Abstract
1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes, containing p-t-butyl (DIM-C-pPhtBu) and phenyl (DIM-C-pPhC6H5) substituents, are peroxisome proliferator-activated receptor γ (PPARγ) agonists; however, DIM-C-pPhtBu-induced growth inhibition and cell death in human HEC1A endometrial cancer cells is PPARγ-independent. DIM-C-pPhtBu decreased mitochondrial membrane potential (MMP) and promoted the release of cytochrome c and caspase activation and nuclear uptake of endonuclease G leading to apoptosis of HEC1A cells. DIM-C-pPhtBu specifically targeted the mitochondrial permeability transition pore complex (PTPC) because the DIM-C-pPhtBu-induced proapoptotic responses were inhibited by atractyloside (Atra), a compound that specifically interacts with the inner mitochondrial membrane adenine nucleotide transport (ANT) proteins. At the dose of Atra used in this study (300 µM), this compound alone did not alter the PTPC but inhibited the mitochondriotoxic effects of DIM-C-pPhtBu. DIM-C-pPhtBu/DIM-C-pPhC6H5 and Atra also differentially affected the ability of eosin-5-maleimide (EMA) to alkylate Cys160 in the ANT protein and Atra, but not DIM-C-pPhtBu, inhibited the exchange of ATP/ADP in isolated mitochondria suggesting that these pharmacophores act on different sites on the ANT protein. Results of this study show that the receptor-independent proapoptotic activity of DIM-C-pPhtBu and DIM-C-pPhC6H5 were related to novel mitochondriotoxic activities involving inner mitochondrial ANT proteins.
Keywords: ANT, C-DIM, apoptosis, mitochondria, HEC1A cells
INTRODUCTION
Peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-activated transcription factor and a member of the nuclear receptor (NR) superfamily that includes steroid hormone, thyroid hormone, vitamin D and retinoic acid receptors, and orphan receptors for which endogenous ligands have not yet been identified [1–6]. PPARγ agonists have been developed for treatment of metabolic diseases, and compounds such as thiazolidinediones (TZDs) are used extensively for clinical treatment of insulinresistant Type II diabetes [7,8]. Wild-type PPARγ is highly expressed in tumors from multiple sites and in cancer cell lines, and the chromosomal location of PPARγ indicates that this receptor “could be a tumor suppressor gene” [9]. PPARγ agonists such as 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2), TZDs, and other synthetic PPARγ agonists inhibit growth of tumors (in vivo) and cancer cell lines through several pathways including inhibition of G0/G1 to S phase progression, modulation of kinase pathways, induction of differentiation, and apoptosis [10–23].
Different structural classes of PPARγ agonists induce one or more genes/pathways that activate growth inhibition or cell death, and there is evidence that these responses may be receptor-dependent or -independent. For example, PGJ2 and the novel triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) induce apoptosis in leukemia cells, and these responses are inhibited by dominant negative PPARγ expression or a PPARγ antagonist [22]. In contrast, inhibition of ovarian cancer cell growth by CDDO was unaffected by cotreatment with a PPARγ antagonist [23]. Studies in this laboratory have identified a series of 1,1-bis(3′-indolyl)-1-(p-substituted phenyl)methanes [methylene-substituted diindolylmethanes (C-DIMs)] containing p-trifluoromethyl (DIM-C-pPhCF3), p-t-butyl (DIM-C-pPhtBu), or phenyl (DIM-C-pPhC6H5) substituents that activate PPARγ-dependent transactivation in breast, colon, and pancreatic cancer cell lines [24–26]. C-DIMs induce PPARγ-dependent expression of caveolin-1 in colon cancer cell lines [25] and the cyclin-dependent kinase inhibitor p21 in Panc-28 pancreatic cancer cells [26]. In contrast, these compounds downregulate cyclin D1 and induce apoptosis in breast and other cancer cell lines through PPARγ-independent pathways [24,27–32].
PPARγ is expressed in HEC1A endometrial cancer cells and PPARγ-active C-DIMs also induce transactivation in cells transfected with a construct (PPRE-luc) containing three tandem PPARγ response elements. These compounds also inhibit growth and induce apoptosis in HEC1A cells; however, these responses were receptor-independent, and the major objective of this study was to further investigate receptor-independent apoptosis pathways induced by these compounds. Using DIM-C-pPhtBu as a model, the results show that this compound activates novel proapoptotic pathways. DIM-C-pPhtBu is mitochondriotoxic and decreases mitochondrial membrane potential (MMP), induces apoptosis, release of mitochondrial cytochrome c, and caspasedependent activation of endonuclease G through specific targeting of inner mitochondrial membrane adenine nucleotide transport (ANT) proteins. In addition to their essential role in transporting newly synthesized ATP from the mitochondrial matrix to the cytosol, ANT proteins form part of the permeability transition pore complex (PTPC), a supramolecular protein structure that mediates the release of death promoting proteins from the intermembrane space of the mitochondria. Results of this study show that C-DIMs are a new class of uncharged small molecules that induce apoptosis in cancer cells through modulation of ANT and PTPC function.
MATERIALS AND METHODS
Cell Lines and Reagents
The HEC1A and HEC1B endometrial, Panc1 and Panc28 pancreatic, and HeLa cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA). DME/F12 with phenol red, 100× antibiotic/antimycotic solution, propidium iodide, atractyloside (Atra), cyclosporin A (CsA), 2-deoxyglucose (2-DG), and aurintricarboxylic acid (ATA) were purchased from Sigma-Aldrich (St. Louis, MO), and fetal bovine serum (FBS) was obtained from Intergen (Purchase, NY). Antibodies for AIF, PPARγ, PARP, bcl-2, VDAC, histone H3, ANT, and β-tubulin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); endonuclease G antibodies were purchased from Prosci (Poway, CA), and cytochrome c and bax antibodies were obtained from BD Pharmingen (San Diego, CA). Lysis buffer, luciferase reagent, and RNase were obtained from Promega Corp. (Madison, WI). Z-VAD(OMe)-FMK was obtained from Alexis (Lausen, Switzerland). TMRM and Hoechst 33258 were obtained from Molecular Probes (Eugene, OR). All other chemicals and biochemicals were of the highest quality available from commercial sources. DIM-C-pPhCF3, DIM-C-pPhtBu, and DIM-C-pPhC6H5 were synthesized in our laboratory [24], and a solution of 1.0 × 10−2 M was prepared in DMSO. The PPARγ agonist GW9662 was synthesized by condensation of the corresponding acid chloride and aromatic amine, purified by thin layer chromatography, and structures were confirmed by gas chromatography-mass spectrometry. Small inhibitory RNAs and nonspecific siRNA (iScr) were prepared by IDT (Coralville, IA) and PPARγ iRNA was targeted to the coding region of PPARγ as previously described [24–26].
Cell Survival Studies
HEC1A cells were maintained in DME/F12 supplemented with 10% FBS at 37°C in a humidified atmosphere of 5% CO2 in air. For cell proliferation experiments, HEC1A cells were seeded at a density of 1–2 × 105 cells in 6-well plates. After 24 h, cells were treated with various concentrations of DIM-C-pPhCF3, DIM-C-pPhtBu, or DIM-C-pPhC6H5 for 96 h. Adherent cells were collected and cell numbers were determined with a Z1 Dual Coulter Counter (Beckman, Coulter, Palatine, IL). In the cell survival assays, the percent cell survival after treatment with C-DIMs is determined from the ratio of the number of adherent cells in the treatment versus the control (DMSO) group. In experiments using the PPARγ antagonist GW9662, HEC1A cells were pretreated with 5 mM GW9662 for 1 h and then treated with various concentrations of PPARγ-active C-DIMs. In experiments using Atra, CSA and BA, HEC1A cells were cotreated with various concentrations of compounds. Both floating and adherent cells were counted after the indicated times. All treatments were replicated at least three times and results were confirmed in at least two or more separate experiments. HEC1B, Panc1, Panc28, and HeLa cells were maintained in Eagle’s minimum essential medium supplemented with 10% FBS, and the experiments were carried out essentially as described above.
Assay of Intracellular ATP Levels
Intracellular ATP levels were determined using a luciferase ATP assay kit (Perkin Elmer, Wellesley, MA) according to the manufacturer’s instructions. The amount of ATP-driven light produced was measured with a BMG luminometer (BMG Labtechnologies, Germany). The amount of total cellular protein was determined with protein assay reagents kit (Pierce) with bovine serum albumin (BSA) as a standard.
In Vitro Assay of ANT Exchange Activity
The adenine nucleotide exchange activity of ANT was measured by a slight modification of the inhibitor-stop method. Briefly, mitochondria were suspended in ice-cold reaction buffer (110 mM KCl, 20 mM Tris-HCl, 1 mM EDTA, pH 7.4) and 50 µM of ADP was added to a reaction medium consisting of the buffer and 0.5 mg/mL of mitochondrial proteins. The reaction was carried out on ice and stopped after 10 s by addition of 100 mmoles Atra. The reaction mixture was then centrifuged at 10 000g for 5 min and the supernatant was harvested. The ADP-induced release of ATP was then quantified by luciferase ATP assay kit.
Measurement of Mitochondrial Membrane Potential
MMP was measured with 3,3′-tetraethylbenzimidazolylcarbo-cyanine iodide (JC-1) (Molecular Probe, Eugene, OR). HEC1A cells (5 × 104) were exposed to different concentrations of DIM-C-pPhtBu as indicated for 24 h. After treatment, cells were incubated in 50 nM JC-1 at 37°C for 30 min and washed (3X) with phosphate-buffered saline (PBS). The red JC-1 fluorescence was measured at 530 nm excitation (Ex)/590 nm emission (Em) and the green cytoplasmic JC-1 fluorescence was measured at 485 nM Ex/530 nm Em with a fluorescence-activated fluorescence reader (BMG Labtechnologies, Offenburg, Germany). After subtraction of background values obtained from wells containing JC-1 but devoid of cells, red/green fluorescence ratios were calculated.
TMRM Staining
HEC1A cells were grown on coverslips in 12-well plates and were treated as indicated. After treatment for 24 h, TMRM and Hoechst 33258 were added to the medium at the concentration of 50 nM and 1 µg/mL, respectively, and incubated for 10 min at 37°C. The fluorescence of TMRM was monitored with Zeiss 410 confocal microscope (Carl Zeiss, Jena, Germany).
Isolation of Mitochondrial and Nuclear Fractions
Mitochondria were extracted using a Mitochondria Isolation Kit (Pierce, Rockford, IL) according to the manufacturer’s instructions. Briefly, 2 × 107 cells were harvested and resuspended in Mitochondria Isolation Reagent A containing a protease inhibitor mixture (Roche, Basel, Switzerland). After 60 strokes in a Dounce homogenizer, mitochondria isolation reagent C was added. The unbroken cells and nuclei were centrifuged at 700g for 10 min, and the mitochondrial fraction was further centrifuged at 12 000g for 10 min. Nuclear proteins were extracted using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL). HEC1A cells (2 × 106) were harvested and resuspended in 200 µL of ice-cold CER I containing a protease inhibitor mixture. After incubation on ice for 10 min, 11 µL ofice-cold CER II was added. Cells were incubated on ice for 5 min and vortexed vigorously for 5 s every minute. After centrifugation at 14 000g for 5 min, the precipitated nuclei were recovered and the supernatant (cytoplasmic extract) was transferred to another prechilled tube and both fractions were either used immediately or stored at −20°C prior to use.
