Abstract
Promoting the degradation of Hsp90 client proteins by inhibiting Hsp90, an important protein chaperone, has been shown to be a promising new anticancer strategy. In this study, we show that an oxazoline analogue of apratoxin A (oz-apraA), a cyclodepsipeptide isolated from a marine cyanobacterium, promotes the degradation of Hsp90 clients through chaperone-mediated autophagy (CMA). We identify a KFERQ-like motif as a conserved pentapeptide sequence in the kinase domain of epidermal growth factor receptor (EGFR) necessary for recognition as a CMA substrate. Mutation of this motif prevents EGFR degradation by CMA and promotes the degradation of EGFR through the proteasomal pathway in oz-apraA–treated cells. Oz-apraA binds to Hsc70/Hsp70. We propose that apratoxin A inhibits Hsp90 function by stabilizing the interaction of Hsp90 client proteins with Hsc70/Hsp70 and thus prevents their interactions with Hsp90. Our study provides the first examples for the ability of CMA to mediate degradation of membrane receptors and cross talks of CMA and proteasomal degradation mechanisms.
Introduction
The molecular chaperone Hsp90 (heat shock protein 90) maintains the conformation, stability, and activity of several key oncogenic proteins, such as EGF receptor (EGFR), cyclin-dependent kinases, Raf, and p53, which are collectively known as Hsp90 client proteins (Whitesell and Lindquist, 2005). The chaperone function of Hsp90 requires the formation of a multichaperone complex, which is dependent on the hydrolysis of ATP and ADP/ATP exchange. A geldanamycin (GA) analogue, which inhibits Hsp90 by blocking its intrinsic ATPase activity, was the first in class inhibitor of Hsp90 in human clinical trials for cancer therapy. Inhibition of Hsp90 function by GA disrupts the interaction of Hsp90 with its client proteins and leads to their proteasome pathway-dependent degradation (Zhang and Burrows, 2004).
Autophagy mediates the bulk degradation of intracellular components such as macromolecule complexes and subcellular organelles through lysosomal-dependent mechanisms and is essential for the maintenance of cellular homeostasis (Meijer, 2003). Three modes of autophagy have been identified in mammals (Levine and Klionsky, 2004). Macroautophagy cargo is sequestered into a specialized double-membrane structure called an autophagosome. Autophagosomes then fuse with lysosomes, and its content is subsequently degraded by hydrolytic enzymes. Microautophagy cargo is generated by direct budding of the lysosomal membrane. In chaperone-mediated autophagy (CMA), substrates are recognized by a chaperone complex containing Hsc70 (heat shock cognate of 70 kD) and delivered into lysosomes through interactions with LAMP-2A (lysosome-associated membrane protein 2A). CMA is activated during prolonged nutrient deprivation, which results in the degradation of cytosolic proteins in a molecule by molecule fashion (Dice, 2007). The relevance of CMA for mediating the degradation of noncytosolic proteins, if any, remains unknown.
Apratoxin A, a cyclodepsipeptide isolated from the marine cyanobacterium Lyngbya majuscula, exhibits cytotoxic activity against several cancer cell lines (Luesch et al., 2006) and has shown to have some activity against colon adenocarcinoma C38-derived tumors in an in vivo study (Luesch et al., 2001). Apratoxin A was found to induce G1 cell cycle arrest and apoptosis (Luesch et al., 2006). However, the cellular and molecular mechanisms of apratoxin A remain unknown. Using the clues suggested by the gene expression signatures of apratoxin A (Luesch et al., 2006), we explored its mechanism of action using a synthetic oxazoline analogue of apratoxin A (oz-apraA). In this study, we demonstrate that oz-apraA modulates degradation of Hsp90 client proteins (e.g., EGFR and ErbB2) through the CMA pathway. We show that EGFR contains a KFERQ-like motif in its kinase domain, and oz-apraA–induced EGFR degradation is dependent on LAMP-2A. Our study demonstrates a CMA-mediated mechanism for degradation of multiple Hsp90 client proteins, including membrane receptors in oz-apraA–treated cells.
