Abstract
Cellular signal transduction machinery integrates information from multiple inputs to actuate discrete cellular behaviors. Interaction complexity exists when an input modulates the output behavior that results from other inputs. To address whether this machinery is iteratively complex, that is, whether increasing numbers of inputs produce exponential increases in discrete cellular behaviors, we examined the modulated secretion of six cytokines from macrophages in response to up to 5-way combinations of an agonist of Toll-like receptor 4 with 3 cytokines and the cyclic adenosine monophosphate pathway. Although all of the selected ligands demonstrated synergy in paired combinations, few examples of nonadditive outputs were found in response to higher-order combinations. This suggests that most potential interactions are not realized and that unique cellular responses are limited to discrete subsets of ligands and pathways that enhance specific cellular functions.
INTRODUCTION
The outcomes of signal transduction pathways are usually ascertained by studying the effects of specific, single ligands on isolated cells in culture; however, cells are rarely exposed to a single stimulus in vivo, and cellular behavior can be highly dependent on the integration of responses to a combination of specific stimuli and on the context in which they occur. This is clearly evident in the immune system. During the response to infection, the interaction of viral or bacterial components with T helper (TH) cells and macrophages causes the production of cytokines and chemokines, which in turn form a network of chemoattractants and activators for circulating leukocytes in vivo. The nature of an inflammatory response is determined by the primary signaling pathways evoked and by interactions among these pathways. For example, the production of cytokines in response to initiators of inflammation can be influenced by synergy between individual Toll-like receptors (TLRs). Mice treated with TLR agonists that stimulate myeloid differentiation marker 88 (MyD88)-dependent pathways in combination with those TLR agonists that activate MyD88-independent pathways produce elevated, nonadditive concentrations of tumor necrosis factor α (TNF-α), macrophage inflammatory protein-1α (MIP-1α), and interleukin 6 (IL-6) in their serum compared to the predicted secretion of the cytokines based on mice treated with each TLR agonist individually1. Environmental factors and secondary responses also modulate output. Thus, production of chemoattractants is often strongly regulated by synergy between secreted cytokines2. For example, intra-articular injection of submaximal doses of TNF-α and IL-1 into the knees of rabbits results in a nonadditive increase in the numbers of polymorphonuclear cells in the synovial lining as compared to that elicited by either cytokine alone, which provides evidence for a biological role of synergy in cell recruitment3, 4.
Regulation of signal transduction through synergy or anergy among pathways provides a simple and plausible mechanism to dramatically change the nature of downstream responses to a stimulus depending on the presence of other relevant stimuli. From an experimental standpoint, these signaling regimes are unique in that they are not inherently predictable from their individual components. The first step in elucidating such synergies is a profiling approach, which may also provide the initial insight needed for the mechanistic deconstruction of the signaling architecture in the cells of interest and the manner in which such signaling is mediated. For example, elucidation of cytokine secretion profiles in response to multiple, simultaneous stimuli and knowledge of their input-output relationships can be a precursor to understanding complex physiological processes mediated by macrophages.
Initial profiling of crosstalk between signal transduction pathways in a macrophage-like cell line (RAW 264.7) by the Alliance for Cellular Signaling (AfCS) included quantification of cytokines secreted in response to stimulation by 22 individual ligands and by all 231 pair-wise combinations5. Global analysis of this dual ligand screen revealed both known and novel synergistic interactions between these ligands. For example, these nonadditive, synergistic effects could result in 20- to 40-fold increases in the output of a given second messenger and they reflected novel patterns of pathway interactions for subsequent elucidation6, 7. However, approximately 90% of the pairwise combinations of ligands resulted in predictable additive responses. This suggests that only a small discrete number of the many possible interactions between signaling pathways are selected and actuated, whereas the vast majority of possible interactions are unrealized or ineffective.
An essential question that follows is whether the same pattern will hold in a biologically relevant process with responses to higher-order combinations of ligands. Thus, will only a small subset of combinations lead to nonadditive, unique responses or will increased complexity of inputs produce exponential increases in discrete cellular behaviors? As with dual ligand stimulations, the potential for synergies among multiple pathways makes simple extrapolation from the dual ligand instance insufficient; direct testing is required. Because the testing of higher-order combinations of ligands increases exponentially the number of experiments to be performed, a subset of stimuli and outputs were chosen from the dual ligand screen to identify additional interactions in response to 3-, 4-, and 5-way combinations of ligands.
Lipopolysaccharide (LPS) and its receptor TLR4 are thought to be crucial arbiters of sepsis. Observations that mice devoid of TLR4 or its associated protein MD-2 are hyporesponsive to LPS8, 9 and resistant to endotoxic shock10 emphasize the biological importance of this receptor complex in vivo. We chose to profile modulation of the cytokine responses of RAW 264.7 cells to stimulation of TLR4, the best characterized receptor of the TLR family11–14, by four ligands: interferon-β (IFN-β), transforming growth factor β-1 (TGF-β), IL-6, and isoproterenol (ISO) or a nonhydrolyzable analog of its second messenger, 8-Bromo-cAMP (8Br). Similarly, the outputs measured were limited to 4 cytokines, granulocyte colony–stimulating factor (G-CSF), IL-6, IL-10, and TNF-α and 2 members of the cysteine-cysteine (CC) family of chemokines, MIP-1α and regulated upon activation, normal T cell expressed and secreted (RANTES), which were readily detectable in secretions from RAW 264.7 cells. Higher-order synergies between the ligands tested were elucidated through a new approach to determine statistical significance in multifactorial experiments based on established methodologies.
Two major sets of findings were revealed through the analysis of this data set. The first relates to the architecture of signaling networks for generating unique signaling regimes. Despite the fact that almost all of the pairs of ligands used were capable of nonadditive interactions that caused unique signaling regimes, only a modest subset of higher-order combinations resulted in unique paradigms of secretion of individual cytokines. The inference to be made from these data is that complex nonlinear interactions are limited in these signaling networks despite the multitude of possibilities. Second, our analysis comprehensively documented that discrete combinations of a TLR agonist, IFN-β, ISO or 8Br, and TGF-β could simultaneously regulate multiple cytokines through parallel mechanisms and provide a basis for future elucidation of novel pathways and modeling of specific context-dependent behaviors.
RESULTS
Profiles of ligand-induced cytokine secretion by RAW 264.7 cells
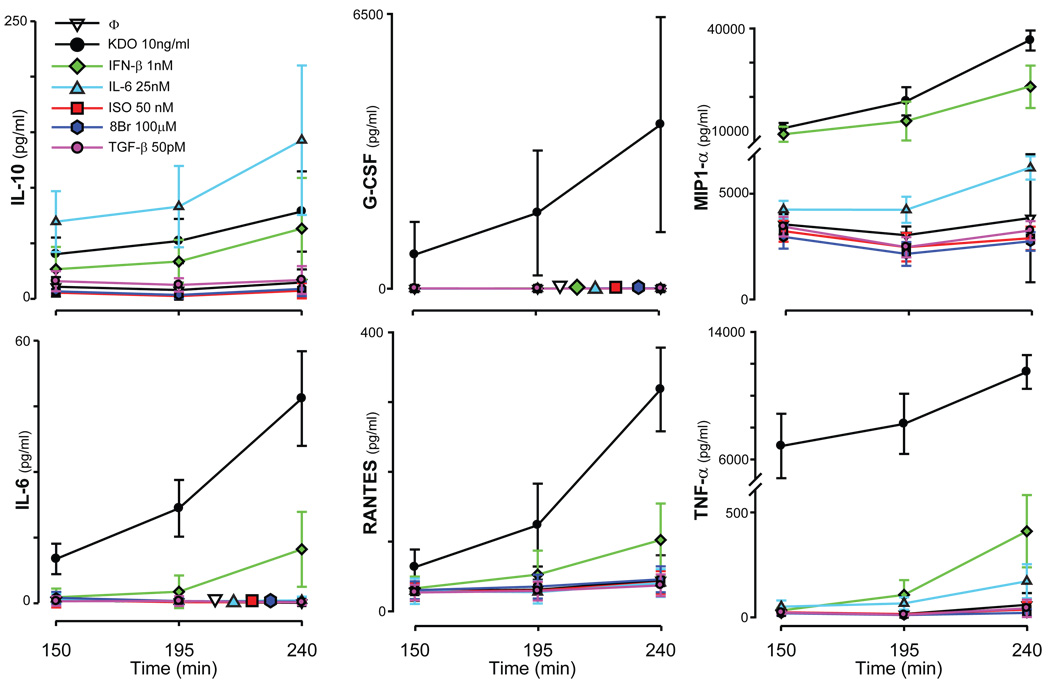
Secretion of IL-10, IL-6, RANTES, MIP-1α, G-CSF, and TNF-α was examined after cells were stimulated through TLR4 with a chemically defined LPS, 3-deoxy-D-manno-octulosonic acid (KDO)2-Lipid A15, 16, in all possible combinations with the ligands ISO (or 8Br), TGF-β, IL-6, or IFN-β. All of the cytokines and chemokines measured in this study were readily secreted by RAW 264.7 cells in response to KDO (Fig. 1), which was chosen as the stimulus instead of LPS to reduce variability in measurements.