Cell Death ELISA
Apoptoic and necrotic cell death were quantitated using an ELISA method according to instructions provided by Roche Applied Sciences. Briefly, 104 cells were seeded into a streptoavidin-coated microtiter plate. After treatment for 24 h, the cell culture supernatants were removed from the cell before lysis, and cells were then resuspended in incubation buffer. A mixture of anti-histone-biotin and anti-DNA-peroxidase was added and incubated for 90 min at 25°C. During incubation, the antihistone antibody binds to the histone components of the nucleosomes and simultaneously captures the immunocomplex to the streptavidin via its biotinyl groups. In addition, the anti-DNA-peroxidase reacts with the DNA components of the nucleosomes. Color development was carried out by adding ABTS substrate solution to each sample. The absorbances at 405 nm and 495 nm were measured to give the relative magnitude of apoptotic and necrotic cell death.
Protein Isolation and Western Blot Analysis
HEC1A cells (1.5 × 105) were seeded in 6-well plates and treated with compounds for the indicated time periods. Cells were washed (2X) with cold PBS and collected in 200 µL of SDS sample buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 1% SDS, 5% 2-mercaptoethanol, and 0.025% bromophenol blue). The protein extract (i.e., whole cell lysate) was boiled for 3 min and loaded onto 10% polyacrylamide gel, electrophoresed, and transferred to PVDF membrane (Immobilon™-P, Millipore Corporation, Bedford, MA). Whole cell lysates and nuclear and mitochondrial proteins were separated by SDS–gel electrophoresis as described in Reference [26]. Protein bands detected by incubation with polyclonal primary antibodies to AIF (SC-9416), endonucleaseG (3035), bcl-2 (SC-492), VDAC (SC-8828), PPARγ (SC-7196), Bax (SC-493) (54104), ANT (SC-9299), histone H3 (SC 10809), and β-tubulin (SC-9104) (all 1:1000 dilution) followed by blotting with horseradish peroxidase-conjugated secondary antibody (1:5000 dilution). Immune complexes were visualized with chemiluminescent substrate (Santa Cruz Biotechnology, Santa Cruz, CA) and luminescent signal recorded in Kodak X-Omat AR autoradiography film (Eastman Kodak, Rochester, NY).
Interaction of DIM-C-pPhC6H5 With HEC1A Cell Mitochondria
HEC1A cells (2 × 106) were treated with DMSO, 10 µM or 20 mM DIM-C-pPhC6H5 for 3 h, and mitochondria were extracted as described above. After washing with ice-cold reaction buffer (200 nM sucrose, 110 mM KCl, 20 mM Tris-HCl, 1 mM EDTA, pH 7.4) (3X), mitochondria were resuspended in 100 mL DMSO, vortexed for 5 min, and the supernatant was recovered after centrifugation for 10 min at 12 000g. For in vitro studies, isolated mitochondria were resuspended in ice-cold reaction buffer to a concentration of 0.5 mg/mL mitochondrial protein. Mitochondria were incubated with different amounts of DIM-C-pPhC6H5 at 0°C for 10 min and were centrifuged at 12 000g at 4°C. The supernatant was removed and mitochondria were resuspended in 100 µL DMSO, and mitochondrial protein was isolated as described above. The fluorescence was measured at 355 nm excitation/460 nm emission using a fluorescence-activated fluorescence reader (BMG Labtechnologies, Offenburg, Germany).
Immunofluorescent Staining
HEC1A cells were washed (3X) in PBS, fixed on coverslips with 2.5% paraformaldehyde (Sigma) for 10 min at room temperature, rinsed twice with PBS, and treated with 0.5% Triton X-100 (Roche Molecular Biochemicals, Indianapolis, IN) for 10 min at room temperature. Cells were then blocked with 100 µL of 3% BSA for 1 h followed by addition of 100 µL of endonuclease G or cytochrome c antibodies diluted 1:500 in 3% BSA overnight at 4°C, and then washed with PBS (3 × 5 min). The FITC-conjugated goat anti-rabbit antibody or goat antimouse (Molecular Probes, Eugene, OR) was diluted 1:1000 and 100 µL of the antibody solution was placed on each coverslip for 1 h at room temperature, and cells were then washed with PBS (3 × 5 min). Nuclei were stained with 0.1 µg/mL propidium iodide. The coverslips were mounted face down on microscope slides with mounting medium (Vector Laboratories, Inc. Burlingame, CA) to be viewed on a Zeiss 410 confocal microscope (Carl Zeiss, Germany). The slides were stored in a light proof black box.
RNA Isolation and Northern Blot Analysis by RT-PCR
Cells (1.5 × 105) were seeded in 6-well plates, and after 24 h, iRNA duplexes were transfected with TransFast Transfection Reagent (Promega Corp., Madison, WI). RNA was harvested with the SV Total RNA Isolation System (Promega Corp.), as per the manufacturer’s instructions, 24 h after transfection. Ten percent of the total RNA was used for cDNA synthesis using the RT system (Promega Corp.), and amplification of ANT and glyceraldehyde-3-phosphate dehydrogenase(GAPDH) cDNAs was carried out in multiplex reactions in the presence of 3% DMSO. The primer sequences used were: GAPDH: forward, 5′-GGT CTC CTC TGA CTT CAA CAG CG-3′; and reverse, 5′-GGT ACT TTA TTG ATG GTA CAT GAC-3′. The primer sequences of ANT are referred to Reference [33]. ANT1: forward, 5′-CTG AGA GCG TCG AGC TGT CA-3′; and reverse, 5′-CTC AAT GAA GCA TCT CTT C-3′. ANT2: forward, 5′-CCG CAG CGC CGT AGT CAA A-3′; and reverse, 5′-AGT CTG TCA AGA ATG CTC AA-3′; ANT3: forward, 5′-CCG TTC TCC GGC TGT CTT CC-3′; and reverse, 5′-GCC GGC CGC ACC GCC GGA GGC-3′. PCR products were run on 1.2% agarose gels prestained with ethidium bromide and visualized in a UV transluminator; digital images of PCR products were captured with the Kodak EDAS 290 Electrophoresis Documentation and Analysis System (Eastman Kodak Co., New Haven, CT).
Cell Proliferation Experiments With iANT
ANT1, ANT2, and ANT3 inhibitory RNAs were prepared by DHARMACON (Chicago, IL) and targeted to the coding region of ANT1, ANT2, and ANT3, respectively. The iRNA duplexes used in this study are summarized below.
ANT1 5′-UGACACUGUUCGUCGUAGATT-3′ 5′-UCUACGACGAACAGUGUCATT-3′
ANT2 5′-AGACUGCGUGGUCCGUAUUTT-3′ 5′-AAUACGGACCACGCAGUCUTT-3′
ANT3 5′-UGUACGACGAGCUCAAGAATT-3′ 5′-UUCUUGAGCUCGUCGUACATT-3′
iScr 5′-ACUCUAUCUGCACGCUGACTT-3′ 5′-GUCAGCGUGCAGAUAGAGUTT-3′
For the cell proliferation experiments, HEC1A cells were seeded at a density of 2–4 × 104 cells in 48-well plates. Twenty-four hours after seeding, iRNA duplexes were transfected with TransFast Transfection Reagent (Promega Corp., Madison, WI). Cells were treated with 10 mM DIM-C-pPhtBu 1 h after transfection. Floating cells and adherent cells were collected after 48 h, and cell numbers were determined with a Z1 Dual Coulter Counter (Beckman Coulter, Palatine, IL).
Detection of Cytochrome c
Isolated mitochondria were incubated with compounds at 25°C in PT buffer (10 mM Tris-MOPS, pH 7.4, 200 mM sucrose, 5 mM succinate, 10 µM EGTA) for 20 min and centrifuged at 13 000g for 10 min at 48C. For detecting cytochrome c released from mitochondria, mitochondrial pellet and supernatant fractions were subjected to SDS–PAGE (15%) and Western blot analysis.
Preparation of Submitochondrial Particles
Submitochondial particles were prepared from Panc1 cells as follows. Cell mitochondria stored at −80°C were thawed and suspended at 20 mg of protein/mL in medium consisting of 250 mM sucrose, 1 mM sodium succinate, 5 mM MgCl2, and 10 mM Tris-HCl buffer, pH 7.4. The suspension was sonicated at 100 watts in an Ultrasonic Processor (Sonics, Newtown, CT) five times for 30 s (5X) with 1 min intervals in an ice bath. Unbroken mitochondria were removed by centrifugation at 3000g for 8 min at 4°C, and then submitochondrial particles were collected by recentrifugation at 14 000g for 30 min at 4°C.
Labeling of ANT by EMA
Submitochondrial particles were suspended in medium consisting 250 mM sucrose, 0.5 mM EDTA, and 10 mM Tris-HCL buffer, pH 7.2 to a final concentration 5 mg/mL, and then preincubated with the compounds indicated for 10 min in a total volume of 100 µL at 0°C in the dark. Eosin-5-maleimide (EMA) was added to a final concentration of 100 µM and the reaction was terminated by addition of 1 M DTT to a final concentration of 50 mM. Mitochondrial proteins were then analyzed by SDS–PAGE in 15% polyacrylamide gel. The fluorescence intensity of the ANT labeled with EMA was measured by fluorography.
Statistical Analysis
Statistical significance was determined by analysis of variance and the levels of probability are noted. The results are expressed as mean ±SD for at least three separate (replicate) experiments for each treatment group.
RESULTS
PPARγ-Active C-DIMs Decrease Survival and Induce Apoptosis in HEC1A and Other Cancer Cell Lines
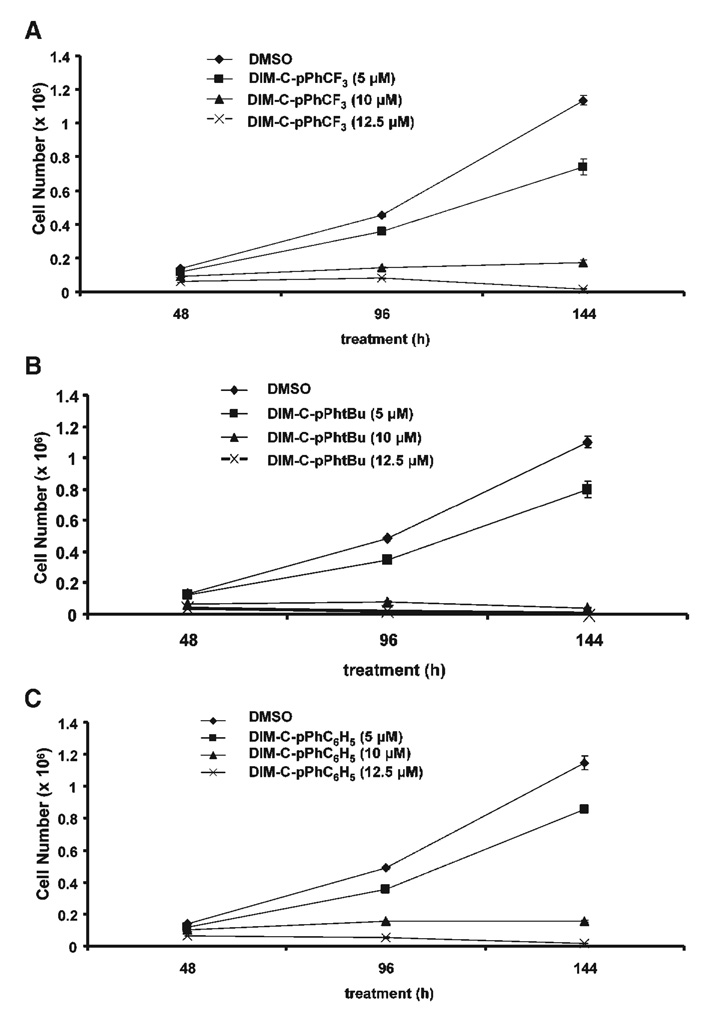
Recent studies in this laboratory have identified a series of C-DIMs that activate PPARγ, and the most active compounds include DIM-C-pPhCF3, DIM-C-pPhtBu, and DIM-C-pPhC6H5 [24–26]. Results illustrated in Figure 1A–C show that the PPARγ-active C-DIMs also decreased HEC1A endometrial cancer cell proliferation. IC50 values for HEC1A cell growth inhibition were in between 5 µM and 10 µM, and cell death was generally observed using 10–15 µM concentrations.