Results
Impact of apratoxin A and its oxazoline analogue on Hsp90 chaperone complexes
To study the molecular mechanism of apratoxin A (Fig. 1 A), we performed retrospective data mining by identifying the gene expression signature of apratoxin A (Fig. S1 A) from a previous study (Luesch et al., 2006) and compared it with gene expression signatures of different compounds in the Connectivity Map database (Lamb et al., 2006). The apratoxin A signatures from 3- and 6-h treatment exhibited a high degree of similarity to the gene expression profile from the well-established Hsp90 inhibitor 17-allylamino-GA (n = 18, P < 0.001; Fig. S1 B). Consistent with an effect on protein chaperones, apratoxin A significantly induced expression of several heat shock–responsive genes, including HSPA1B, DNAJB1, HSPA6, DDIT4, DNAJB9, and BAG3 (Fig. S1 A and Tables S1 and S2). We reperformed the analysis of apratoxin A signatures by removing heat shock–responsive genes to exclude toxicological effects (Meyer et al., 1995; Schiaffonati and Tiberio, 1997). The signature of apratoxin A treatment with heat shock–responsive genes subtracted also showed high similarity to that of Hsp90 inhibitors (Fig. S1 C). Because immediate cellular responses are more likely to be the result of direct interaction with the primary targets, we hypothesize that the mechanism of apratoxin A action might share similarity to that of GA and act by modulating Hsp90 complex formation with its client proteins.
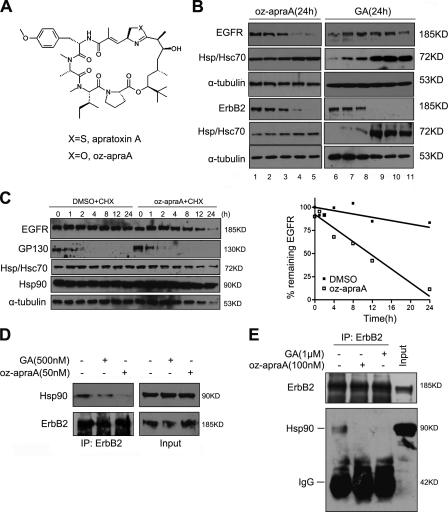
Figure 1.
Apratoxin A and its analogue inhibit the Hsp90 pathway. (A) Chemical structure of the compounds. (B) Oz-apraA reduces the levels of Hsp90 client proteins in different cell lines. Oz-apraA and GA induce concentration-dependent decreases of EGFR levels in A549 cells, ErbB2 levels in MDA-MB-453 cells at 24 h, and increases in Hsp70 levels. Lanes 1–5: 0, 1, 10, 100, and 500 nM; lanes 6–11: 0, 1, 10, 100, 500, and 1,000 nM. (C) Half-lives of EGFR and short-lived protein GP130 were analyzed in HeLa cells treated with 100 µg/ml CHX in the presence of DMSO or 100 nM oz-apraA for the indicated periods of time. The levels of EGFR were normalized to that of α-tubulin expression, and results were plotted against inhibitor treatment time points. (D) Immunoprecipitation (IP) of endogenous ErbB2-containing protein complexes from MDA-MB-453 cells after treatment of 500 nM GA or 50 nM oz-apraA for 6 h. (E) In vitro Hsp90-binding assay. SKoV3 cell lysates were incubated with 1 µM GA or 100 nM oz-apraA at 4°C for 2 h. ErbB2 was immunoprecipitated, and bound Hsp90 was detected using Western blotting.
Investigation of the apratoxin A mechanism of action has been hampered by the scarcity of natural material. Because our previous work demonstrated that oz-apraA, which has similar potency with respect to tumor toxicity as apratoxin A, could be obtained through total synthesis (Ma et al., 2006), we next examined the impact of oz-apraA on activation of the Hsp90 pathway.