Fig. 1.
Secretion of cytokines in response to single ligands. RAW 264.7 cells were treated with KDO (black circle), IFN-β (green diamond), IL-6 (light-blue triangle), ISO (red square), 8Br-cAMP (dark-blue hexagon), TGF-β (pink circle), or were left untreated (open inverted triangle) for 150, 195, or 240 minutes. The supernatants were removed, and the concentrations of IL-10, IL-6, G-CSF, TNF-α, RANTES, and MIP-1α were measured (pg/ml). The normalized traces [mean ± standard deviation (s.d.), n>10) are shown. For traces in which the lines overlap, additional staggered symbols are shown for easy identification.
IFN-β also induced the production of IL-6, IL-10, MIP-1α, RANTES, and TNF-α, and IL-6 induced the production of IL-10. Although TGF-β and ISO did not have any measurable effects on cytokine release individually during this time course (Fig. 1), they were included because of their substantial modulation of cytokine secretion induced by TLRs5. As will be shown, the combination of ISO, 8Br, TGF-β, IL-6, or IFN-β with KDO potentiated or inhibited the secretion of multiple cytokines induced by KDO, data that are consistent with previously published findings that show crosstalk between LPS and a subset of biologically diverse ligands that regulate the secretion of cytokines5. These studies are extended here to examine the extent of crosstalk among pathways, that is, to determine what higher-order interactions could be observed when cells were stimulated with KDO and multiple combinations of the interacting ligands.
Evaluation of interactions among multiple ligands
Interactions between cytokines were quantified through an approach similar to previous methodologies5 wherein a z-score-like parameter estimated the difference between the expected outcome and the observed outcome as weighted by the propagated errors (see Materials and Methods). Note that a nonadditive response to two or more ligands simply indicates that the downstream signal transduction pathways share a signaling component; the mechanism of how the interaction occurs is not revealed. The z-score parameter simply quantifies the significance of nonadditivity in the positive (synergy) or negative (antagonism) direction.
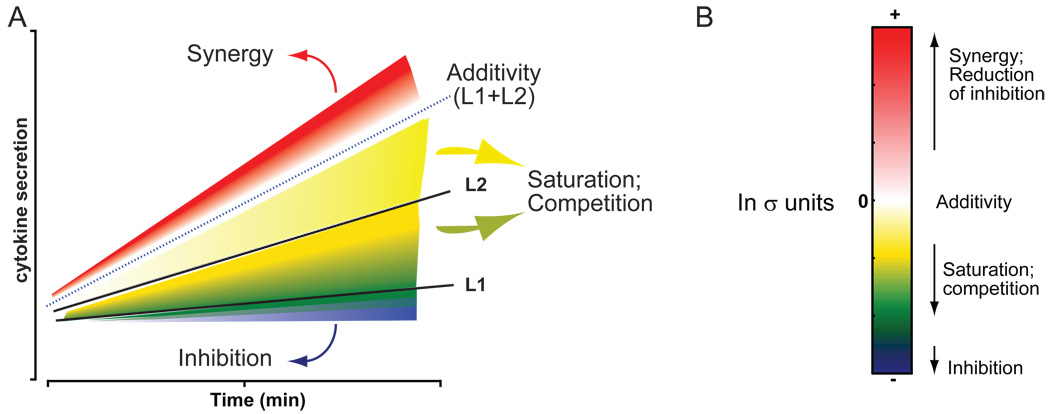
A graphic illustration shows how different signaling regimes are possible (Fig. 2). The combination of two ligands with the cytokine responses L1 and L2 can elicit: (i) significant synergy—a response greater than the sum of both responses (>L1+L2), indicated by the region in red; (ii) significant inhibition—a response less than the least response to any individual ligand as indicated by the region in blue, that is <L1 (an exception occurs when L1 or L2 has no effect alone, in which case any significant decrease is clear inhibition); (iii) additivity—a response that is not significantly different from the additive sum of all the responses, indicated by the region in white around the dotted line that represents the sum of all responses (~L1+L2). In addition to these simple cases, there is also a region that is below additivity but above inhibition (that is, L1+L2 > response > L1) that may be a result of any one of several factors, such as partial inhibition, saturation of a signaling component, antagonism or competition, for example (shaded in a range from light yellow to dark green). With the inclusion of more ligands in the experiment, the predicted additivity is not simply the sum of the responses to individual ligands but also includes nonadditive contributions between them. For example, for 4-way combinations, the nonadditive contributions of all 2-way and 3-way combinations in addition to the responses to the four individual ligand treatments are factored in (see Methods and Materials).
Fig. 2.
Classification of interactions between ligands. Two ligands, L1 and L2, are capable of eliciting cytokine secretion individually (A, solid black lines). When applied in combination, the two ligands may act in any one of several ways: they may act independently of each other yielding an additive response (dotted black line, surrounding white region); they could synergize to yield a greater-than-additive response (red); they may inhibit each other to yield a response lower than the individual responses (blue); they could elicit some intermediate response between the responses of the individual ligands and that of the predicted additivity (yellow to green). The last set of responses may be mechanistically generated by any one of several processes, including saturation and noncompetitive antagonism, among others. (B) The significance of nonadditivity can be calculated and displayed by this colorimetric scheme in a heat map.
More complex scenarios are likely to exist with the larger combinations of ligands examined in this study. For instance, in the event that some ligands are excitatory whereas other ligands are inhibitory, the addition of another ligand could relieve the net inhibition, which is numerically calculated as a positive z-score, yet the final amount of cytokine that is secreted in response to the combination may be much smaller than some of the responses to individual ligands. Extensive experimentation is likely necessary to decipher the mechanisms of action in all such cases. We have characterized all signaling outcomes with the above coloring scheme to provide an easy visual method of identifying significant changes. Note that a z-score does not have a one-to-one relationship with color intensity for less-than-additive values. If the observed value is lower than the least response to any individual ligand or to an n-1 subset of combinations, we can then clearly classify the response as inhibitory and its significance is coded in blue. For all other less-than-additive cases, we cannot distinguish between inhibition, saturation, or competition without further experimentation. Therefore these are explicitly color-coded yellow-green. Thus, the actual color code in the less-than-additive responses is not dependent purely upon the magnitude of the z-score (significance of difference), but is also dependent upon the least response within each experiment and is thus relative. Thus, it is possible to have a scenario in which a large negative z-score may be colored green in one experiment whereas a smaller negative z-score may be colored blue in a different experiment. A cumulative summary of responses is presented in the final figure for easy comparison across combinations and for use by the signaling community at large. We describe and characterize some of the strong synergistic and inhibitory responses in this study below.
Examples of complex signaling regimes induced by stimulation with multicombinatorial ligands
Examples of nonadditivity between ligands on the secretion of IL-10 are first discussed (Fig. 3). Individually, IFN-β, IL-6, and KDO induced the secretion of IL-10 from RAW 264.7 cells (Fig. 1, Fig. 3, A and B). When either IFN-β or IL-6 was combined with KDO and added simultaneously to RAW 264.7 cells, the expected secretion of IL-10 (the additive sum of responses from stimulation with each ligand alone) was far less than the experimentally observed responses to the combinations (Fig. 3, A and B). The dramatic increase in the amount of IL-10 secreted suggests that IFN-β and IL-6 can cooperatively interact with KDO to regulate the secretion of IL-10. The large synergies with the dual ligands were partially blunted when all three ligands were applied to the cells. Thus, the observed effect was significantly less than the additive prediction (Fig. 3D) and saturation of common elements in the pathway or an antagonism between IL-6 and IFN-β is likely. An interesting hypothesis derives from the observation that the combination of KDO and IFN-β also synergistically induced the secretion of IL-6 (Fig. 3C).
Fig. 3.
Secretion of IL-10 in response to KDO, IFN-β, and IL-6. RAW 264.7 cells were treated with single, double, or triple combinations of ligands. The results for each cytokine were normalized and the statistical significance was determined as described in the Materials and Methods. Examples of secretion of IL-10 are shown for the dual (A and B) and triple (D) combinations of ligands. (C) Secretion of IL-6 can also be synergistically induced by the combination of KDO with IFN-β. Panels A to C display the single ligand responses, the predicted response of combinations of ligands (expected), and the observed values for combinations (observed). Panel D shows only the predicted and observed values for the 3-way combinations. Statistical significance for nonadditivity of the combinations is shown at the right.