Figure 1.
Growth inhibition of HEC1A cells by PPARγ-active C-DIMs. HEC1A cells were treated with different concentrations of DIM-C-pPhCF3 (A), DIM-C-pPhtBu (B), and DIM-C-pPhC6H5 (C). The number of cells was determined after 48, 96, or 144 h as described in the Section ″Materials and Methods.″ Significant (P < 0.05) growth inhibition was observed at concentrations ≥ 5.0 µM.
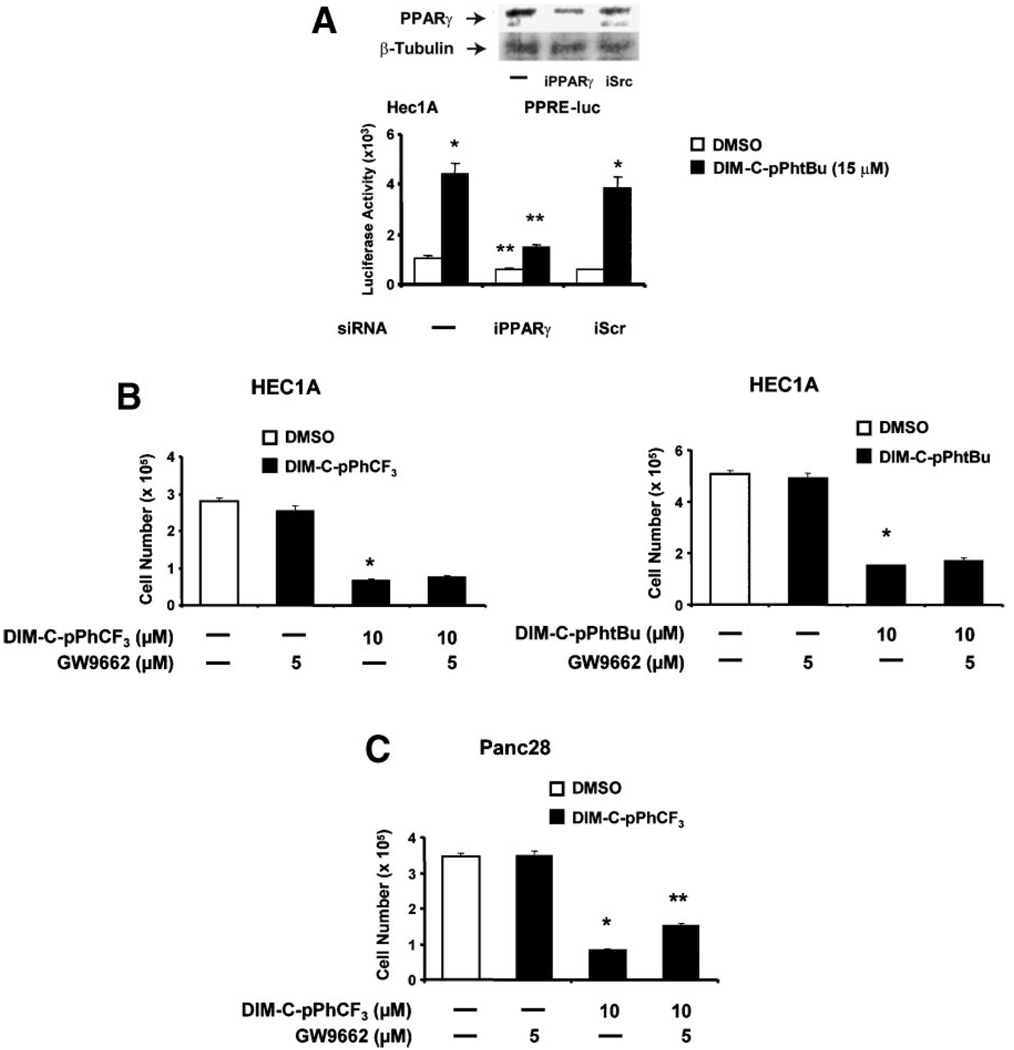
HEC1A cells express PPARγ (Figure 2A), and DIM-C-pPhtBu induced luciferase activity in cells transfected with a PPRE-luc construct as previously reported for this compound in Panc28 cells. In cells cotransfected with PPRE-luc and nonspecific iScrRNA or small inhibitory RNA for PPARγ (iPPARγ), there was a significant decrease in DIM-C-pPhtBu-induced luciferase activity in the latter treatment group. This corresponded to the decreased expression of PPARγ protein in these cells (Figure 2A). Results in Figure 2B show that 10µM DIM-C-pPhCF3 and DIM-C-pPhtBu alone decreased cell survival (96 h) and in HEC1A cells cotreated with the PPARγ antagonist GW9662, the cytotoxic effects of the C-DIM compounds were not affected, suggesting that these responses were PPARγ-independent. In parallel experiments, GW9662 blocks induction of luciferase activity by C-DIM compounds in cells transfected with a PPARγ-responsive (PPRE-luc) construct (data not shown) and similar results have previously been reported [25–31]. The results in Figure 2C show that GW9662 only partially blocked DIM-C-pPhCF3-induced inhibition of Panc28 cell growth; however, even in this cell line, a significant fraction of this response was PPARγ-independent. Similar results have been reported for other PPARγ agonists, indole-3-carbinol, and DIM which induce receptor-independent growth inhibition and apoptosis in cancer cells [24,26–32,34–41].
Figure 2.
Decreased cell survival by PPARγ-active C-DIM compounds and role of PPARγ. (A) Expression and activation of PPARγ in HEC1Acells. Cells were treated with DMSOor 15µM DIM-C-pPhtBu, transfected with PPRE-luc alone or cotransfected with iPPARγ or iScr (nonspecific) and luciferase activity determined as described in the Section ″Materials and Methods.″ Results are expressed as mean-± SE for three replicate determinations for each treatment group, and significant induction by DIM-C-pPhtBu (*) or inhibition by iPPARγ (**) is indicated. PPARγ protein expression relative to β-tubulin (loading control) is also indicated. Similar inhibition of DIM-C-pPhtBu-induced luciferase activity was also observed in cells cotreated with 5 µM GW9662 (PPARγ antagonist) (data not shown). Effect of GW9662 on inhibition of HEC1A (B) and Panc28 (C) cell growth by DIM-C-pPhtBu or DIM-C-pPhCF3. The inhibition of cell growth by 10 µM DIM-C-pPhtBu or DIM-C-pPhCF3 alone or in combination with 5 µM GW9662 after treatment for 48 h was determined as described in the Section ″Materials and Methods.″ Significantly (P < 0.05) decreased cell proliferation compared to solvent control (*) and inhibition of this response by GW9662 (**) are indicated.