We first determined the effect of oz-apraA on the protein levels of the Hsp90 client proteins. Treatment with oz-apraA led to concentration- and time-dependent reductions in the protein levels of known Hsp90 clients, EGFR, ErbB2 (Fig. 1 B and Fig. S1 D), and RIP-1 (see Fig. 4 D). In A549 cells, wild-type EGFR is more sensitive to oz-apraA than to GA. Although, in MDA-MB-453 cells, the treatment with 10 nM oz-apraA or GA for 24 h led to a similar reduction in the levels of ErbB2 (Fig. 1 B, bottom). Oz-apraA treatment also led to a dose-dependent increase in the levels of Hsp70 (Hsp/Hsc70), as detected by an antibody recognizing both Hsp70 and Hsc70, but to a lesser extent than that with GA treatment (Fig. 1 B). We further tested whether oz-apraA had any effect on the half-life of EGFR (Fig. 1 C). Although the levels of EGFR did not change appreciably over 24 h in the presence of cycloheximide (CHX) alone, in the presence of oz-apraA and CHX, a reduction in the levels of EGFR was first detected at 8 h. By 24 h, <20% remained. In contrast, oz-apraA had no effect on the turnover of interleukin 6 signal transducer GP130 (Fig. 1 C) or the turnover of pEGFP-CL1 (see Fig. 3 D), suggesting that oz-apraA does not directly inhibit proteasome activity. Furthermore, ∼500 nM oz-apraA or up to 1 µM GA had no effect on the levels of FGFR1, a non-Hsp90 client membrane receptor (Fig. S1 E; Citri et al., 2006). From these results, we conclude that oz-apraA specifically reduces the levels of at least a subset of Hsp90 client proteins.
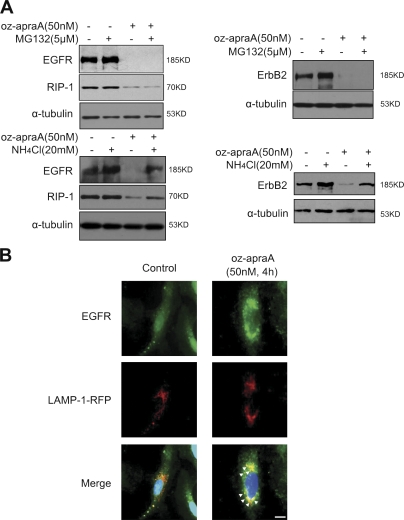
Figure 4.
Oz-apraA induces Hsp90 client protein degradation through lysosomal pathway. (A) Lysosomal inhibitor abrogates oz-apraA–induced Hsp90 client protein degradation. A549 cells (left) and MDA-MB-453 cells (right) were pretreated with the indicated compounds for 2 h followed by treatment of 50 nM oz-apraA for 24 h. Cell lysates were separated by SDS-PAGE. The Western blot was probed with anti-EGFR, anti-RIP1, and anti-ErbB2 antibodies. Anti–α-tubulin was used as a loading control. (B) Immunolocalization of EGFR and lysosomal marker LAMP-1 in A549 cells before and after treatment with oz-apraA in the presence of 20 mM NH4Cl. A549 cells were transiently transfected with LAMP-1–RFP expression plasmid, and after a 24-h transfection, cells were treated with or without 50 nM oz-apraA and 20 mM NH4Cl for 6 h, fixed, permeabilized, and stained with anti-EGFR rabbit polyclonal antibody. Arrows indicate the colocalization of EGFR and LAMP-1. Bar, 15 µm.
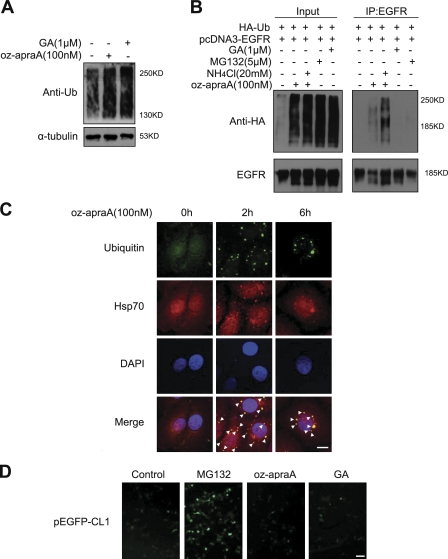
Figure 3.