The latter result suggests the hypothesis that synergistic production of IL-6 by KDO and IFN-β may have acted in an autocrine feedback loop to contribute to the observed synergistic increase in the secretion of IL-10 evoked by KDO and IFN-β. If so, the three-way combination of KDO, IFN-β, and IL-6 must have resulted in a less-than-additive response, because any synergy from the combination of KDO and IL-6 would already be represented, at least in part, from the IL-6 produced by the combination of KDO and IFN-β. This would most likely be observed as a time-dependent decrease in synergy or loss of additivity as IL-6 accumulated in the assay. A clear trend to support this was not observed (Fig 3D). In order to assess whether secreted IL-6 was playing a substantial role, the secretion of IL-10 was examined at different doses of IL-6. The threshold concentration of IL-6 required for stimulating the secretion of IL-10 was 0.2 nM (> 4 ng/ml, a concentration above the maximal 2 ng/ml of IL-6 produced by the stimulating conditions at 4 hr. Whereas it is possible that the secreted IL-6 might have been more effective than that added in bulk to initiate incubations, it is probable that the large synergy in secretion of IL-10 seen with KDO and IFN-β and the less than additive response seen with the combination of KDO, IFN-β and IL-6, were not due to an autocrine effect of secreted IL-6 (see Discussion).
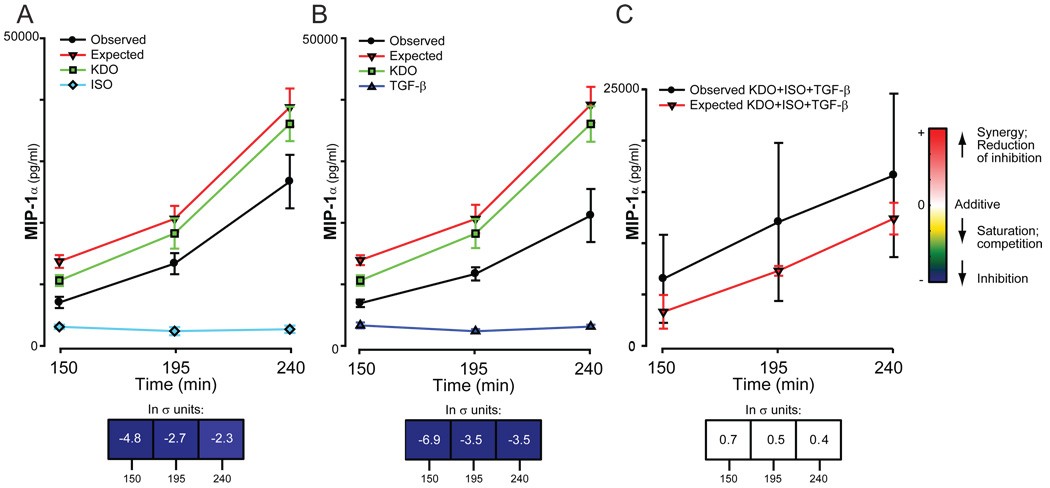
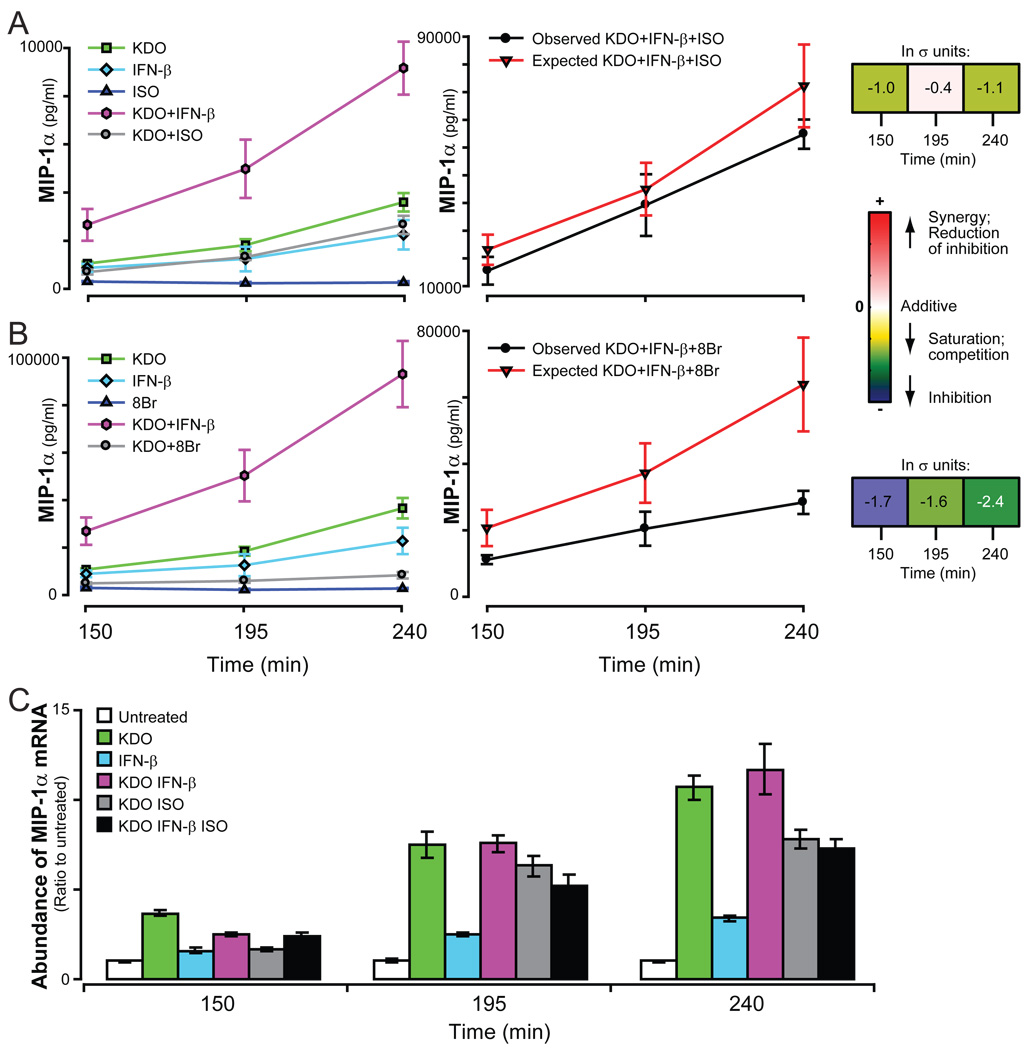
In the instance where nonadditive responses to two or more combinations of ligands are mediated through completely independent mechanisms, a multifactorial combination of these same ligands should yield additive responses. One such instance is now discussed (Fig. 4). ISO and TGF-β independently inhibited the amount of MIP-1α secreted in response to KDO (Fig. 4, A and B). These results suggest that ISO and TGF-β each individually share a common regulatory element with KDO in the regulation of the secretion of MIP-1α. However, the essentially additive response to the triple ligand combination (σ < ±1, Fig. 4C) suggests that although the combinations of KDO-ISO and KDO-TGF-β each share a common stimulating component to regulate the secretion of MIP-1α, the two interaction elements and subsequent pathways are probably different from each other. If this is the case, the data suggest that there are at least two distinct mechanisms by which the secretion of MIP-1α can be regulated by KDO.
Fig. 4.
Secretion of MIP-1α in response to KDO, IFN-β, and IL-6. RAW 264.7 cells were treated with single, double, or triple combinations of ligands and the secretion of MIP-1α was measured as described. The results from multiple experiments were normalized and statistical significance was determined as described in the Materials and Methods. Each panel shows the single ligand responses and the observed and predicted (expected) responses for dual (A and B) and triple (C) ligand combinations. The predicted response is based on the additive effects of the ligands; the bar underneath the graphs quantitates the nonadditivity of the ligand combinations.