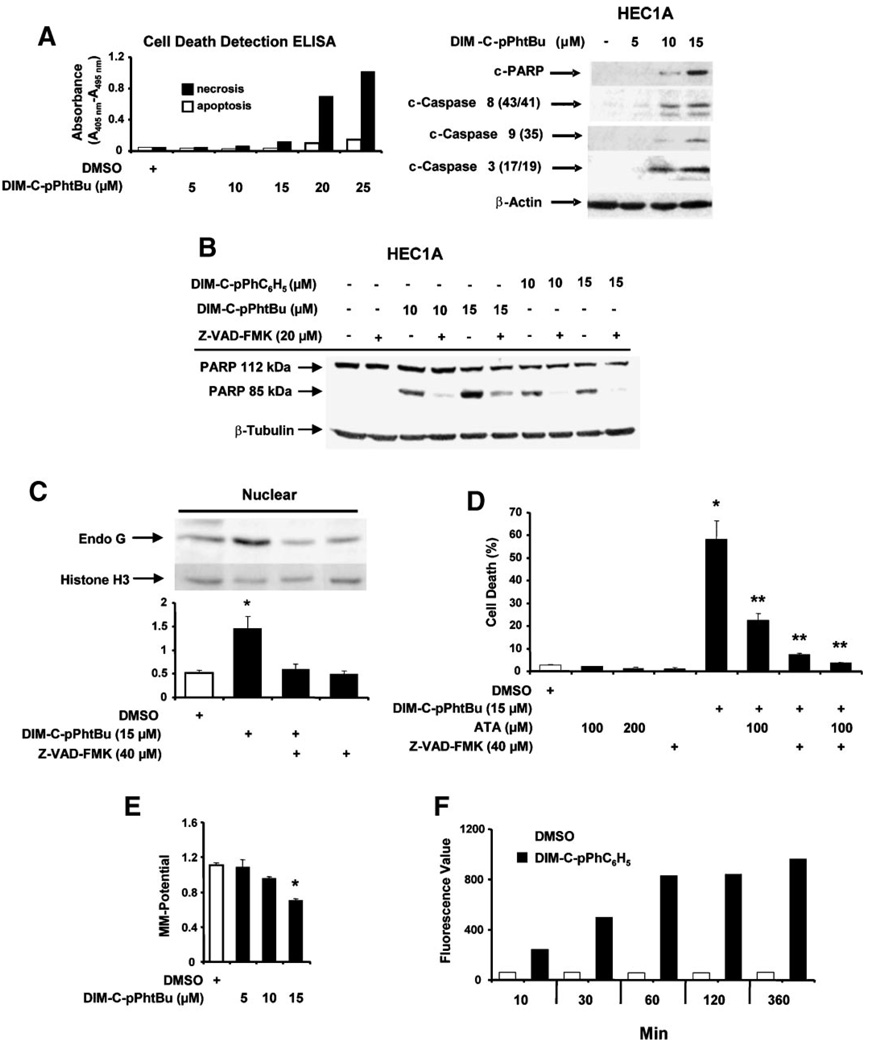
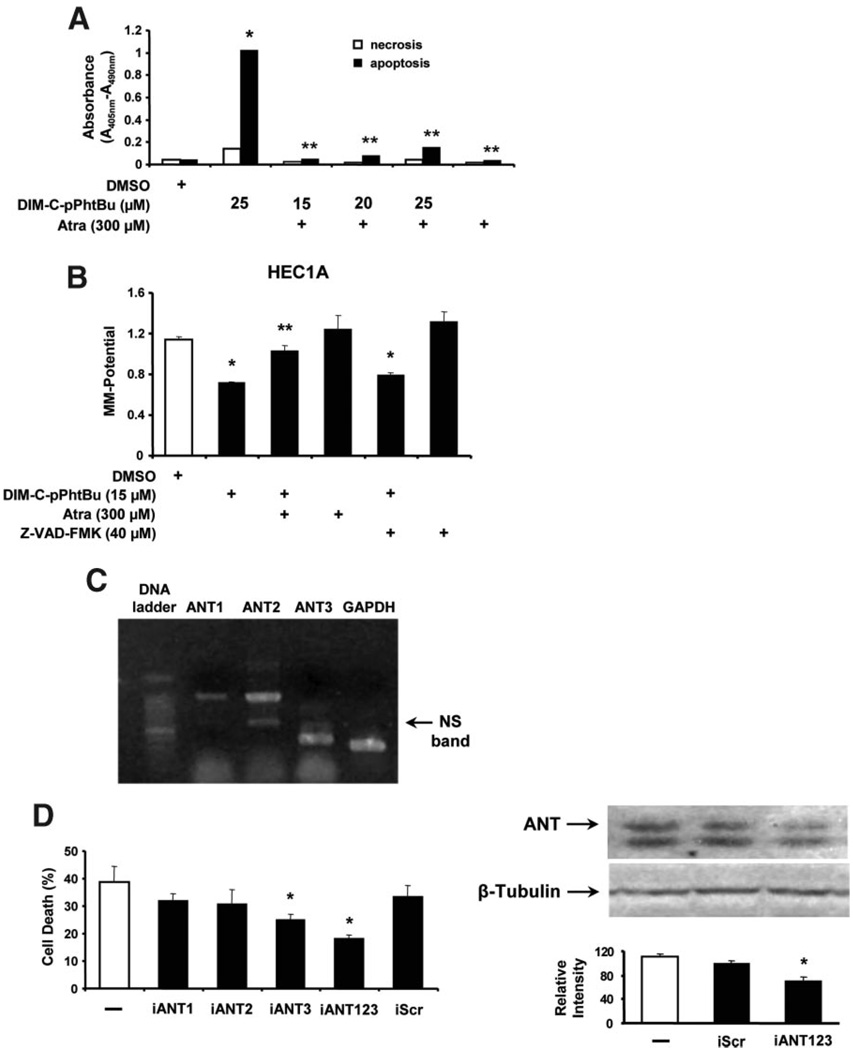
Induction of apoptosis by the C-DIM compounds was further investigated in HEC1A cells using a cell death ELISA assay which detects cytoplasmic histoneassociated DNA fragments. The results (Figure 3A) show that 10–20 µM DIM-C-pPhtBu primarily induced apoptotic cell death, whereas cell necrosis was only minimally changed by the treatments. In addition, treatment of HEC1A cells with DIM-C-pPhtBu for 24 h induced cleavage of PARP, caspases-3, −8, and −9. DIM-C-pPhtBu and DIM-C-pPhC6H5 induced PARP cleavage and DNA laddering (data not shown) in HEC1A cells, and PARP cleavage was blocked in cells cotreated with the pancaspase inhibitor Z-VAD-FMK (Figure 3B). Apoptosis in cancer cells can also be induced by activation of several pathways including release of apoptosisinducing factor (AIF) or endonuclease G which directly cleave DNA. The results in Figure 3C show that treatment of HEC1A cells with DIM-C-pPhtBu induces accumulation of endonuclease G in the nucleus, whereas this was not observed for AIF. Endonuclease G and pancaspase inhibitors ATA and Z-VAD-FMK, respectively, block DIM-C-pPhtBuinduced cell death and the pancaspase inhibitor Z-VAD-FMK alone or in combination with ATA was a more effective inhibitor than ATA alone (Figure 3D). These observations are in accord with a recent report showing that mitochondrial release of endonuclease G was also caspase-dependent [42].
Figure 3.
Effects of C-DIM compounds on cell death and MMP. (A) Induction of apoptosis. HEC1A cells were treated with DMSO or different concentrations of DIM-C-pPhtBu for 24 h and cell death/ necrosis were determined with a cell death detection ELISA as described in the Section ″Materials and Methods,″ and cleavage of PARP and caspases were determined by Western blot analysis of whole cell lysates. (B) Induction of PARP cleavage. HEC1A cells were treated with 10 or 15 µM DIM-C-pPhtBu and DIM-C-pPhC6H5 alone or in the presence of Z-VAD-FMK for 24 h, and whole cell lysates were analyzed for PARP cleavage by Western blot analysis as described in the Section ″Materials and Methods.″ (C) DIM-C-pPhtBu induces nuclear uptake of endonuclease G. Cells were treated with the various reagents, and nuclear levels of endonuclease G protein were determined as described in the Section ″Materials and Methods.″ Significant (P < 0.05) increases in nuclear levels of endonuclease G are indicated as (*). Results are expressed as mean ± SE for three replicate determinations for each treatment group. (D) Inhibition of DIM-C-pPhtBu-induced cell death by ATA or Z-VAD-FMK. HEC1A cells were treated with cytotoxic dose of DIM-C-pPhtBu alone in the presence or absence of various inhibitors, and cell numbers in each treatment group were determined after 24 h as described in the Section ″Materials and Methods.″ Significant (P < 0.05) induction of cell death by DIM-C-pPhtBu (*) and inhibition by ATA or Z-VAD-FMK (**) are indicated. (E) DIM-C-pPhtBu decreases MMP. HEC1A cells were treated with 5–15 µM DIM-C-pPhtBu and after 24 h, the loss of MMP was determined fluorimetrically using the JC-1 dye as described in the Section ″Materials and Methods.″ (F) Mitochondrial uptake of DIM-C-pPhC6H5. HEC1A cells were treated with DMSO or 10 µM DIM-C-pPhC6H5 for 10 min to 3 h, and mitochondria were then isolated, resuspended in DMSO, and fluorescence intensity was determined as described in the Section ″Materials and Methods.″ The results are an average of duplicate determinations. Results in (D) and (E) are expressed as mean ± SE for three separate experiments.
DIM and other apoptosis-inducing agents can directly act on mitochondria by decreasing MMP and results in Figure 3E show that DIM-C-pPhtBu significantly decreased MMP in HEC1A cells using 3,3′-tetraethylbenzimidazolylcarbo-cyanine iodide (JC-1), a red fluorescent dye that accumulates and oligomerizes in the mitochondrial matrix. Since we have observed that DIM-C-pPhC6H5 fluoresces, this compound interferes with measurement of changes in JC-1 fluorescence and changes in MMP; however, this compound can be used as a model to investigate the mitochondrial uptake of C-DIMs in HEC1A cells by measuring the fluorescence of DIM-C-pPhC6H5. The results showed accumulation of DIM-C-pPhC6H5 in the mitochondrial fraction within 10 min after treatment and maximal accumulation was observed with 60 min (Figure 3F). Similar results were observed with mitochondrial particles in which DIM-C-pPhC6H5 was detected in mitochondria within minutes after treatment (data not shown).
Inhibition of C-DIM-Induced Decreased MMP and Cell Death by Atractyloside (Atra)
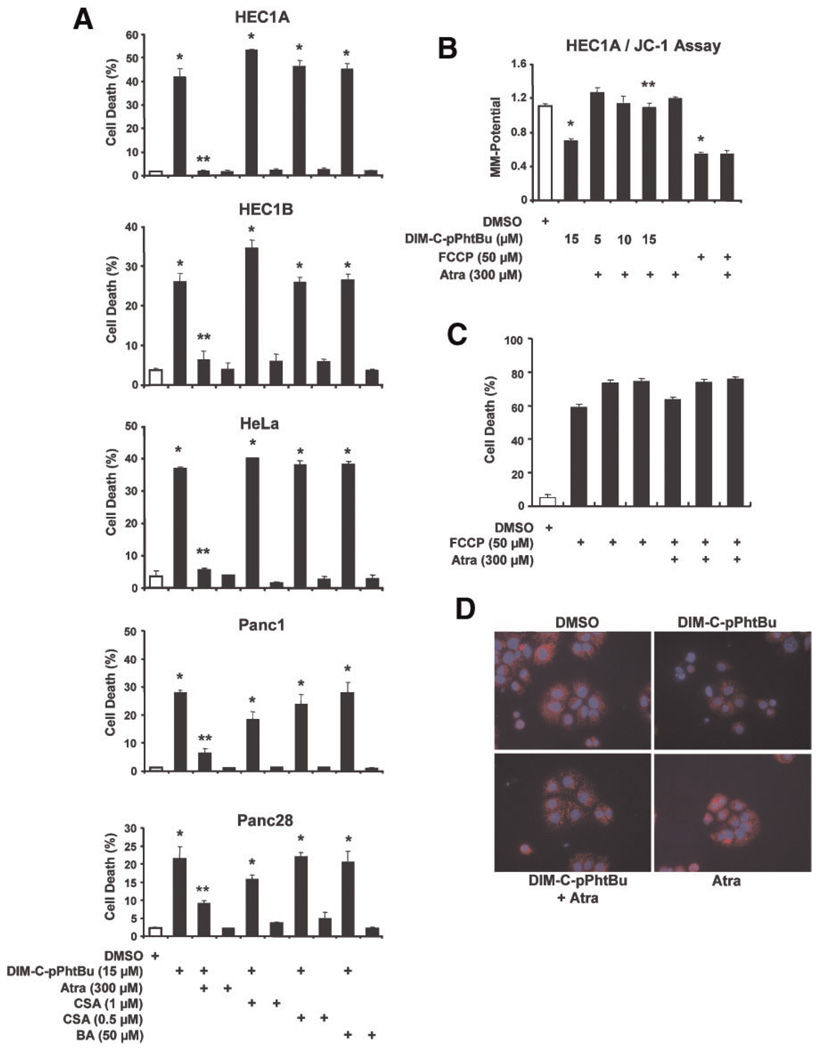
Since C-DIMs directly interacted with mitochondria and decreased MMP, we investigated the effects of three reagents, namely bongkrekic acid (BA), Atra, and CSA [43–48] on C-DIM-induced HEC1A1 cell death. BA induces the m conformation of the ANT protein and closes the PTPC; CSA inhibits apoptosis by interacting with cyclophilin D and inhibits cyclophilin D-ANT-dependent membrane permeabilization; and Atra fixes ANT in the c conformation which facilitates mitochondrial permeabilization in some cells using relatively high(mM) concentrations of Atra. The appropriate concentrations of CSA, Atra, and BA were determined in preliminary studies to ensure that the concentrations used were not cytotoxic. Results in Figure 4A show that treatment of HEC1A cells with 15 µM DIM-C-pPhtBu for 24 h resulted in>40% cell death and cell morphology was also consistent with apoptosis (data not shown). In HEC1A cells cotreated with DIM-C-pPhtBu plus Atra, CSA, or BA, only Atra significantly (>90%) inhibited DIM-C-pPhtBu-induced cell death. This cellular response to DIM-C-pPhtBu is not limited to HEC1A cells because cell death is also induced by DIM-C-pPhtBu in HEC1B endometrial (data not shown), HeLa cervical adenocarcinoma, Panc1 and Panc28 pancreatic cancer cells, and Atra, but not CSA or BA, significantly protected against the cell killing response (Figure 4A). The specificity of Atra–DIM-C-pPhtBu interactions was further investigated in HEC1A cells treated with FCCP, a known mitochondriotoxic compound that uncouples mitochondrial electron transport. Like DIM-C-pPhtBu, FCCP decreased MMP in HEC1A cells (Figure 4B), and FCCP also induced HEC1A cell death (Figure 4C). However, these responses were not affected after cotreatment with Atra, indicating that interactions between Atra and C-DIM compounds are highly specific. DIM-C-pPhtBu-induced loss of MMP was also determined in HEC1A cells treated with tetramethylrhodamine (TMRM) which accumulates in active mitochondria and exhibits a bright red color as indicated in solvent-treated cells (Figure 4D, upper left). Treatment with DIM-C-pPhtBu dramatically decreased the TMRM red staining (upper right); however, treatment with Atra plus DIM-C-pPhtBu (lower left) restored the intense red mitochondrial staining, and this was also observed in cells treated with Atra alone (lower right) which does not affect MMP.