Oz-apraA induces the accumulation of ubiquitinated proteins and the formation of aggresomes. (A) 100 nM oz-apraA and1 µM GA induce the accumulation of endogenous ubiquitinated proteins in HeLa cells. (B) Oz-apraA promotes the ubiquitination of EGFR. HEK293 cells were transfected with 2 µg pcDNA3-EGFR and 2 µg wild-type pcDNA-HA-ubiquitin (Ub) followed by treatment with the indicated compounds for 6 h. EGFR was immunoprecipitated from the lysates, and ubiquitinated EGFR were detected with anti-HA antibody by Western blotting. IP, immunoprecipitation. (C) Oz-apraA promotes the accumulation of ubiquitinated proteins. HeLa cells were treated with 100 nM oz-apraA for the indicated times followed by immunostaining with antiubiquitin (green), anti-Hsp/Hsc70 (red), and DAPI (blue) as indicated. Superimposed confocal images (merge) demonstrate the colocalization of ubiquitin with Hsp/Hsc70. Arrowheads indicate the colocalization of ubiquitin and Hsp/Hsc70. Bar, 10 µm. (D) Oz-apraA does not inhibit proteasome activity. pEGFP-CL1–transfected HEK293 cells were treated with or without 100 nM oz-apraA, 5 µM MG132, or 1 µM GA for 12 h. Bar, 50 µm.
The GA class of Hsp90 inhibitors binds to the N-terminal ATP-binding site of Hsp90 with higher affinity than that of natural nucleotides and thus abrogates the formation of Hsp90 chaperone–client protein complexes by preventing exchange of ADP for ATP (Whitesell and Lindquist, 2005). To determine whether oz-apraA also disrupts the interaction of Hsp90 with its client proteins, we examined the interaction of Hsp90 with ErbB2 by coimmunoprecipitation. We found that like GA, treatment of MDA-MB-453 cells with oz-apraA reduced the interaction of Hsp90 with its client protein, ErbB2 (Fig. 1 D). Furthermore, in the presence of 100 nM oz-apraA, the interaction of Hsp90 with ErbB2 in SKoV3 cell lysates was inhibited to the same extent as in cells treated with 1 µM GA (Fig. 1 E). From these data, we conclude that oz-apraA can inhibit the physical interactions of Hsp90 with at least a subset of its clients.
Oz-apraA–binding proteins
To identify oz-apraA target proteins, we generated a biotin derivative of oz-apraA (bio–oz-apraA) with a spacer at position 26 of O-methyl-tyrosine (Fig. 2 A). This compound was assembled by a click reaction of a biotin-embodied azide with alkynyl–oz-apraA; a new analogue of oz-apraA was synthesized based on our previous procedure. Our early study indicated that modification of the tyrosine tail might have little influence to the biological activities (Ma et al., 2006). As desired, bio–oz-apraA remains as active as unsubstituted oz-apraA and induces the degradation of EGFR in a concentration-dependent manner (Fig. S1 F). Using bio–oz-apraA conjugated with streptavidin-agarose, affinity pull-down assays were performed with lysates of HeLa or HEK293 cells. Proteins with an apparent molecular mass of ∼70 kD were specifically pulled down with bio–oz-apraA, and this 70-kD protein was competed away by excess oz-apraA (Fig. 2 B, lanes 2 and 3). Using mass spectrometry, the 70-kD band was identified as proteins of the Hsp70 family, including HSPA8 (which encodes an Hsc70) and HSPA1B (which encodes an inducible Hsp70; Fig. 2 B). Other identified oz-apraA–binding proteins include RARS (arginyl-tRNA synthetase protein) and SRP68 (signal recognition particle protein 68). RARS, a member of the class I aminoacyl-tRNA synthetase family, has been reported to be associated with Hsp90 (Kang et al., 2000). SRP68, a ribonucleoprotein in the signal peptide recognition particle, is involved in docking the nascent polypeptide–ribosome complex at receptors on the ER (Politz et al., 2000). We concentrated further analysis on Hsp70/Hsc70 because they are cochaperones for Hsp90 and are directly involved in folding of Hsp90 client proteins.