Regulation of secretion of G-CSF by KDO, IFN-β, and cAMP pathways
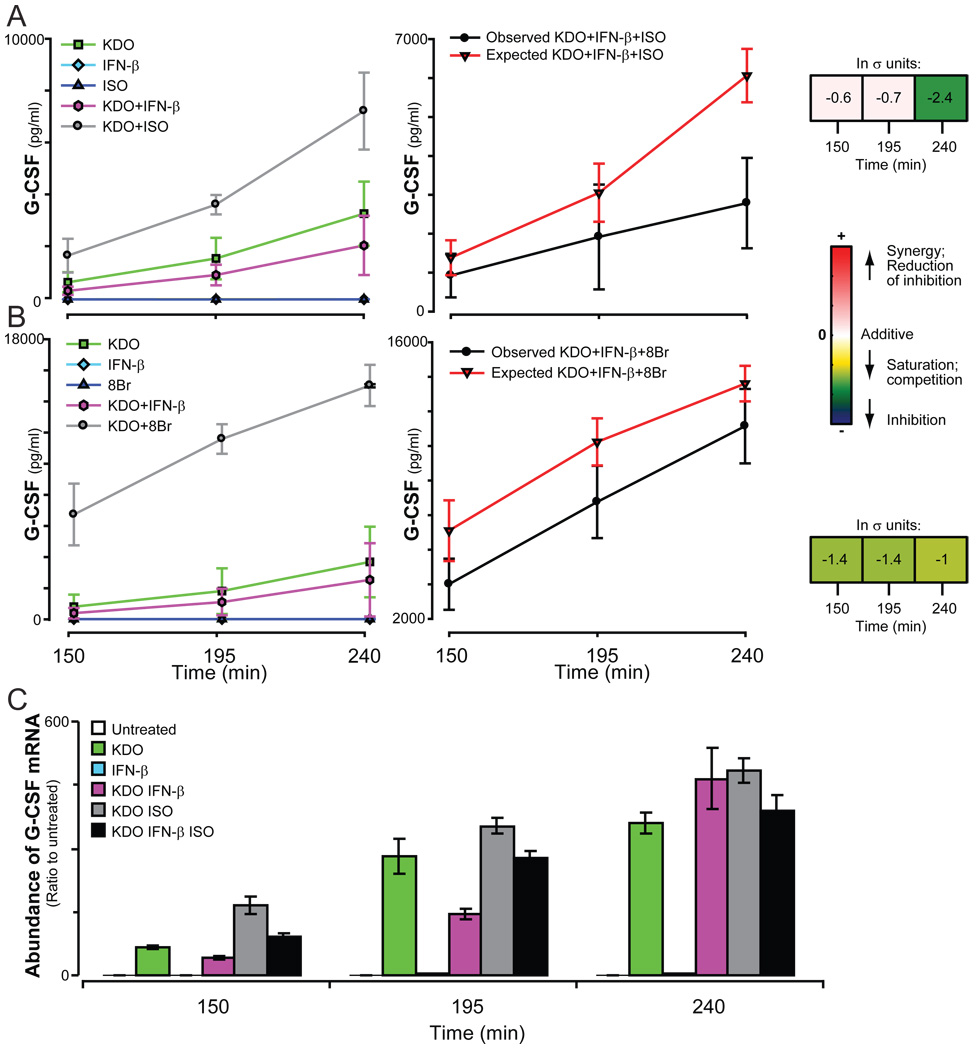
Our multiligand screen has also identified previously uncharacterized interactions among KDO, IFN-β, and cAMP pathways that simultaneously regulated the secretion of 3 cytokines. Both ISO or 8Br and IFN-β regulated KDO-induced secretion of G-CSF, but in opposing directions; cAMP, either stimulated by ISO or added directly as 8Br amplified KDO-dependent secretion of G-CSF, whereas IFN-β modestly inhibited secretion of the cytokine. When cells were treated with the combination of KDO, IFN-β, and ISO or KDO, IFN-β, and 8Br, secretion of G-CSF was less than that of the additive prediction (Fig. 5, A and B). Indeed, IFN-β almost eliminated the enhancement effected by ISO, although it was less dominant against the more efficacious 8Br. These results suggest a shared mode of regulation for G-CSF among the three pathways.
Fig. 5.
Secretion of G-CSF in response to KDO, IFN-β, and ISO. The secretion of G-CSF (A and B) by RAW 264.7 cells was measured in response to KDO, IFN-β, and either ISO or 8Br. (C) The relative abundance of G-CSF mRNA was quantified by qRT-PCR. A single representative experiment is shown. Error bars show the range between duplicate samples. All samples were normalized to time-matched untreated samples. All columns except the responses to IFN-β alone were significantly different from the untreated sample (p<0.05).
This interaction among the pathways was also observed in the relative abundance of G-CSF messenger RNA (mRNA). The addition of ISO increased the abundance of G-CSF mRNA, whereas the inclusion of IFN-β reduced the amount of G-CSF mRNA induced by KDO at early times or by KDO and ISO at all times (Fig. 5C). The similarity in the pattern of expression of G-CSF mRNA and the secretion of G-CSF protein indicates that the point at which these pathways intersect is at, or upstream of, the production of mRNA. The data also suggest that regulation of G-CSF production by these ligands is not dominated by a single pathway and that responses will be dictated by the relative strength of the different stimuli.
Regulation of MIP-1α secretion by KDO, IFN-β, and cAMP pathways
The second cytokine that was regulated by the combination of KDO, IFN-β, and cAMP pathways was MIP-1α (Fig. 6). KDO and IFN-β individually induced increases in the abundance of MIP-1α mRNA (Fig. 6C) and in the secretion of MIP-1α protein (Fig. 1). The combination of KDO and IFN-β resulted in potentiated secretion of MIP-1α (Fig. 6), a result that contrasted with the inhibitory effect of IFN-β on KDO-induced secretion of G-CSF (Fig. 5). ISO interacted with the KDO pathway to antagonize secretion of MIP-1α (Fig. 6), which is also a contrast to its potentiation of G-CSF secretion by the TLR pathway. When IFN-β, ISO, and KDO were simultaneously added to RAW 264.7 cells, the competing effects on the secretion of MIP-1α were less than additive. This pattern of interaction among the three pathways was further substantiated by the observations with 8Br, which maximally stimulated the cAMP pathway (Fig. 6B). The less-than-additive effects obtained with ISO and IFN-β on stimulating the secretion of MIP-1α by KDO may be explained by the inhibitory effects of ISO on secretion of the cytokine induced by IFN-β alone5. Similar to G-CSF, regulation of MIP-1α did not appear to be dominated by KDO, IFN-β, or the cAMP pathway.
Fig. 6.
Secretion of MIP-1α in response to KDO, IFN-β, and ISO. The secretion of MIP-1α by RAW 264.7 cells was measured in response to KDO, IFN-β, and either ISO (A) or 8Br (B). Results are the average of 2 to 5 normalized experiments each containing duplicate samples. (C) The relative abundance of G-CSF mRNA was quantified by qRT-PCR. A single representative experiment is shown. Error bars show the range between duplicate samples. All samples were normalized to time-matched untreated samples. All columns were statistically significantly different from the untreated sample (p<0.05).
Unlike that of G-CSF, changes in the secretion of MIP-1α did not mirror changes in the abundance of its mRNA (Fig 6C). When KDO and ISO were combined, the abundance of MIP-1α mRNA and the secretion of MIP-1α protein were both lower than with KDO alone. However, the combination of KDO and IFN-β produced less than or additive effects on the abundance of MIP-1α mRNA while potentiating secretion of the protein. This suggests that the synergy between KDO and IFN-β in the regulation of MIP-1α secretion occurs through a posttranscriptional regulatory mechanism. This complex regulation was further observed when all three ligands were combined; the less-than-additive response in the secretion of the cytokine was not accompanied by a significant reduction in the abundance of MIP-1α mRNA beyond that observed when ISO alone was added with KDO. These observations likely represent nonredundant mechanisms (transcriptional and posttranscriptional) of regulating the secretion of MIP-1α by KDO and IFN-β because the observed synergy was not detected at the level of transcription.
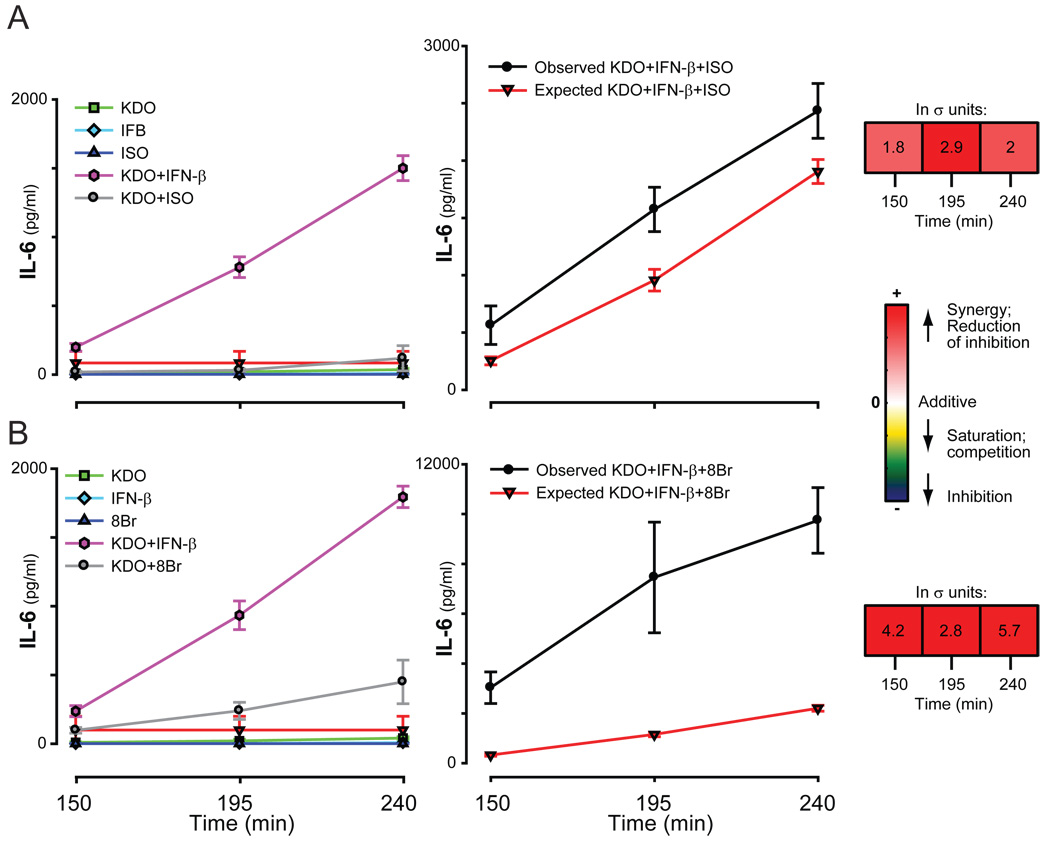
Potentiation of the secretion of IL-6, but not IL-10, by KDO, IFN-β, and cAMP pathways
Although the triple ligand combination resulted in intermediate quantities of secreted G-CSF and MIP-1α, secretion of IL-6 was greatly potentiated when IFN-β, KDO, and the cAMP pathway were combined (Fig. 7), which suggests that these pathways share common mechanisms to simultaneously regulate secretion of this cytokine. Interestingly, IL-10, which was secreted in response to IL-6, did not show similar potentiation (see Fig. 9). Presumably, the amount of IL-6 secreted was insufficient to support an autocrine response, as discussed earlier. Whereas the synergism of IFN-β and KDO observed for both IL-6 and IL-10 may share a common mechanism, the data indicate that synergy of the cAMP pathway is achieved at a branch unique to regulation of IL-6 and probably downstream from the interaction of IFN-β and KDO.