Figure 4.
Atractyloside (Atra) inhibits DIM-C-pPhtBu-induced growth inhibition and effects on MMP. (A) Effects of inhibitors on DIM-C-pPhtBu-induced HEC1A, HeLa, Panc1 and Panc28 cell death. Cells were treated with DIM-C-pPhtBu alone, or the mitochondrial channel modulators cyclosporin A (CSA), Atra or bongkrekic acid (BA) alone or in combination with 15 µM DIM-C-pPhtBu for 24 h. The percentage of dead cells was determined for each treatment group as described in the Section ″Materials and Methods.″ Significant (P < 0.05) induction of cell death by DIM-C-pPhtBu alone or in combination with channel modulators (*) and inhibition by the cotreatments (**) are indicated. (B) Atra blocks effects of DIM-C-pPhtBu on MMP. HEC1A cells were treated with DIM-C-pPhtBu alone or in combination with FCCP and Atra, and loss of MMP was determined fluorimetrically using the JC-1 dye as described in the Section ″Materials and Methods.″ Significant (P < 0.05) inhibition of MMP (*) and reversal of this effect (*) are indicated. (C) Effects of Atra on FCCP-induced cell death. HEC1A cells were treated with various compounds/combinations as indicated for 24 h. Cell death was determined as described in the Section ″Materials and Methods,″ and data (in triplicate) were analyzed as described above. FCCP (10–50 µM) significantly decreased cell death which was not significantly reversed by Atra. (D) Decreased MMP and staining with Hoechst 33258 dye. HEC1A cells were treated with DMSO, DIM-C-pPhtBu alone, DIM-C-pPhtBu plus Atra, and Atra alone for 24 h. TMRM and Hoechst 33258 dye were added, and TMRMfluorescence was determined as described in the Section ″Materials and Methods.″ [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Role of the Adenine Nucleotide Translocator (ANT) Proteins in C-DIM-Induced Responses in Cancer Cells
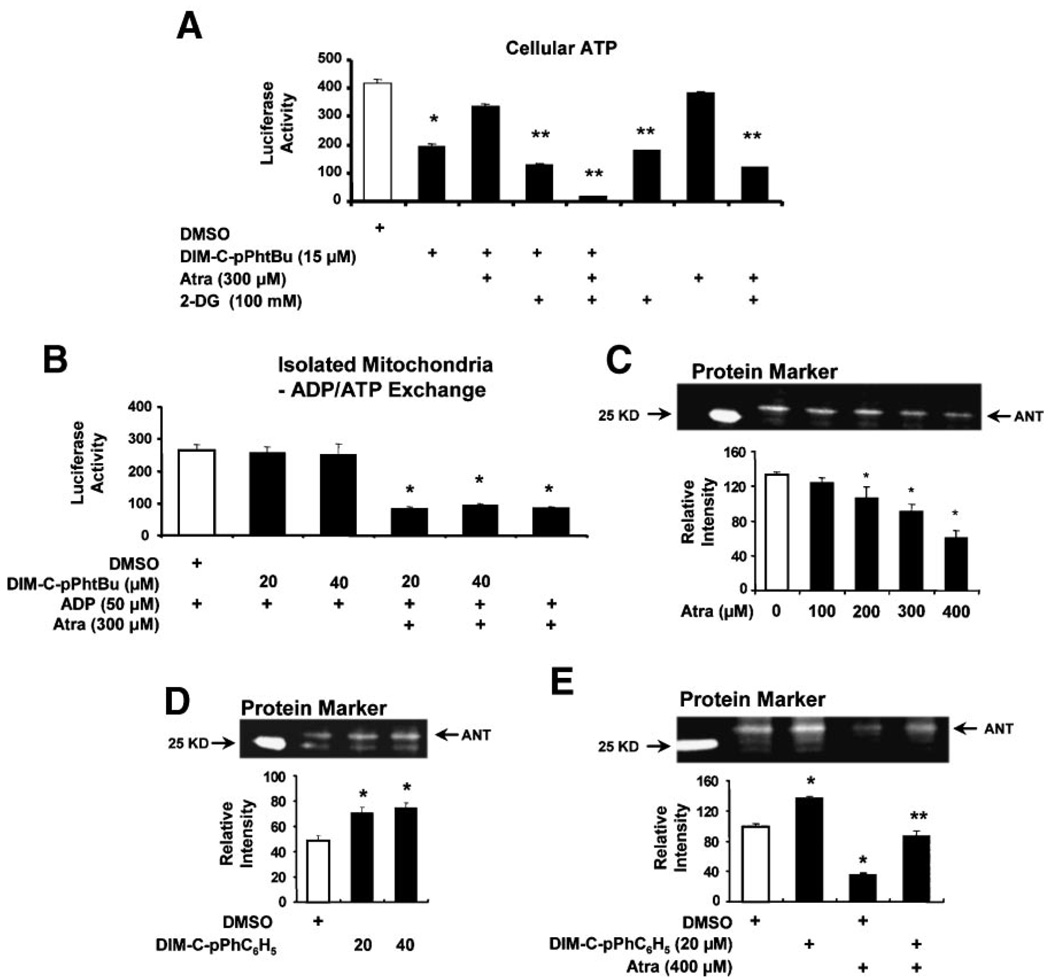
The cytotoxicity of Atra and effects of this compound on the PTPC are usually observed in cells treated with mM concentrations, and a report by Brown and coworkers showed that 300 µM Atra was not cytotoxic to HeLa and other cancer cell lines [49] and this was also observed in endometrial and pancreatic cancer cells (Figure 4). TheANTchannel is associated with the mitochondrial matrix and facilitates the import of cytosolic ADP and the export of ATP which is synthesized during aerobic metabolism, and it is possible that DIM-C-pPhtBu-induced responses may be related to ATP transport and levels. We therefore investigated the effects of DIM-C-pPhtBu alone and in combination with Atra on ATP levels in HEC1A cells cotreated with 2-DG, an inhibitor of glycolytic ATP production. Inhibition of glycolytic ATP synthesis facilitates examination of ATP levels associated with mitochondrial production and transport via the ANT channel. DIM-C-pPhtBu alone decreased total cellular ATP levels and this response was reversed by Atra, whereas Atra alone did not decrease ATP (Figure 5A). 2-DG alone inhibited glycolytic ATP production and represented levels of ATP produced by mitochondria. ATP levels were slightly decreased in cells cotreated with 2-DG plus DIM-C-pPhtBu or 2-DG plus Atra (compared to 2-DG alone), suggesting that both DIM-C-pPhtBu and Atra decreased mitochondrial ATP production. 2-DG plus both DIM-C-pPhtBu and Atra almost abolished cellular ATP levels, whereas this same combination had minimal effect on HEC1A cell death (data not shown). Thus, although both Atra and DIM-C-pPhtBu decreased mitochondrial production of ATP, this response was not correlated with cell death. Moreover, in isolated mitochondria from HEC1A cells, we observed that Atra but not DIM-C-pPhtBu inhibited ADP/ATP exchange in isolated mitochondria (Figure 5B). These results demonstrate that DIM-C-pPhtBu does not affect Atra-dependent inhibition of ATP/ADP exchange in mitochondria, whereas Atra inhibited DIM-C-pPhtBu-induced cell death and decreased MMP.
Figure 5.
Mitochondrial interactions of Atra and DIM-C-pPhtBu. (A) ATP levels. HEC1A cells were treated with the various compounds for 24 h, and cellular ATP levels were determined as described in the Section ″Materials and Methods.″ Significant (P < 0.05) decreases in ATP levels relative to DMSO in the absence (*) or presence (**) of 2-DG after treatment with various compounds are indicated. (B) ATP transport in isolated mitochondria. Mitochondrial preparations from HEC1A cells were incubated with ADP, treated with various compounds, and ATP levels were determined as described in the Section ″Materials and Methods.″ Significant (P < 0.05) inhibition of ATP levels is indicated by an asterisk. Fluorescent labeling of ANT by EMA in presence of Atra alone (C), DIM-C-pPhC6H5 alone (D), or in combination (E). Submitochondrial particles were incubated with the fluorescent probe (EMA) and various compounds, and analyzed by Western blot analysis as described in the Section ″Materials and Methods.″ Significant (P < 0.05) inhibition or enhancement of ANT labeling is indicated (*), and attenuation of the labeling in cells cotreated with Atra plus DIM-C-pPhC6H5 is also indicated (**). Results are expressed as mean ± SE for three replicate determinations for each treatment group.
Since inhibition of ATP transport by Atrais because of interactions of Atra with ANT proteins and induction of the c conformation of ANT, we further investigated the effects of Atra alone or in combination with DIM-C-pPhC6H5 on ANT using isolated mitochondria incubated with the fluorescent thiol alkylating reagent EMA which specifically labels cysteine 160 of ANT [50–52]. Mitochondrial suspensions were preincubated with different concentration of Atra for 10 min, EMA was added for 30 s, and the fluorescent intensity of the ANT-EMA complex was determined after SDS–PAGE (Figure 5C). Western blot analysis confirmed that the fluorescent bands were ANT proteins (data not shown). The results show that Atra significantly decreased fluorescent labeling of ANT as previously described [51]. In contrast, DIM-C-pPhC6H5 (20 or 40 µmol/mg) slightly enhanced fluorescent labeling of ANT (Figure 5D), suggesting that the C-DIM compound affects the conformation of ANT so that cysteine 160 is more readily alkylated by EMA. Coincubation of suspended mitochondrial particles with Atra plus DIM-C-pPhC6H5 resulted in labeling of ANT by EMA that was between that observed for both compounds alone (Figure 5E). These results suggest that the interactive effects of Atra and C-DIMs are due, in part, to their effects on different regions of ANT proteins and this is consistent with their differential affects on alkylation of cysteine 160 by EMA. The results also parallel the opposing effects of DIM-C-pPhtBu and Atra on cell death, ATP transport, and MMP.