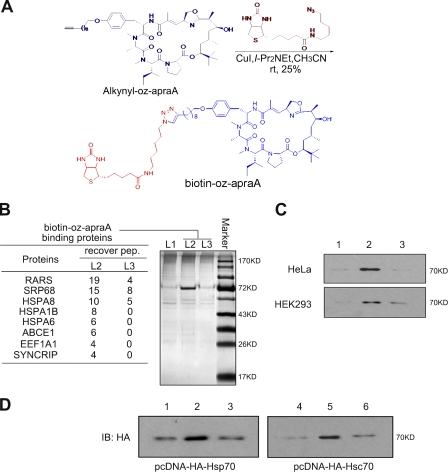
Figure 2.
Oz-apraA–binding proteins. (A) Brief synthetic steps of bio–oz-apraA. CuI, cuprous iodide; I-Pr2NEt, diisopropyl-ethyl amine; CH3CN, acetonitrile. (B) The pull-down products from bio–oz-apraA using HeLa cells were separated by SDS-PAGE and visualized by silver staining. The data from mass spectrometry analysis are shown in the table. Lane 1, streptavidin-agarose beads; lane 2, bio–oz-apraA incubation followed by pulling down by streptavidin-agarose beads; lane 3, the presence of excess oz-apraA. recover pep, recovery peptides of the enzyme digestion. (C) Confirmation of bio–oz-apraA endogenous binding protein (Hsp/Hsc70) in HeLa and HEK293 cell lysates by Western blotting. Lane 1, DMSO; lane 2, first incubation with DMSO followed by incubation with 2 µM bio–oz-apraA; lane 3, first incubation with 20 µM oz-apraA followed by incubation with 2 µM bio–oz-apraA. (D) Bio–oz-apraA binds to both Hsp70 and Hsc70 proteins. HEK293 cells were transiently transfected with the indicated plasmids, and the pull-down assay was performed as described in Materials and methods after Western blotting using anti-HA antibody. Lanes 1 and 4, DMSO; lanes 2 and 5, first incubation with DMSO followed by incubation with 2 µM bio–oz-apraA; lanes 3 and 6, first incubation with 20 µM oz-apraA followed by incubation with 2 µM bio–oz-apraA. IB, immunoblotting.
The identity of the 70-kD protein was further confirmed by Western blotting using an antibody recognizing Hsp70/Hsc70 after the pull-down assay using HeLa cell or HEK293 cell lysates (Fig. 2 C). The binding of bio–oz-apraA with Hsp70/Hsc70 binding is specific, as it can be competed away with excessive oz-apraA. To directly analyze the interaction of bio–oz-apraA with Hsp70 and Hsc70, we next transiently transfected HEK293 cells with pcDNA-HA-Hsp70 or pcDNA-HA-Hsc70 and repeated the pull-down assay followed by Western blotting with an anti-HA antibody. As shown in Fig. 2 D, bio–oz-apraA can bind to both Hsp70 and Hsc70, and this binding can be competed away by excessive oz-apraA, which is consistent with the results of the pull-down assay for endogenous Hsp70/Hsc70. However, no direct interaction of Hsp90 with bio–oz-apraA was detected using a similar assay (unpublished data). Thus, we conclude that oz-apraA can bind to Hsp70/Hsc70 but not Hsp90. Interestingly, binding of oz-apraA to Hsp70/Hsc70 may also disrupt the interaction of Hsp70/Hsc70 with Hsp90 (Fig. S2).
