81–90. doi: 10.4161/pri.2.2.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo J.T., Wetzel R., Wu Y. Molecular modeling of the core of Aβ amyloid fibrils. Proteins. 2004;57:357–364. doi: 10.1002/prot.20222. [DOI] [PubMed] [Google Scholar]
- 59.Nagar B., Hantschel O., Young M.A., Scheffzek K., Veach D. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 60.Brautigan C., Sun S., Piccirilli J., Steitz T. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′-5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry. 1999;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- 61.Shindyalov I., Bourne P. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 62.Baker D., Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 63.Zagrovic B., Lipfert J., Sorin E.J., Millett I.S., van Gunsteren W.F. Unusual compactness of a polyproline type II structure. Proc. Natl. Acad. Sci. USA. 2005;102:11698–11703. doi: 10.1073/pnas.0409693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wells M., Tidow H., Rutherford T.J., Markwick P., Jensen M.R. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grishaev A., Wu J., Trewhella J., Bax A. Refinement of multidomain protein structures by combination of solution small-angle x-ray scattering and NMR data. J. Am. Chem. Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- 66.Grishaev A., Tugarinov V., Kay L.E., Trewhella J., Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. J. Biomol. NMR. 2008;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- 67.Chattopadhyay R., Meador W.E., Means A.R., Quiocho F.A. Calmodulin structure refined at 1.7 Å resolution. J. Mol. Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 68.Heidorn D.B., Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- 69.Vigil D., Gallagher S.C., Trewhella J., Garcia A.E. Functional dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophys. J. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bushnell G.W., Louie G.V., Brayer G.D. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 71.Wilkens S., Dunn S.D., Chandler J., Dahlquist F.W., Capaldi R.A. Solution structure of the terminal domain of the δ-subunit of the E. coli ATPsynthase. Nat. Struct. Biol. 1997;4:198–201. doi: 10.1038/nsb0397-198. [DOI] [PubMed] [Google Scholar]
- 72.Manion M.K., O'Neill J.W., Giedt C.D., Kim K.M., Zhang K.Y.Z. Bcl-XL mutations suppress cellular sensitivity to antimycin A. J. Biol. Chem. 2004;279:2159–2165. doi: 10.1074/jbc.M306021200. [DOI] [PubMed] [Google Scholar]
- 73.Usón I., Pohl E., Schneider T.R., Dauter Z., Schmidt A. 1.7 Å structure of the stabilized REIv mutant T39K. Application of local NCS restraints. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1158–1167. doi: 10.1107/s0907444999003972. [DOI] [PubMed] [Google Scholar]
- 74.Ulaganathan, V., L. Buetow, and W. Hunter. Nucleotide substrate recognition by UDP-n-acetylglucosamine acyltransferase (LPXA) in the first step of lipid A biosynthesis. Accepted. [DOI] [PubMed]
- 75.Thoden J.B., Kim J., Raushel F.M., Holden H.M. The catalytic mechanism of galactose mutarotase. Protein Sci. 2003;12:1051–1059. doi: 10.1110/ps.0243203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumann U., Bode W., Huber R., Travis J., Potempa J. Crystal structure of cleaved equine leukocyte elastase inhibitor determined at 1.95Å resolution. J. Mol. Biol. 1992;226:1207–1218. doi: 10.1016/0022-2836(92)91062-t. [DOI] [PubMed] [Google Scholar]
- 77.Douglas A.E., Corbett K.D., Berger J.M., McFadden G., Handel T.M. Structure of M11L: a myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci. 2007;16:695–703. doi: 10.1110/ps.062720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeffrey P., Bewley M., MacGillivray R., Mason A., Woodworth R. Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin. Biochemistry. 1998;37:13978–13986. doi: 10.1021/bi9812064. [DOI] [PubMed] [Google Scholar]
- 79.MacGillivray R., Moore S., Chen J., Anderson B., Baker H. Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry. 1998;37:7919–7928. doi: 10.1021/bi980355j. [DOI] [PubMed] [Google Scholar]
- 80.Meador W., Means A., Quiocho F. Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 81.van den Ent F., Moller-Jensen J., Amos L.A., Gerdes K., Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]