Role of ANT Proteins in Mediating C-DIM-Induced Apoptosis
The pancaspase inhibitor Z-VAD-FMK blocks DIM-C-pPhtBu-induced cell death (Figure 3D),andsimilar results were obtained for Atra in HEC1A and other cancer cell lines treated with DIM-C-pPhtBu (Figure 4A). We also investigated the effects of Atra on DIM-C-pPhtBu-induced apoptos is inHEC1A cells using a cell death ELISA assay (Figure 6A) and the results show that Atra blocked the effects of DIM-C-pPhtBu. Thus, bothZ-VAD-FMK and Atra block DIM-C-pPhtBu-induced apoptosis; however, only Atra but not Z-VAD-FMK inhibited the decrease of MMP in HEC1A cells treated with DIM-C-pPhtBu (Figure 6B). These results are consistent with a mechanism in which DIM-C-pPhtBu initially induces decreased MMP through interaction with the inner mitochondrial protein ANT followed by activation of the intrinsic apoptosis pathway. The interactions of Atra and DIM-C-pPhtBuareconsistentwitharoleforANT proteins as initial mitochondrial targets for both compounds. This was further investigated by RNA interference using siRNAs for ANT1, ANT2, and ANT3. Results in Figure 6C show that ANT1 (A1), ANT2 (A2), and ANT3 (A3) mRNA are expressed in HEC1A cells, and expression of ANT2/ANT3 was higher than ANT1. Results of ANT knockdown on DIM-C-pPhtBu-induced HEC1A cell death showed that transfection of individual siRNAs for ANT proteins (iANT1, iANT2, and iANT3) inhibited the effects of DIM-C-pPhtBu; however, significant inhibition was only observed with iANT3 (Figure 6D). A combination of iANT1, iANT2, and iANT3 was the most effective treatment for inhibiting DIM-C-pPhtBu-induced cell death. Thus, knockdown of ANTexpression significantly protected the cells from DIM-C-pPhtBu-induced apoptosis, suggesting that the ANT proteins are a target for this compound. ANT antibodies do not distinguish between the different forms of these proteins; however, in cells cotransfected with iANT123 (combined siRNAs), there was a significant decrease in the immunoreactive ANT protein band (Figure 6D). These results suggest that the initial effects of DIM-C-pPhtBu involve interactions of DIM-C-pPhtBu with inner mitochondrial membrane ANT proteins which results in decreased MMP and activation of apoptosis.
Figure 6.
Role of ANT protein in DIM-C-pPhtBu-induced cell death and decreased MMP. (A) Cell death ELISA. HEC1A cells were treated with DMSO, Atra, DIM-C-pPhtBu alone, or in combination, and necrosis/apoptosis was determined in an ELISA assay (in duplicate) as described in the Section ″Materials and Methods.″ Significant induction of apoptosis (*) and inhibition after cotreatment with Atra (**) are indicated. (B) Effects of Atra on decreased MMP induced by DIM-C-pPhtBu. HEC1A cells were treated with DMSO, DIM-C-pPhtBu, Atra, Z-VAD-FMK or combinations, and MMP was determined using the fluorescence JC-1 probe as described in the Section ″Materials and Methods.″ Significantly (P < 0.05) decreased MMP (*) induced by DIM-C-pPhtBu and inhibition by Atra (**) are indicated. Results in (A) and (B) are expressed as mean ± SE for three replicate determinations for each treatment group. (C) Expression of ANT mRNA levels. Expression of ANT1, ANT2, and ANT3 mRNA was determined by RT-PCR from RNA isolated from HEC1A cells as described in the Section ″Materials and Methods.″ GAPDH served as a reference control. Effects of ANT knockdown on the percentage of DIM-C-pPhtBu-induced dead cells. (D) Effects of ANT knockdown on DIM-C-pPhtBu-induced cell death. HEC1A cells were transfected with iANT1, iANT2, iANT3, or their combination, and cells were treated with 10 µM DIM-C-pPhtBu alone or in combination with iScr or siRNAs for ANT. The percentage of dead cells was determined after 48 h as described in the Section ″Materials and Methods.″ ANT protein expression in HEC1A cells transfected with siRNAs was determined by Western blot analysis as described in the Section ″Materials and Methods.″ Significant (P < 0.05) inhibition of cell death and ANT protein expression is indicated by an asterisk (*).
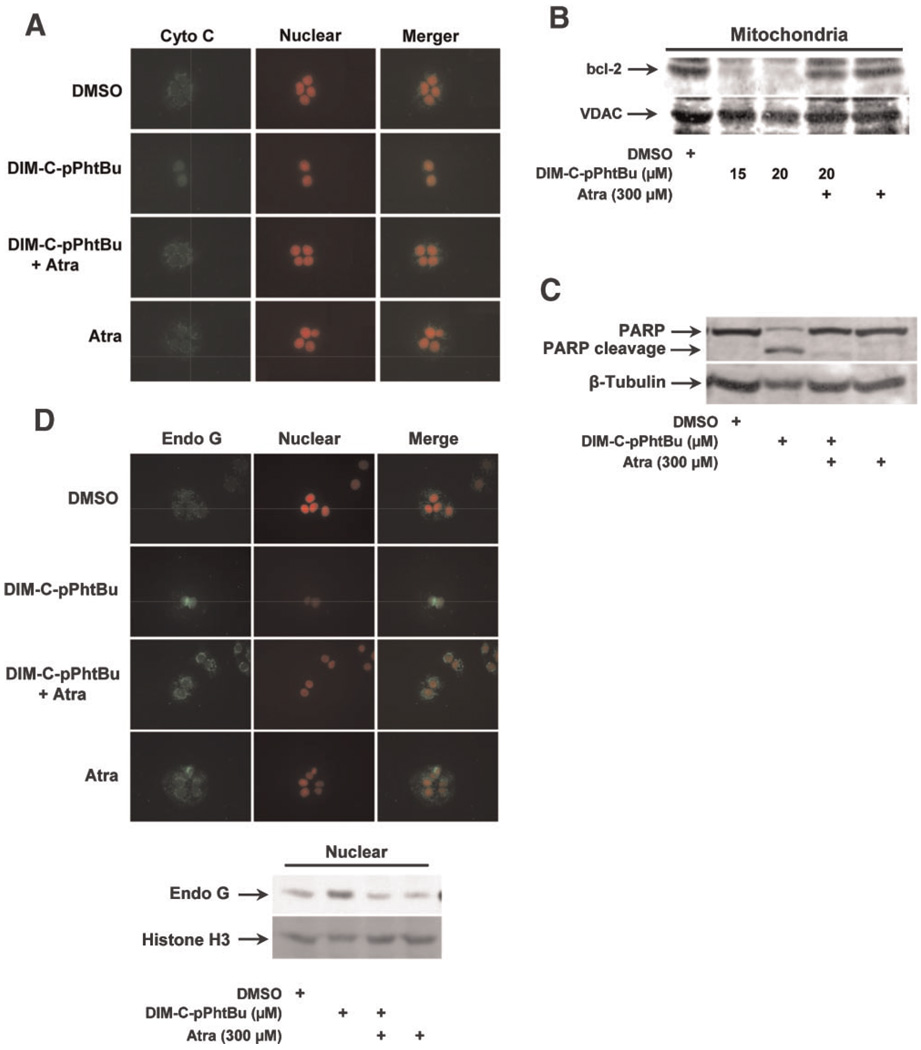
The role of ANT proteins in mediating proapoptotic responses induced by DIM-C-pPhtBu was further investigated by taking advantage of the inhibitory effects of Atra. In HEC1A cells treated with DMSO or Atra alone, immunostained cytochrome c was observed primarily localized in the mitochondria. In contrast, after treatment with DIM-C-pPhtBu alone, there was diffuse cytochrome c staining in these cells and this staining was also observed in cell nuclei, and cotreatment with Atra reversed the release of cytochrome c from the mitochondria (Figure 7A). This pattern of mitochondrial cytochrome c staining and subsequent release into the cytosol after treatment with DIM-C-pPhtBu has previously been reported with other apoptosisinducing agents [53,54] and is consistent with activation of the intrinsic pathway of apoptosis by DIM-C-pPhtBu in HEC1A cells. In common with other mitochondriotoxic agents, DIM-C-pPhtBu induced loss of bcl-2 from HEC1A mitochondria (Figure 7B), and this was accompanied by caspasedependent PARP cleavage (Figure 7C) and all of these responses were inhibited in HEC1A cells after cotreatment with Atra. In contrast, the mitochondrialvoltage-dependention channel(VDAC) protein which is a target of mitochondriotoxic arsenic trioxide [55] is not affected by DIM-C-pPhtBu (Figure 7B). DIM-C-pPhtBu also induced immunostaining of endonuclease G in nuclei of HEC1A cells, whereas minimal nuclear staining was observed in HEC1A cells treated with DMSO, Atra alone, or Atra plus DIM-C-pPhtBu confirming that induced nuclear uptake of endonuclease G was associated with the disruption of the ANT proteins by DIM-C-pPhtBu (Figure 7D). Cell nuclei were also stained with propidium idodide, and colocalization experiments confirm the treatment-dependent effects on nuclear localization of endonuclease G. Similar results were observed in Western blot analysis of endonuclease G in nuclear extracts from HEC1A cells (Figure 7D). These results demonstrate that Atra protects against activation of the intrinsic pathway of apoptosis by DIM-C-pPhtBu and suggests that the inner mitochondrial protein ANT is a major target for this compound.
Figure 7.
Atra inhibits DIM-C-pPhtBu-induced apoptosis in HEC1A cells. (A) Immunostaining of cytochrome c. HEC1A cells were treated with DMSO, 15 µM DIM-C-pPhtBu, 300 µM Atra, or Atra plus DIM-C-pPhtBu for 24 h. Each treatment group was immunostained with antibodies to cytochrome C as described in the Section ″Materials and Methods.″ Atra inhibits DIM-C-pPhtBu-induced mitochondrial bcl-2 loss (B) and PARP cleavage (C). HEC1A cells were treated for 36 h and protein levels were determined by Western blot analysis as described in the Section ″Materials and Methods.″ (D) Immunostaining of endonuclease G in HEC1A cell nuclei. Cells were treated with DMSO, DIM-C-pPhtBu, Atra or their combinations for 24 h, and immunostaining and colocalization were determined as described in the Section ″Materials and Methods.″ Western blot analysis of HEC1A whole cell lysates also showed that Atra inhibited nuclear uptake of endonuclease G in cells treated with DIM-C-pPhtBu. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
C-DIMs are a new class of PPARγ agonists which inhibit growth and/or induce death of several cancer cell lines [24–32]. Caveolin-1, a growth inhibitory tumor suppressor gene in colon cancer cells, is induced by C-DIMs and this response was reversed by PPARγ antagonists [25].However, PPARγ-active C-DIMs and other PPARγ agonists, including PGJ2, CDDO, and TZDs, induce both receptor-dependent and -independent responses in cancer cell lines, and activation of more than one cell growth inhibitory/ cell death pathway may be an important chemotherapeutic advantage for these compounds as anticancer drugs [24–36,41,56].