Oz-apraA induces the accumulation of ubiquitinated proteins and the formation of aggresome
Because the inhibition of Hsp90 by GA has been shown to promote ubiquitination of Hsp90 client proteins (Xu et al., 2002), we asked whether there was also an increase in the levels of ubiquitinated proteins in oz-apraA–treated cells. Like GA, treatment with oz-apraA significantly induced accumulation of endogenous or transfected polyubiquitin-positive proteins (Fig. 3, A and B). Importantly, as shown in Fig. 3 B, oz-apraA significantly induced ubiquitination of EGFR. Because the lysosomal inhibitor ammonium chloride (NH4Cl) inhibited oz-apraA–mediated EGFR degradation (see Fig. 4 A), we observed that combining treatment of oz-apraA and NH4Cl further enhanced oz-apraA–mediated EGFR ubiquitination. In comparison, although both GA and MG132 treatments induced total cellular polyubiquitin accumulation, they were much less effective at inducing polyubiquitination of EGFR (Fig. 3 B).
Oz-apraA–induced ubiquitin accumulation was also supported by an immunofluorescence experiment using antiubiquitin. As shown in Fig. 3 C, oz-apraA led to the appearance of ubiquitin-positive protein aggregation around the nuclear membrane, which partially colocalized with Hsp70/Hsc70. The prominent single juxtanuclear body (Fig. 3 C, 6-h treatment) resembled an aggresome, which forms in the proximity of the microtubule-organizing center as a result of accumulating misfolded proteins (Xu et al., 2002). The colocalization of Hsp70 and γ-tubulin in protein aggregates induced by oz-apraA provided a further confirmation of aggresome (Fig. S3).
Because oz-apraA treatment for 2 h was sufficient to induce the accumulation of aggregated ubiquitin (Fig. 3 C), we asked whether oz-apraA directly inhibited proteasome activity, which might be responsible for the accumulation of ubiquitinated proteins. We used a reporter consisting of a short degron, CL1, fused to the C terminus of EGFP (pEGFP-CL1; Bence et al., 2001). HEK293 cells transiently transfected with pEGFP-CL1 were treated with MG132, oz-apraA, or GA individually. As expected, the presence of MG132 increased the fluorescence of pEGFP-CL1 in transfected HEK293 cells, whereas oz-apraA and GA had no effect (Fig. 3 D). Because oz-apraA also had no effect on the half-life of short-lived protein GP130 (Fig. 1 C), we conclude that oz-apraA does not inhibit the proteasomal pathway.
Oz-apraA–induced degradation of EGFR is independent of proteasome or macroautophagy
Because the proteasomal pathway is critical for the degradation of most of the known Hsp90 client proteins when the Hsp90 complex function is abrogated by Hsp90 inhibitors (Zhang and Burrows, 2004; Qing et al., 2006), we investigated whether the enhanced turnover of Hsp90 client proteins in oz-apraA–treated cells also involves the proteasome. In contrast to GA, MG132 failed to inhibit the degradation of Hsp90 client proteins induced by oz-apraA in different cell lines, whereas NH4Cl could block oz-apraA–induced Hsp90 client protein degradation (Fig. 4 A). Consistent with a function of lysosomes in this context, a significantly increased amount of EGFR was colocalized with LAMP-1–RFP upon treatment of oz-apraA in A549 cells (Fig. 4 B). However, the presence of MG132 but not NH4Cl inhibited the degradation of ErbB2 induced by GA (Fig. S4 A), as reported previously (Peng et al., 2005). Collectively, these results suggest that contrary to GA, the degradation of Hsp90 clients induced by oz-apraA is dependent on lysosomal, not proteasomal, mechanisms.