Fig. 7.
Secretion of IL-6 in response to KDO, IFN-β, and ISO. The secretion of IL-6 by RAW 264.7 cells was measured in response to KDO, IFN-β, and ISO (A) or 8Br (B). The average of the normalized values of 2 to 5 experiments, each with at least duplicate samples, are shown.
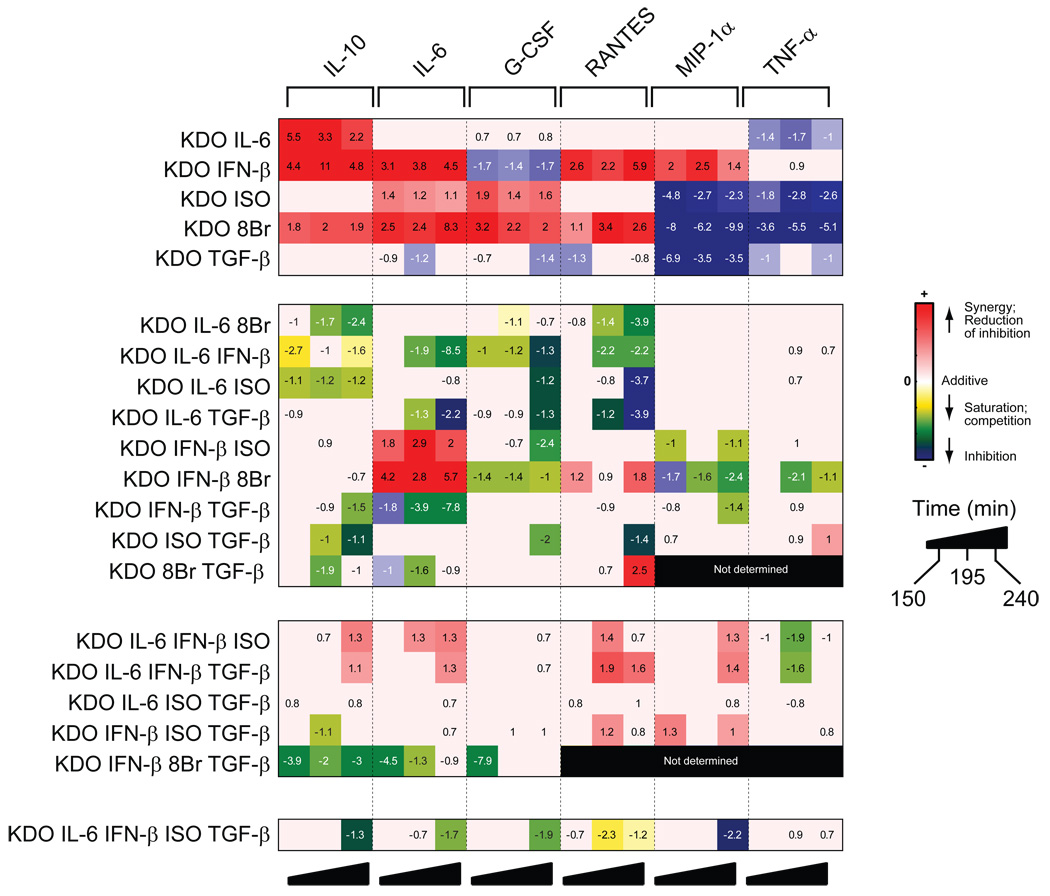
Fig. 9.
Quantified, synergistic cytokine responses to stimulation with multiple ligands. A summary matrix of the cytokine responses in RAW 264.7 cells is shown for all treatments performed with single, double, triple, quadruple, and quintuple combinations of ligands. The results for each cytokine were normalized and the statistical significance was determined as described in the Materials and Methods.
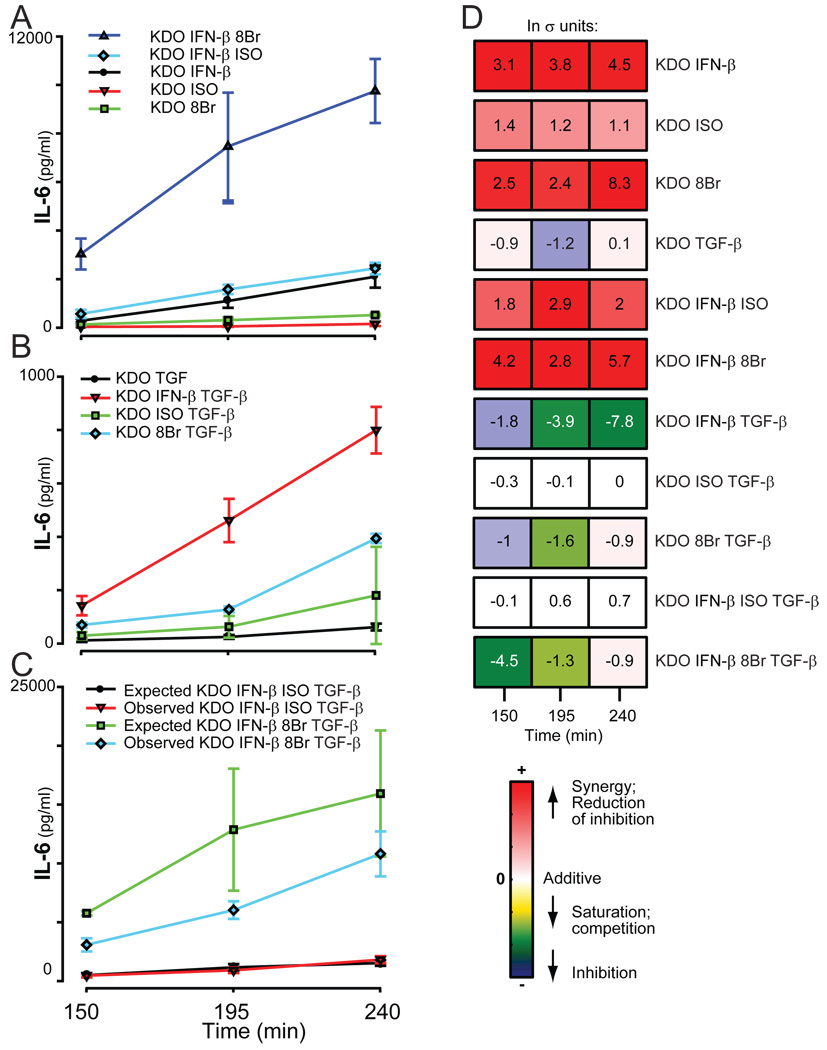
TGF-β regulates secretion of IL-6 stimulated by the KDO, IFN-β, and cAMP pathways
TGF-β by itself had no measurable effect on the secretion of IL-6 from RAW 264.7 cells; however, it attenuated the two-way synergistic combination of KDO and IFN-β (Fig 8). Whereas TGF-β did not appear to attenuate the synergy between KDO and ISO, anergy was observed when TGF-β was combined with the more efficacious combination of 8Br and KDO (Fig. 7B, Fig. 8). This demonstrates that TGF-β strongly inhibited the synergistic responses of these ligand pairs. Although the four-way combination with KDO, IFN-β, ISO, and TGF-β was largely additive, the much larger synergy in IL-6 production by KDO, IFN-β and 8Br was significantly attenuated by TGF-β beyond the additive attenuation expected (Fig. 8B). This suggests that not only could TGF-β inhibit the synergistic stimulation of IL-6 by KDO and cAMP pathways or by KDO and IFN-β, it also suppressed the three-way synergy observed with KDO, IFN-β and mobilization of cAMP-dependent pathways.
Fig. 8.
Secretion of IL-6 in response to KDO, IFN-β, TGF-β, and the cAMP pathway. The secretion of IL-6 by RAW 264.7 cells was measured after treatment of cells with KDO, IFN-β, TGF-β, and either ISO or 8Br for the indicated times. The normalized values of 2 to 5 experiments with samples prepared in at least duplicate were averaged and the errors determined.