In this study, we investigated the effects of PPARγ-active C-DIMs in HEC1A endometrial cancer cells which express PPARγ (Figure 2A). These compounds activated PPARγ-dependent transactivation in HEC1A cells and inhibited their growth (Figure 1); however, the latter response was PPARγ-independent (Figure 2B). We also observed that the PPARγ antagonist had only minimal effects on the cytotoxicity of C-DIMs in Panc28 cells in which C-DIMs induce PPARγ-dependent activation of p21 [26]. The results in Figure 3 clearly demonstrate that C-DIM compounds preferentially induced apoptotic cell death in HEC1A cells, and this was accompanied by decreased MMP and the rapid uptake of DIM-C-pPhC6H5 into HEC1A cell mitochondria. These results suggest that C-DIMs and the prototypical DIM-C-pPhtBu (used for most experiments) may initiate their cytotoxicity by disrupting mitochondrial function.
Several anticancer drugs including PPARγ agonists are mitochondrial poisons, and many of these compounds generally decrease MMP and perturb the mitochondrial PTPC resulting in activation of proteins associated with the cell death pathways [29,30,35–40,56,57]. Our results show that DIM-C-pPhC6H5 or DIM-C-pPhtBu accumulated in the mitochondria (Figure 3F), decreased MMP (Figure3A and Figure4D), enhanced fluorescent labeling of ANT-cysteine 160 by EMA (Figure 5D), and induced cell death/ growth inhibition (Figure 3A and Figure 6A), and Atra significantly blocked all of these induced responses. Atra has been characterized as a specific agent that binds to the inner-membrane side of ANT proteins which regulate mitochondrial ATP/ADP transport [43,45]. During this transport process, ANT undergoes m- and c-state conformational changes, and Atra induces the c-state conformation. In some cells, Atra alone induces ANT-mediated mitochondrial pore formation [43–48]; however, in this study with endometrial, pancreatic cancer, and HeLa cells, up to 300 mM Atra alone did not disrupt the PTPC or cause cell death but clearly protected cells from DIM-C-pPhtBu-induced cell death. However, in isolated mitochondria (Figure 3F) and in HEC1A cells (data not shown), there was evidence that some of the reported pharmacological properties of Atra were functional. For example, Atra decreased mitochondrial ATP release (Figure 5A) and also inhibited EMA labeling of ANT (cysteine 160) in submitochondrial particles (Figure 5C). These results, coupled with the interactive effects of DIM-C-pPhC6H5 and Atra on EMA labeling of ANT (Figure 5E), suggest that ANT may be the common mitochondrial target for both compounds.
The role of ANT in mediating the effects of DIM-C-pPhtBu on HEC1A cell death was confirmed in RNA interference studies which suggest that all three forms of ANT may play a role (Figure 6D). Cotransfection with iANT1, iANT2, and iANT3 decreased the ANT protein band using an antibody that recognizes all three ANT proteins, and this was accompanied by protection from DIM-C-pPhtBu-induced cell death. These responses were highly significant, even though transfection efficiencies were 60–75%, and whole cell lysates were used for quantitative Western blot analysis.
The mechanisms associated with induction of cell death by drugs that target the ANT proteins are diverse and dependent on the specific agent and celltype suggesting that these compounds may act on multiple sites within the PTPC. For example, agents that induce oxidative stress, including thiol oxidation by alkylating compounds, can compromise the function of ANT proteins through modification of key thiol groups [45–47,57]. The anticancer drug MT-21 also targets ANT proteins in leukemia cells, and both MT-21 and Atra induce mitochondrial cytochrome c release without swelling and MT-21 induces activation of caspase 3 and apoptosis [58,59]. Like Atra, MT-21 appears to modulate the conformation of ANT; however, treatment of leukemia cells with MT-21 did not affect MMP in these cells, but induced ROS and apoptosis [58] and these responses were inhibited after cotreatment with the antiox-idant N-acetylcysteine (NAC). DIM-C-pPhtBu also induced apoptosis and release of mitochondrial cytochrome c (Figure 7); however, in contrast to MT-21, DIM-C-pPhtBu-induced responses were not related to ROS or reversed by NAC (data not shown). Moreover, all DIM-C-pPhtBu-induced responses were inhibited by Atra, suggesting that DIM-C-pPhtBu-dependent loss of MMP, cell death, and associated pathways (Figure 4B–D) are related to initial perturbation of ANT proteins and clearly differed from MT-21-induced responses which did not affect MMP. A recent report shows that ANT2 knockdown sensitizes some cancer cell lines to lonidamine a mitochondrial-targeted antitumor drug [60], whereas the mitochondriotoxicity of DIM-C-pPhtBu is ANT-dependent and is decreased after ANT knockdown (Figure 6D).
In summary, this study has identified the ANT proteins as specific mitochondrial targets for DIM-C-pPhtBu in HEC1A cells, and preliminary studies also show that other C-DIMs exhibit comparable effects in other cancer cell lines. Our results also indicate that many C-DIM compounds including DIM-C-pPhtBu target mitochondria, and their mechanism of action may involve direct interactions with ANT proteins. The crystal structure of bovine ANT, which is highly homologous with human ANT, exhibits at least one hydrophobic pocket that could accommodate the lipophilic DIM-C-pPhtBu and related compounds [61]. Preliminary studies on the structural specificity of the effects of C-DIMs on apoptosis show that methylation of the indole nitrogen significantly decreased mitochondriotoxicity of these compounds (data not shown), suggesting that a free indole group was required for maximal mitochondriotoxic activity. Current studies are investigating the mitochondriotoxic effects of C-DIMs and other substituted DIMs agonists on mitochondrial function in different cancer cell lines and determining the role of individual ANT proteins as critical mitochondrial targets that mediate the anticancer activity of C-DIMs and related compounds.
ACKNOWLEDGMENTS
The financial assistance of the National Institutes of Health (ES09106 and CA108178) and the Texas Agricultural Experiment Station is gratefully acknowledged.
Abbreviations
- PPARγ
peroxisome proliferator-activated receptor γ
- TZDs
thiazolidinediones
- CDDO
2-cyano-3,12-dioxooleana-1, 9-dien-28-oic acid
- MMP
mitochondrial membrane potential
- ANT
adenine nucleotide transport
- PTPC
permeability transition pore complex
- Atra
atractyloside
- CsA
cyclosporin A
- 2-DG
2-deoxyglucose
- ATA
aurintricarboxylic acid
- BSA
bovine serum albumin
- EMA
eosin-5-maleimide
REFERENCES
- 1.Tsai MJ, O′Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: Many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 4.Kliewer SA, Lehmann JM, Willson TM. Orphan nuclear receptors: Shifting endocrinology into reverse. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 5.Giguere V. Orphan nuclear receptors: From genetofunction. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 6.Mohan R, Heyman RA. Orphan nuclear receptor modulators. Curr Top Med Chem. 2003;3:1637–1647. doi: 10.2174/1568026033451709. [DOI] [PubMed] [Google Scholar]
- 7.Escher P, Wahli W. Peroxisome proliferator-activated receptors: Insight into multiple cellular functions. Mutat Res. 2000;448:121–138. doi: 10.1016/s0027-5107(99)00231-6. [DOI] [PubMed] [Google Scholar]
- 8.Lee CH, Olson P, Evans RM. Minireview: Lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 9.Ikezoe T, Miller CW, Kawano S, et al. Mutational analysis of the peroxisome proliferator-activated receptor g gene in human malignancies. Cancer Res. 2001;61:5307–5310. [PubMed] [Google Scholar]
- 10.Motomura W, Okumura T, Takahashi N, Obara T, Kohgo Y. Activation of peroxisome proliferator-activated receptor gamma by troglitazone inhibits cell growth through the increase of p27KiP1 in human pancreatic carcinoma cells. Cancer Res. 2000;60:5558–5564. [PubMed] [Google Scholar]
- 11.Sato H, Ishihara S, Kawashima K, et al. Expression of peroxisome proliferator-activated receptor (PPAR)g in gastric cancer and inhibitory effects of PPARg agonists. Br J Cancer. 2000;83:1394–1400. doi: 10.1054/bjoc.2000.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi N, Okumura T, Motomura W, Fujimoto Y, Kawabata I, Kohgo Y. Activation of PPARg inhibits cell growth and induces apoptosis in human gastric cancer cells. FEBS Lett. 1999;455:135–139. doi: 10.1016/s0014-5793(99)00871-6. [DOI] [PubMed] [Google Scholar]
- 13.Kubota T, Koshizuka K, Williamson EA, et al. Ligand for peroxisome proliferator-activated receptor g (troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Res. 1998;58:3344–3352. [PubMed] [Google Scholar]
- 14.Elstner E, Muller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptor gamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TH, Szabo E. Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor g in non-small cell lung cancer. Cancer Res. 2000;60:1129–1138. [PubMed] [Google Scholar]
- 16.Suh N, Wang Y, Williams CR, et al. A new ligand for the peroxisome proliferator-activated receptor-g (PPAR-g), GW7845, inhibits rat mammary carcinogenesis. Cancer Res. 1999;59:5671–5673. [PubMed] [Google Scholar]
- 17.Qin C, Burghardt R, Smith R, Wormke M, Stewart J, Safe S. Peroxisome proliferator-activated receptor g (PPARg) agonists induce proteasome-dependent degradation of cyclin D1 and estrogen receptor a in MCF-7 breast cancer cells. Cancer Res. 2003;63:958–964. [PubMed] [Google Scholar]
- 18.Clay CE, Monjazeb A, Thorburn J, Chilton FH, High KP. 15-Deoxy-D(12,14)-prostaglandin J2-induced apoptosis does not require PPARg in breast cancer cells. J Lipid Res. 2002;43:1818–1828. doi: 10.1194/jlr.m200224-jlr200. [DOI] [PubMed] [Google Scholar]
- 19.Brockman JA, Gupta RA, DuBois RN. Activation of PPARg leads to inhibition of anchorage independent growth of human colorectal cancer cells. Gastroenterology. 1998;115:1049–1055. doi: 10.1016/s0016-5085(98)70072-1. [DOI] [PubMed] [Google Scholar]
- 20.Gupta RA, Sarraf P, Mueller E, et al. Peroxisome proliferatoractivated receptor g-mediated differentiation: A mutation in colon cancer cells reveals divergent and cell type-specific mechanisms. J Biol Chem. 2003;278:22669–22677. doi: 10.1074/jbc.M300637200. [DOI] [PubMed] [Google Scholar]
- 21.Gupta RA, Brockman JA, Sarraf P, Willson TM, DuBois RN. Target genes of peroxisome proliferator-activated receptor g in colorectal cancer cells. J Biol Chem. 2001;276:29681–29687. doi: 10.1074/jbc.M103779200. [DOI] [PubMed] [Google Scholar]