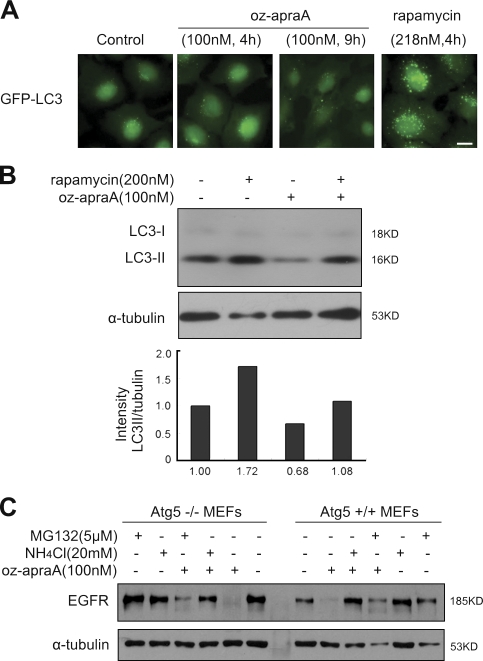
Because macroautophagy and microautophagy are also lysosomal-dependent protein degradation mechanisms, we first examined whether oz-apraA had any effects on macroautophagy. LC3 (microtubule-associated protein 1 light chain 3) is the mammalian orthologue of yeast Atg8 (Kabeya et al., 2000). The conversion of cytosolic LC3-I to lipidated LC3-II, which is associated with autophagosome membrane, is a well-established marker for macroautophagy (Kabeya et al., 2000). Vesicle-localized GFP-LC3 is a sensitive assay to detect autophagosome formation (Kabeya et al., 2000). In contrast to rapamycin, the treatment with oz-apraA did not increase the number of LC3-positive autophagic vesicles in H4 cells (Fig. 5 A, 4 h and 9 h). More importantly, the treatment of oz-apraA alone did not increase the levels of LC3-II (Fig. 5 B). In fact, as predicted by the microarray analysis (Fig. S1 B, 6-h treatment), oz-apraA inhibited autophagy, as evident by the reduced basal levels of LC3-II in HeLa cells (Fig. 5 B, lane 3). Furthermore, pretreatment with oz-apraA abrogated rapamycin-induced LC3-II increases (Fig. 5 B, lane 4), suggesting that oz-apraA inhibits macroautophagy. We further confirmed that oz-apraA–induced degradation of EGFR is not through macroautophagy by using Atg5-deficient mouse embryonic fibroblast (MEF) cells. Atg5 is essential for autophagosome formation, and Atg5 deficiency blocks macroautophagy (Kuma et al., 2004). Oz-apraA induced degradation of EGFR as efficiently in Atg5−/− MEFs in an NH4Cl−-sensitive but MG132-insensitive manner as that in the Atg5+/+ MEFs (Fig. 5 C, left).
Figure 5.
Oz-apraA does not induce Hsp90 client protein degradation through macroautophagy. (A) Stable GFP-LC3–expressed H4 cells were treated with 100 nM oz-apraA for 4 or 9 h or with 218 nM rapamycin for 4 h. Bar, 10 µm. (B) Oz-apraA does not increase the levels of LC3-II. HeLa cells were treated with 200 nM rapamycin or 100 nM oz-apraA for 3 h except that in lane 4, cells were pretreated with 100 nM oz-apraA for 1 h followed by 200 nM rapamycin for an additional 3 h. The endogenous LC3-II levels were detected with anti-LC3 antibody and quantified relative to α-tubulin. (C) Atg5 wild-type (+/+) or knockout (−/−) MEFs were treated with 100 nM oz-apraA, 5 µM MG132, and/or 20 mM NH4Cl as indicated for 24 h. Protein levels were detected with anti-EGFR and anti–α-tubulin antibodies.
Finally, we found that oz-apraA did not increase the amount of acidic lysosomes, whereas the positive control, nigericin, could significantly increase the numbers of acidic lysosome (Fig. S4 B). Thus, oz-apraA does not act by stimulation of lysosomal activity per se. Collectively, these findings indicate that it is unlikely that oz-apraA–induced degradation of Hsp90 client proteins is mediated through the proteasome pathway, macroautophagy, or by increasing number of lysosomes.
We also monitored the endocytosis of rhodamine-labeled EGF upon treatment with oz-apraA and GA (Fig. S4 C), and neither of them affected EGF internalization. Thus, we further excluded a possible effect of oz-apraA on endocytosis.
Oz-apraA induces the degradation of Hsp90 client proteins through CMA
Because bio–oz-apraA was able to bind Hsp70/Hsc70, we next examined the interaction of Hsp70/Hsc70 with EGFR in cells treated with oz-apraA. Interestingly, although the total levels of Hsp70/Hsc70 in oz-apraA–treated cells were not increased as dramatically as in GA-treated cells, an increased association of Hsp70/Hsc70 with EGFR or ErbB2 was observed after oz-apraA treatment (Fig. 6, A and B).