A systematic compilation of non-additivity across all combinations
As the calculation of nonadditivity by σ units is inherently dimensionless, it enables us to compile, and therefore compare, responses to all combinations of ligands to determine the extent of nonadditivity in response to all of the 2-, 3-, 4-, and 5-way combinations examined in this study (Fig. 9). This approach allowed us to easily visualize the effects of compounding nonadditive interactions from smaller combinations of ligands to higher-order combinations—a tool whose value cannot be underestimated when compiling such complex datasets. Secondly, this allows us to estimate the extent of nonadditivity with increases in input complexity.
DISCUSSION
This study simultaneously examines the secretion of 6 factors (G-CSF, IL-6, IL-10, MIP-1α, RANTES, and TNF-α) by RAW 264.7 cells in response to simultaneous application of multiple stimuli. The multi-cytokine responses were used to examine two issues. As a platform to identify instances of significant nonadditivity, the multi-factorial experiments addressed the general question of complexity and crosstalk in signal transduction. This same profiling process was used to identify previously uncharacterized and specific higher-order interactions that regulated cytokine secretion. With regard to the first point, does greater information complexity, in this case larger numbers of stimuli encountered by a cell, equate to greater complexity in output? In other words, if a cell is capable of responding to a number of individual stimuli that result in a unique set of behaviors, does presenting the total ensemble or various subsets of inputs result in unique sets of responses? Moreover, does such behavior scale with an increase in the number of inputs? The current literature presents contrasting possibilities. High-throughput measurements of protein-protein interactions, such as those obtained through two-hybrid experiments, and the large number of components now identified in cellular signaling systems, lead to speculation that the networks of signal transduction machinery are inordinately complex17, 18. However, such observations often reflect connections that are possible in an in vitro context and are not necessarily of physiological consequence in vivo. Other studies indicate that cells may have evolved elaborate mechanisms to limit interaction complexity, and may have invested heavily in the macromolecular organization of signaling machinery to limit both the spatial and temporal spreading of signaling events in cells19, 20. The mechanisms that limit interactions are more difficult to define than those that encourage synergy.
Our study examined this issue in a system in which a number of different factors are known to interact in simple combinations of inputs, and thus allowed us to ask in a meaningful context whether novel signaling regimes arise from complex higher-order combinations of elements. Although our own work5 and other studies21–25 have profiled the extent and context-dependence of nonadditive cellular responses to pairs of stimulators, this is the first study that systematically examines whether complex signaling responses can be mediated in response to stimulation by higher-order combinations of ligands. Importantly, the results show that such higher-order combinations of ligands can induce unique signaling regimes, but that such instances are limited when compared to the total number of combinations possible (~33%, with the majority in the 3-way combinations). This limitation is considered important because all of the ligands used in this study were known to be interactive through their production of nonadditive responses in two-way combinations.
Although we cannot generalize signaling architectures from one set of results, the following conclusions can be drawn in this instance. First, complexity of output responses is “encoded” or “hard-wired” into the system by the presence of interactions between pathways (as evinced by the nonadditivity of the two-way combinations). Increasing the number of stimuli to engage more of these interactions between pathways did not, in general, result in a concomitant, iterative increase in output complexity. Second, the interactions that are realized can uniquely shape overall cellular behavior in response to complex stimuli. Almost every higher-order combination of ligands, (that is, looking at each row in Fig.9 for all combinations of ligands 3- or higher, 14/15 rows) did elicit a significant, nonadditive response in at least one cytokine; however, nonadditive changes among individual cytokines are often not observed even though certain pairs of ligands involved were highly interactive [see responses for MIP-1α and TNF-α (Fig. 9)]. Because such unrealized interactions to ligand combinations varied with individual cytokines, correlations in nonadditive responses among various cytokines observed with dual ligands5 (Fig. 9, top panel) were not maintained with higher-order combinations of the same ligands. Thus, higher-order combinations of ligands gave rise to limited, unique behaviors based on restricted interactions among the signaling pathways.
The estimate of nonadditivity does not inform us about the mechanism by which it happens. Although some cytokines appear to trend together in their responses to the two-ligand perturbations, this does not imply a similarity in how this nonadditivity is mediated for these cytokines. This becomes especially evident when the correlation is lost with higher-order combinations of ligands. Indeed, this suggests that the mechanisms for the correlated behavior with dual ligands are different. By comparing and contrasting responses across combinations of ligands, we can gain some insight into the requirements for the nonadditivity observed, and this will help shape future studies to elucidate the molecular mechanisms involved.
Thus, even in a multifactorial system such as the cytokine network in which multiple crosstalk and feedback loops are present, information complexity is encoded in sparse and discrete instances. This suggests that context-dependence, or the ability of the system to recognize two stimuli in conjunction and to respond differently from either stimulus taken individually, is encoded through select interactions or pathways that are not extensively or promiscuously activated. Several other studies have argued that such limitation of interaction complexity is evolved in architectures that enhance “robustness” or dependability of signal transduction19, 20. If this is true, it stands to reason that the few higher-order interactions observed are very likely to indicate biologically important phenotypes.
A second objective of this study was to identify any novel higher-order interactions between the signaling mechanisms downstream of the receptor for LPS (a major antigenic factor of gram-negative bacteria) and the 4 families of receptors used. Although the molecular mechanisms by which these pathways exert their effects cannot be definitively determined by this approach, our data are highly consistent with published mechanisms that regulate cytokine secretion (as outlined below) and allow for tractable generation of hypotheses for future experimentation.
We focused specifically on the novel observation that the downstream pathways from IFN-β and cAMP interacted to simultaneously both amplify and inhibit G-CSF and MIP-1α produced in response to KDO. Furthermore, secretion of IL-6 was greatly potentiated when all three ligands were combined. These data demonstrate the multiple regulatory nature of each pathway to simultaneously and differentially modulate multiple cytokines as discussed below.
The secretion of many cytokines is regulated through de novo synthesis of their nascent transcript. Stimulation of the secretion of G-CSF by KDO was inhibited by IFN-β, but was potentiated by cAMP-dependent pathways. When all three ligands were combined, a less-than-additive, intermediate amount of G-CSF was secreted. The relative pattern of secreted G-CSF obtained with single, double, and triple combinations of ligands mirrored the pattern observed in the expression of its mRNA (Fig. 5), which suggests that these interacting pathways regulated secretion of this cytokine by regulating the expression of its mRNA.
Regulation of MIP-1α by the same ligands showed opposing effects and suggests the existence of additional mechanisms. Unlike its effect on secretion of G-CSF, cAMP-dependent pathways suppressed secretion of MIP-1α induced by KDO and this was paralleled by attenuation of the KDO-induced expression of MIP-1α mRNA (Fig. 6). This modulation of mRNA expression for both cytokines indicates that the intersection between the TLR4 and cAMP pathways for regulation of secretion either resides upstream of transcription or at the regulation of transcript production or stability. One putative node of intersection could be in the regulation of nuclear factor κB (NF-κB), a key mediator of the TLR-induced secretion of many cytokines; however, the opposing action of cAMP on G-CSF and MIP-1α indicates that this cannot uniquely account for both effects.
In contrast to G-CSF, the greater-than-additive amounts of MIP-1α secreted in the presence of KDO and IFN-β was accomplished while the abundance of MIP-1α mRNA either did not change or showed a less-than-additive response. This result has also been observed in purified human monocytes cultured with IFN-α and LPS26. Thus, regulation by IFN-β appears to occur through two pathways. Stimulation of MIP-1α secretion by IFN-β alone correlated with a modest increase in the abundance of MIP-1α mRNA and predicts a transcriptional mechanism of regulation. However, a lack of increased abundance of mRNA in contrast to the greater-than-additive secretion obtained with the combination of IFN-β and KDO predicts an additional mechanism that is not transcriptional in nature, such as an increase in the rate of translation, trafficking through secretory vesicles, or both. Whereas regulation of the secretory pathway for newly synthesized cytokines, such as MIP-1α, is not well understood, potential mechanisms are suggested by regulation of the secretion of TNF-α. Recent evidence suggests roles for vesicle-associated membrane protein 3 (VAMP3) and Rab11, a small guanosine triphosphatase (GTPase) implicated in membrane trafficking, in the secretion of TNF-α. LPS induces the production of both VAMP3 and Rab11. Macrophages that overexpress VAMP3 or are transfected with a constitutively active form of Rab11 show increased delivery of TNF-α to the cell surface27. In contrast, the delivery of TNF-α to the cell surface, but not the synthesis of TNF-α, is blocked in macrophages that express dominant-negative forms of VAMP3 or Rab11. Furthermore, production of both VAMP3 and Rab11 is induced more quickly by priming cells with IFN-γ. It is possible that IFN-β, when combined with KDO, induces a similar synergistic increase in VAMP3 and Rab11 or some equivalent pathway.