- 22.Konopleva M, Elstner E, McQueen TJ, et al. Peroxisome proliferator-activated receptor g and retinoid X receptor ligands are potent inducers of differentiation and apoptosis in leukemias. Mol Cancer Ther. 2004;3:1249–1262. [PubMed] [Google Scholar]
- 23.Melichar B, Konopleva M, Hu W, Melicharova K, Andreeff M, Freedman RS. Growth-inhibitory effect of a novel synthetic triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, on ovarian carcinoma cell lines notdependentonperoxisome proliferator-activated receptor-g expression. Gynecol Oncol. 2004;93:149–154. doi: 10.1016/j.ygyno.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Qin C, Morrow D, Stewart J, et al. A new class of peroxisome proliferator-activated receptor g (PPARg) agonists that inhibit growth of breast cancer cells: 1,1-bis(3′-indolyl)-1-(p-substitutedphenyl)methanes. Mol Cancer Therap. 2004;3:247–259. [PubMed] [Google Scholar]
- 25.Chintharlapalli S, Smith R, III, Samudio I, Zhang W, Safe S. 1, 1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes induce peroxisome proliferator-activated receptor g-mediated growth inhibition, transactivation and differentiation markers in colon cancer cells. Cancer Res. 2004;64:5994–6001. doi: 10.1158/0008-5472.CAN-04-0399. [DOI] [PubMed] [Google Scholar]
- 26.Hong J, Samudio I, Liu S, Abdelrahim M, Safe S. Peroxisome proliferator-activated receptor g-dependent activation of p21 in Panc-28 pancreatic cancer cells involves Sp1 and Sp4 proteins. Endocrinology. 2004;145:5774–5785. doi: 10.1210/en.2004-0686. [DOI] [PubMed] [Google Scholar]
- 27.Contractor R, Samudio I, Estrov Z, et al. A novel ringsubstituted diindolylmethane 1,1-bis[3′-(5-methoxyindolyl)]-1-(p-t-butylphenyl)methane inhibits ERK activation and induces apoptosis in acute myeloid leukemia. Cancer Res. 2005;65:2890–2898. doi: 10.1158/0008-5472.CAN-04-3781. [DOI] [PubMed] [Google Scholar]
- 28.Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substitutedphenyl)methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptorindependent activation of early growth response-1 and NAG-1. Mol Pharmacol. 2005;68:1782–1792. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- 29.Abdelrahim M, Newman K, Vanderlaag K, Samudio I, Safe S. 3,3′-Diindolylmethane (DIM) and derivatives induce apopto-sis in pancreatic cancer cells through endoplasmic reticulum stress-dependent upregulation of DR5. Carcinogenesis. 2006;27:717–728. doi: 10.1093/carcin/bgi270. [DOI] [PubMed] [Google Scholar]
- 30.Chintharlapalli S, Papineni S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes inhibit colon cancer cell and tumor growth through PPARg-dependent and PPARg-independent pathways. Mol Cancer Ther. 2006;5:1362–1370. doi: 10.1158/1535-7163.MCT-06-0002. [DOI] [PubMed] [Google Scholar]
- 31.Lei P, Abdelrahim M, Safe S. 1,1-Bis(3′-indolyl)-1-(p-sub-stituted phenyl)methanes inhibit ovarian cancer cell growth through peroxisome proliferator-activated receptor-dependent and independent pathways. Mol Cancer Ther. 2006;5:2324–2336. doi: 10.1158/1535-7163.MCT-06-0184. [DOI] [PubMed] [Google Scholar]
- 32.Kassouf W, Chintharlapalli S, Abdelrahim M, Nelkin G, Safe S, Kamat AM. Inhibition of bladder tumor growth by 1,1-bis(3′-indolyl)-1-(p-substitutedphenyl)methanes: A new class of peroxisome proliferator-activated receptor g agonists. Cancer Res. 2006;66:412–418. doi: 10.1158/0008-5472.CAN-05-2755. [DOI] [PubMed] [Google Scholar]
- 33.Doerner A, Pauschinger M, Badorff A, et al. Tissue-specific transcription pattern of the adenine nucleotide translocase isoforms in humans. FEBS Lett. 1997;414:258–262. doi: 10.1016/s0014-5793(97)01000-4. [DOI] [PubMed] [Google Scholar]
- 34.Palakurthi SS, Aktas H, Grubissich LM, Mortensen RM, Halperin JA. Anticancer effects of thiazolidinediones are independent of peroxisome proliferator-activated receptor g and mediated by inhibition of translation initiation. Cancer Res. 2001;61:6213–6218. [PubMed] [Google Scholar]
- 35.Samudio I, Konopleva M, Hail N, Jr, et al. 2-Cyano-3,12 dioxooleana-1,9 diene-28-imidazolide (CDDO-Im) directly targets mitochondrial glutathione to induce apoptosis in pancreatic cancer. J Biol Chem. 2005;280:36273–36282. doi: 10.1074/jbc.M507518200. [DOI] [PubMed] [Google Scholar]
- 36.Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I, Safe S. 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor g-dependent and -independent pathways. Mol Pharmacol. 2005;68:119–128. doi: 10.1124/mol.105.011437. [DOI] [PubMed] [Google Scholar]
- 37.Rahman KM, Aranha O, Sarkar FH. Indole-3-carbinol, (I3C) induces apoptosis in tumorigenic but not in nontumorigenic breast epithelial cells. Nutr Cancer. 2003;45:101–112. doi: 10.1207/S15327914NC4501_12. [DOI] [PubMed] [Google Scholar]
- 38.Ge X, Yannai S, Rennert G, Gruener N, Fares FA. 3, 3′-Diindolylmethane induces apoptosis in human cancer cells. Biochem Biophys Res Commun. 1996;228:153–158. doi: 10.1006/bbrc.1996.1631. [DOI] [PubMed] [Google Scholar]
- 39.Nachshon-Kedmi M, Yannai S, Fares FA. Induction of apoptosis in human prostate cancer cell line, PC3, by 3,3′-diindolylmethane through the mitochondrial pathway. Br J Cancer. 2004;91:1358–1363. doi: 10.1038/sj.bjc.6602145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong C, Firestone GL, Bjeldanes LF. Bcl–2 family-mediated apoptotic effects of 3,3′-diindolylmethane (DIM) in human breast cancer cells. Biochem Pharmacol. 2002;63:1085–1097. doi: 10.1016/s0006-2952(02)00856-0. [DOI] [PubMed] [Google Scholar]
- 41.Clay CE, Namen AM, Atsumi G, et al. Influence of J series prostaglandins on apoptosis and tumorigenesis of breast cancer cells. Carcinogenesis. 1999;20:1905–1911. doi: 10.1093/carcin/20.10.1905. [DOI] [PubMed] [Google Scholar]
- 42.Arnoult D, Gaume B, Karbowski M, Sharpe JC, Cecconi F, Youle RJ. Mitochondrial release of AIF and EndoG requires caspase activation downstream of Bax/Bak-mediated per-meabilization. EMBO J. 2003;22:4385–4399. doi: 10.1093/emboj/cdg423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gropp T, Brustovetsky N, Klingenberg M, Muller V, Fendler K, Bamberg E. Kinetics of electrogenic transport by the ADP/ ATP carrier. Biophys J. 1999;77:714–726. doi: 10.1016/S0006-3495(99)76926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brustovetsky N, Becker A, Klingenberg M, Bamberg E. Electrical currents associated with nucleotide transport by the reconstituted mitochondrial ADP/ATP carrier. Proc Natl Acad Sci USA. 1996;93:664–668. doi: 10.1073/pnas.93.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broekemeier KM, Dempsey ME, Pfeiffer DR. Cyclosporin A is a potent inhibitor of the inner membrane permeability transition in liver mitochondria. J Biol Chem. 1989;264:7826–7830. [PubMed] [Google Scholar]
- 46.Belzacq AS, Vieira HL, Kroemer G, Brenner C. The adenine nucleotide translocator in apoptosis. Biochimie. 2002;84:167–176. doi: 10.1016/s0300-9084(02)01366-4. [DOI] [PubMed] [Google Scholar]
- 47.Vieira HL, Haouzi D, El Hamel C, et al. Permeabilization of the mitochondrial inner membrane during apoptosis: Impact of the adenine nucleotide translocator. Cell Death Differ. 2000;7:1146–1154. doi: 10.1038/sj.cdd.4400778. [DOI] [PubMed] [Google Scholar]
- 48.Belzacq AS, Jacotot E, Vieira HL, et al. Apoptosis induction by the photosensitizer verteporfin: Identification of mitochondrial adenine nucleotide translocator as a critical target. Cancer Res. 2001;61:1260–1264. [PubMed] [Google Scholar]
- 49.Brown J, Higo H, McKalip A, Herman B. Human papillomavirus (HPV) 16 E6 sensitizes cells to atractyloside-induced apoptosis: Role of p53, ICE-like proteases and the mitochondrial permeability transition. J Cell Biochem. 1997;66:245–255. doi: 10.1002/(sici)1097-4644(19970801)66:2<245::aid-jcb11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 50.Majima E, Koike H, Hong YM, Shinohara Y, Terada H. Characterization of cysteine residues of mitochondrial ADP/ ATP carrier with the SH-reagents eosin 5-maleimide and N-ethylmaleimide. J Biol Chem. 1993;268:22181–22187. [PubMed] [Google Scholar]
- 51.Majima E, Shinohara Y, Yamaguchi N, Hong YM, Terada H. Importance of loops of mitochondrial ADP/ATP carrier for its transport activity deduced from reactivities of its cysteine residues with the sulfhydryl reagent eosin-5-maleimide. Biochemistry. 1994;33:9530–9536. doi: 10.1021/bi00198a019. [DOI] [PubMed] [Google Scholar]
- 52.McStay GP, Clarke SJ, Halestrap AP. Role of critical thiol groups on the matrix surface of the adenine nucleotide translocase in the mechanism of the mitochondrial permeability transition pore. Biochem J. 2002;367:541–548. doi: 10.1042/BJ20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kennedy SG, Kandel ES, Cross TK, Hay N. Akt/Protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Mol Cell Biol. 1999;19:5800–5810. doi: 10.1128/mcb.19.8.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du C, Fang M, Li Y, Li L, Wang X, Smac a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Y, Shi Y, Tian C, et al. Essential role of the voltagedependent anion channel (VDAC) in mitochondrial permeability transition pore opening and cytochrome c release induced by arsenic trioxide. Oncogene. 2004;23:1239–1247. doi: 10.1038/sj.onc.1207205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiau CW, Yang CC, Kulp SK, et al. Thiazolidenediones mediate apoptosis in prostate cancer cells in part through inhibition of Bcl-xL/Bcl-2 functions independently of PPAR-gamma. Cancer Res. 2005;65:1561–1569. doi: 10.1158/0008-5472.CAN-04-1677. [DOI] [PubMed] [Google Scholar]
- 57.Don AS, Kisker O, Dilda P, et al. A peptide trivalent arsenical inhibits tumor angiogenesis by perturbing mitochondrial function in angiogenic endothelial cells. Cancer Cell. 2003;3:497–509. doi: 10.1016/s1535-6108(03)00109-0. [DOI] [PubMed] [Google Scholar]
- 58.Watabe M, Machida K, Osada H. MT-21 is a synthetic apoptosis inducer that directly induces cytochrome c release from mitochondria. Cancer Res. 2000;60:5214–5222. [PubMed] [Google Scholar]
- 59.Machida K, Hayashi Y, Osada H. A novel adenine nucleotide translocase inhibitor, MT-21, induces cytochrome c release by a mitochondrial permeability transition-independent mechanism. J Biol Chem. 2002;277:31243–31248. doi: 10.1074/jbc.M204564200. [DOI] [PubMed] [Google Scholar]
- 60.Le Bras M, Borgne-Sanchez A, Touat Z, et al. Chemosensitization by knockdown of adenine nucleotide translocase-2. Cancer Res. 2006;66:9143–9152. doi: 10.1158/0008-5472.CAN-05-4407. [DOI] [PubMed] [Google Scholar]
- 61.Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]