The potentiation of secretion of IL-6 that resulted from the simultaneous stimulation of cells with IFN-β, KDO, and ISO or 8Br indicates that these three pathways converge at least at one site (Fig. 7). A similar potentiation in the secretion of IL-10 was not observed under the same conditions (Fig. 3). This seems surprising because IL-6 induces the secretion of IL-10 in these cells5 (Fig. 1). In the same experiments, potentiation of IL-10 was observed with the combination of KDO and IL-6 and correlated with dual combinations of ligands that potentiate secretion of IL-6, such as KDO paired with either IFN-β or 8Br (Fig. 3 and Fig. 9). The lack of synergy at the first time point might be expected because the concentration of secreted IL-6 (about 100 pM) was insufficient to induce production of IL-10. In this case, the additive nature of the triple response would suggest that the synergy in IL-10 production produced by the combinations of KDO-IFN-β and the KDO-cAMP pathway stems from separate parallel pathways. However, the amount of IL-6 secreted by the three combined ligands at longer times of incubation exceeded the threshold for stimulating the secretion of IL-10. A potential explanation for a lack of synergy could be that the IL-6 pathway is simply saturated by the other ligands. Alternatively, the absence of potentiated amounts of IL-10 by the increased quantities of extracellular IL-6 reflects the existence of an underlying inhibitory mechanism in which IL-6 can also mediate attenuation of IL-10 secretion, which is synergized by other ligand pairs (Fig. 9). Such established mechanisms of cytokine regulation may include suppressors of cytokine signaling (SOCS) proteins to inhibit signal transduction from the IL-6 receptor, the downregulation of functional IL-6 receptor itself, or the induction of a decoy receptor antagonist in analogy to mechanisms that regulate IL-1 and IL-1028.
TGF-β inhibited the potentiation of IL-6 by KDO in combination with IFN-β, ISO or 8Br. When combined with KDO and IFN-β, more prominent inhibition at later time points suggests that TGF-β may act through a secondary mechanism, which potentially involves secretion of an additional factor to mediate its suppressive effects. In general, the data presented here demonstrate that TGF-β in this experimental paradigm inhibits cytokine secretion and has the potential to reduce powerful stimulatory paradigms. The ability of TGF-β to inhibit is consistent with its known immunosuppressive properties29, 30. However, IL-6 is also a pleiotropic cytokine whose activities have been characterized as anti- and pro-inflammatory31 and the ability of TGF-β to markedly modify its secretion in different venues may produce the divergent effects.
The profiles of cytokine secretion reported here can be applied to well-studied paradigms as well as novel areas where the roles of the cytokines are less defined. For example, the interactions of IFN-β with signaling by TLR4 potentially play a role in defense against viruses. The TLR family of receptors is one of two classes of receptors that can detect viruses within a host organism11, 13, 14, 32, and type I IFNs such as IFN-β are necessary for viral clearance; thus, mice devoid of the type I IFN receptors are highly susceptible to various viral pathogens33–35. Pathways dependent on cAMP have been proposed to mediate aspects of inflammation and fever through their mediation of TLR-dependent cytokines by prostaglandin E2 (PGE2) receptors36, 37 or adenosine38 receptors, which couple to Gs, the stimulatory heterotrimeric guanine nucleotide–binding protein (G protein) that activates adenylyl cyclase and effects increases in the concentration of cAMP.
We have provided a systematic experimental paradigm to investigate the cytokine secretion response by cells in response to stimulation by multiple ligands, a scenario likely to occur in vivo. The higher-order responses uncovered in this study demonstrate both a rich diversity in the interactions that are used to coordinate responses to multiplex stimuli as well as a high degree of constraint in the number of pathway interactions that are realized. Thus, the context in which various stimuli are detected by the cell leads to limited, but unique behaviors. The limitation of unique responses suggests the biological importance of maintained crosstalk and the interactions identified with this limited analysis of TLR4-dependent cytokine secretion in macrophages. This systematic approach should be applicable to other systems and help identify key interactions for regulating other cellular paradigms.
MATERIALS AND METHODS
Cell lines and tissue culture
Detailed information regarding the culture of RAW 264.7 cells can be obtained at www.signaling-gateway.org, including the following Alliance for Cell Signaling (AfCS) Protocols: Culture, PP00000159.1; preparation of frozen stocks, PP00000180; thaw protocol, PP00000160. In brief, RAW 264.7 cells, a macrophage-like, Abelson leukemia virus–transformed cell line derived from BALB/c mice, were obtained from the American Type Culture Collection (ATCC; cat. no. TIB-71; lot no. 2263775). The thawed cells were expanded by maintenance in RAW 264.7 growth medium 1 (RAWGM1) and passaged every two days until sufficient cells were obtained for the preparation of frozen stocks. Numerous aliquots of frozen cells from similar passages provided a uniform source of RAW 264.7 cells for experimentation. Frozen aliquots of cells from the AfCS contain approximately 5 × 106 cells in 1 ml of RAW 264.7 freezing medium [90% RAWGM1, 10% dimethyl sulfoxide (DMSO), PS00000568]. Stock vials of frozen AfCS cells were thawed and then maintained in RAWGM1 at 37°C in a humidified atmosphere with 5% CO2. For routine maintenance in culture (passage), cells were seeded at a confluence of approximately 10% (1 × 106 and 3 × 106 cells in 100-mm and 150-mm plates, respectively) and grown to approximately 80% confluency. This procedure required the cells to be split every two days. For passage over the weekend, 3.3 × 105 and 7.5 × 105 cells are seeded in 100-mm and 150-mm dishes, respectively, to ensure no more than 80% confluence when harvested after three days. Cultures were not maintained beyond three months.
Reagents
The following reagents were used to treat cells singly or in combination: 3-deoxy-D-manno-octulosonic acid (KDO)2-Lipid A (a kind gift from Dr. H. A. Brown, Vanderbilt Univ.), recombinant mouse IFN-β (Biosource International, PMC4024A), recombinant human IL-6 (Biosource International, PHC0064), isoproterenol hydrochloride (Sigma-Aldrich, I5627), recombinant TGF-β1 (R & D Systems, 240-B), 8-Bromoadenosine 3′,5′-cyclic- AMP (Sigma-Aldrich, B7880). Concentrations used to treat cells were as follows: KDO (10 ng/ml), IFN-β (1 nM), hIL-6 (25 nM), ISO (50 nM), TGF-β1 (50 pM), and 8-Bromo cAMP (100 µM). The dose chosen for each stimulation was approximately the EC50 for effects observed with single ligands or in combination with LPS. This single dose provides a practical approach to maintain experimentally manageable parameters and the likelihood of observing both positive and negative effects potentially produced by other inputs. Human IL-6 was used to stimulate cells to avoid interference with measuring secretion of mouse IL-6.
Cytokine measurements
Cells (2 ×105) were seeded into each well of a 24-well tissue culture plate and grown overnight in 0.6 ml of RAWGM1 at 37°C and 5% CO2. After approximately 24 hours, the growth medium was removed and replaced with 0.54ml of fresh, warm RAWGM1. Cells were returned to the incubator for 1 hour while the ligands were prepared. Ligands were diluted in RAWGM1 and added to the cells as 10X stocks (0.06 ml) for treatments. All treatments occurred in the presence of serum, circumventing the need for LPS binding protein (LBP). Cells were then incubated with ligands for the indicated times at 37°C in an atmosphere containing 5% CO2. Incubations were terminated by harvesting the culture media and clarifying by centrifugation for 5 minutes at 300g. Aliquots were prepared and frozen until ready for measurement of cytokines. G-CSF, IL-6, IL-10, MIP-1α, RANES, and TNF-α were quantified with the Bio-Plex™ cytokine array system (Bio-Rad Laboratories, Hercules, CA) with a custom-prepared mouse cytokine assay kit for G-CSF, IL-6, IL-10, RANTES and individual kits for TNF-α (Bio-Rad, 171G12221) and MIP1-α (Bio-Rad, X600004QE0).
Numerical analysis
The methods for the comparison of cytokine responses have been described previously5. In brief, data were collected for each cytokine in mock-stimulated cells to provide a mean value and variance with no applied stimulus. Each data variable collected upon ligand stimulation was then expressed as the number of standard deviations removed from the error model so obtained. Thus, for ligand i generating a value of ai for the experimental variable x, the transformed representation of the value is given by
where is the significance (in σ units) of observing ai given the error model. Multiple repeats of applying ligand i produce a distribution of Z-scores for variable x; thus, the raw data variable is transformed into a mean Z-score with error. This approach provides a dimensionless unit of significance for any particular response.
It is easy to estimate the interactions between any pair of ligands, and these methods have been described extensively in prior work5. For instance, consider the case when any two ligands (1,2) are applied. The observed response of any cytokine x to the combination of both ligands applied is compared to the expected effect if the two ligands act independently , and the difference is used to identify the extent of interaction between the two ligands. Because observations are made multiple times, the interaction observed is calculated as the difference between the two, weighted for the propagated errors of measurement as
Note that the significance of response is critically dependent upon the error measurement; therefore, care was exercised to remove systematic errors in measurement before these calculations were made. As the individual errors of measurement in multiple ligand stimuli can be collectively bundled into the expected sum of ligand responses, the mean area under the curve for the expected trace was first calculated. A scaling factor equal to the ratio of a given expected curve to the mean expected curve was calculated. Each single ligand and pairwise treatment was divided by the scaling factor for its pair-matched expected curve. The resultant dataset was used for generation of mean single ligand and observed combinatorial responses and their respective standard deviations.
When higher-order combinations of ligands were used, the observed response to the combination of n ligands could be similarly decomposed into the individual responses of each of the n ligands acting independently , all interactions of the two way combinations within the n ligands, all interactions of the three way combinations within the n ligands, etc. up to (n) combinations:
To illustrate, the secretion of cytokine x in response to three ligands is simply the effect of each of the three ligands acting individually, the interactions between any two pairs of ligands, and the interactions between all three ligands, and is instantiated as:
To identify the synergy induced by the three-way combination alone, we can simply reconfigure the above as:
In other words, quantification of a synergy or anergy caused by a specific combination of n ligands can be decomposed easily based on knowledge of experimentally observed responses to all (n-1) combinations of ligands. Note that we do not have to calculate the contributions to nonadditivity (and correspondingly the error) at each level of ligand combination—such a process can introduce bias in the error analysis. To further the visualization of these data, the data within each experiment were scaled and represented on a colorimetric scale such as shown in Fig. 2. Note that the color schemes within an experiment corresponded with absolute z-values, but colors were relative across experiments.
Quantitative reverse transcriptase polymerase chain reaction assays
The abundance of mRNAs for G-CSF and MIP-1α in lysates of RAW cells was assessed by real-time, quantitative reverse transcriptase PCR (qRT-PCR) with a Bio-Rad iCycler. Detailed procedures for the assessment of mRNA by qRT-PCR have been described previously39. The sequences of the sense and antisense amplification primers and the probes used were as follows. G-CSF: 5’-CGTTCCCCTGGTCACTGTCA-3’; 5’-GGGTGACACAGCTTGTAGGTG-3’; 5’-TexasRed-CGCTCTGCCACCATCCCTGCCTCT-BHQ2-3’; MIP-1α: 5’-CTCTGTCACCTGCTCAACATCA-3’; 5’-CTAGGAGACACCTGGTCATCTTG-3’; 5’-TexasRed-AGGTCTCCACCACTGCCCTTGCTG-BHQ2-3’; β-actin (reference): 5’-TCCATGAAATAAGTGGTTACAGGA-3’; 5’-CAGAAGCAATGCTGTCACCTT-3’; 5’-HEX-TCCCTCACCCTCCCAAAAGCCACC-BHQ1-3’.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
REFERENCES AND NOTES
- 1.Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin H-S, Valentine C, Hellman J. MyD88-Dependent and MyD88-Independent Pathways in Synergy, Priming, and Tolerance between TLR Agonists. J Immunol. 2007;178:1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- 2.Gouwy M, Struyf S, Proost P, Van Damme J. Synergy in cytokine and chemokine networks amplifies the inflammatory response. Cytokine Growth Factor Rev. 2005;16:561–580. doi: 10.1016/j.cytogfr.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Movat HZ, Burrowes CE, Cybulsky MI, Dinarello CA. Acute inflammation and a Shwartzman-like reaction induced by interleukin-1 and tumor necrosis factor. Synergistic action of the cytokines in the induction of inflammation and microvascular injury. Am J Pathol. 1987;129:463–476. [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson B, Pettipher ER. Arthritogenic actions of recombinant IL-1 and tumour necrosis factor alpha in the rabbit: evidence for synergistic interactions between cytokines in vivo. Clin Exp Immunol. 1989;75:306–310. [PMC free article] [PubMed] [Google Scholar]
- 5.Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 6.Jiang LI, Collins J, Davis R, Lin K-M, DeCamp D, Roach T, Hsueh R, Rebres RA, Ross EM, Taussig R, Fraser I, Sternweis PC. Use of a cAMP BRET Sensor to Characterize a Novel Regulation of cAMP by the Sphingosine 1-Phosphate/G13 Pathway. J. Biol. Chem. 2007;282:10576–10584. doi: 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang LI, Collins J, Davis R, Fraser ID, Sternweis PC. Regulation of cAMP Responses by the G12/13 Pathway Converges on Adenylyl Cyclase VII. J. Biol. Chem. 2008;283:23429–23439. doi: 10.1074/jbc.M803281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 9.Poltorak A, He X, Smirnova I, Liu M-Y, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 10.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 12.Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14:103–110. doi: 10.1016/s0952-7915(01)00304-1. [DOI] [PubMed] [Google Scholar]
- 13.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 15.Moreno E, Pitt MW, Jones LM, Schurig GG, Berman DT. Purification and characterization of smooth and rough lipopolysaccharides from Brucella abortus. J Bacteriol. 1979;138:361–369. doi: 10.1128/jb.138.2.361-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Ribeiro AA, Jr, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, White KP, Jr, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noselli S, Perrimon N. Signal transduction. Are there close encounters between signaling pathways? Science. 2000;290:68–69. doi: 10.1126/science.290.5489.68. [DOI] [PubMed] [Google Scholar]
- 20.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 21.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud Ø, Gjertsen BT, Nolan GP. Single Cell Profiling of Potentiated Phospho-Protein Networks in Cancer Cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A Systems Model of Signaling Identifies a Molecular Basis Set for Cytokine-Induced Apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 23.Janes KA, Albeck JG, Peng LX, Sorger PK, Lauffenburger DA, Yaffe MB. A High-throughput Quantitative Multiplex Kinase Assay for Monitoring Information Flow in Signaling Networks: Application to Sepsis-Apoptosis. Mol Cell Proteomics. 2003;2:463–473. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Janes KA, Reinhardt HC, Yaffe MB. Cytokine-Induced Signaling Networks Prioritize Dynamic Range over Signal Strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 26.Bug G, Aman MJ, Tretter T, Huber C, Peschel C. Induction of macrophage-inflammatory protein 1alpha (MIP-1alpha) by interferon-alpha. Exp Hematol. 1998;26:117–123. [PubMed] [Google Scholar]
- 27.Murray RZ, Kay JG, Sangermani DG, Stow JL. A Role for the Phagosome in Cytokine Secretion. Science. 2005;310:1492–1495. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- 28.D'Amico G, Frascaroli G, Bianchi G, Transidico P, Doni A, Vecchi A, Sozzani S, Allavena P, Mantovani A. Uncoupling of inflammatory chemokine receptors by IL-10: generation of functional decoys. Nat Immunol. 2000;1:387–391. doi: 10.1038/80819. [DOI] [PubMed] [Google Scholar]
- 29.Sporn MB. The early history of TGF-beta, and a brief glimpse of its future. Cytokine Growth Factor Rev. 2006;17:3–7. doi: 10.1016/j.cytogfr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Wrzesinski SH, Wan YY, Flavell RA. Transforming Growth Factor-{beta} and the Immune Response: Implications for Anticancer Therapy. Clin Cancer Res. 2007;13:5262–5270. doi: 10.1158/1078-0432.CCR-07-1157. [DOI] [PubMed] [Google Scholar]
- 31.Moller B, Villiger PM. Inhibition of IL-1, IL-6, and TNF-alpha in immune-mediated inflammatory diseases. Springer Semin Immunopathol. 2006;27:391–408. doi: 10.1007/s00281-006-0012-9. [DOI] [PubMed] [Google Scholar]
- 32.Georgel P, Jiang Z, Kunz S, Janssen E, Mols J, Hoebe K, Bahram S, Oldstone MB, Beutler B. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology. 2007 doi: 10.1016/j.virol.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 33.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 34.Billiau A. Interferon: the pathways of discovery I. Molecular and cellular aspects. Cytokine Growth Factor Rev. 2006;17:381–409. doi: 10.1016/j.cytogfr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Treffkorn L, Scheibe R, Maruyama T, Dieter P. PGE2 exerts its effect on the LPS-induced release of TNF-alpha, ET-1, IL-1alpha, IL-6 and IL-10 via the EP2 and EP4 receptor in rat liver macrophages. Prostaglandins Other Lipid Mediat. 2004;74:113–123. doi: 10.1016/j.prostaglandins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Blatteis CM, Li S, Li Z, Feleder C, Perlik V. Cytokines PGE2 and endotoxic fever: a re-assessment. Prostaglandins Other Lipid Mediat. 2005;76:1–18. doi: 10.1016/j.prostaglandins.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Hasko G, Cronstein BN. Adenosine: an endogenous regulator of innate immunity. Trends Immunol. 2004;25:33–39. doi: 10.1016/j.it.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Fraser I, Liu W, Rebres R, Roach T, Zavzavadjian J, Santat L, Liu J, Wall E, Mumby M. The use of RNA interference to analyze protein phosphatase function in mammalian cells. Methods Mol Biol. 2007;365:261–286. doi: 10.1385/1-59745-267-X:261. [DOI] [PubMed] [Google Scholar]
- 40.Acknowledgements. This work was supported by National Institutes of Health Grant GM 62114 and the Alfred and Mabel Gilman chair in molecular pharmacology (to P.C.S.).