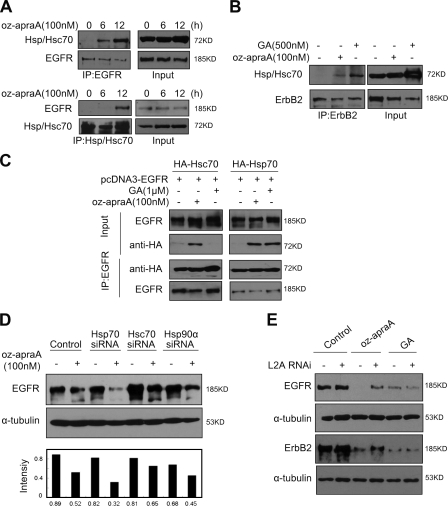
Figure 6.
Oz-apraA induces Hsp90 client membrane protein degradation through CMA. (A) Interaction of EGFR and Hsp/Hsc70 after treatment with oz-apraA. (B) Oz-apraA and GA both increase the interaction of ErbB2 and Hsp/Hsc70. (C) Oz-apraA but not GA induces the interaction between Hsc70 and EGFR. HEK293 cells were transfected with 2 µg pcDNA3-EGFR together with 2 µg pcDNA-HA-Hsc70 or pcDNA-HA-Hsp70 for 24 h followed by treatment with 100 nM oz-apraA or 1 µM GA for an additional 12 h. The cells were lysed, and the lysates were immunoprecipitated with anti-EGFR antibody followed by immunoblotting with anti-HA antibody to detect the interaction of EGFR with Hsc70 or Hsp70. (D) Hsc70 RNAi could partially inhibit oz-apraA–induced EGFR degradation. HeLa cells were transfected with siRNA specifically against Hsc70, Hsp70, or Hsp90 for 48 h followed by treatment with or without 100 nM oz-apraA for an additional 12 h. EGFR levels were quantified to that of α-tubulin. (E) LAMP-2A was required for oz-apraA–induced Hsp90 client protein degradation. A549 cells (top panels) or MDA-MB-453 cells (bottom panels) were transiently transfected with pSuper-LAMP-2A for 48 h and treated with 100 nM oz-apraA or 500 nM GA for 24 h. IP, immunoprecipitation; L2A, LAMP-2A.
Because our antibody does not distinguish Hsp70 or Hsc70, we asked whether oz-apraA can differentiate between Hsp70 and Hsc70 using transgenes. We transfected HEK293 cells with pcDNA-HA-Hsc70 or pcDNA-HA-Hsp70 together with pcDNA3-EGFR. As shown in Fig. 6 C, oz-apraA induced interaction of EGFR with both Hsc70 and Hsp70, whereas GA could only induce interaction of EGFR with Hsp70, which is consistent with a previous study (Doong et al., 2003). However, knocking down the expression of Hsc70 but not Hsp70 or Hsp90 partially abrogated oz-apraA–induced EGFR degradation (Fig. 7 D and Fig. S5 A), suggesting that the interaction of oz-apraA with Hsc70 but not Hsp70 may play an important functional role in promoting the degradation of EGFR.
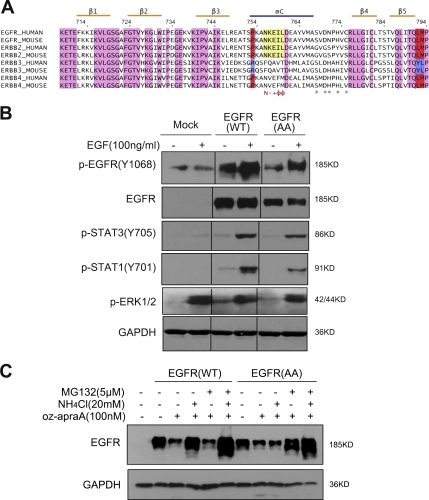
Figure 7.
KFERQ motif is required for EGFR degradation through CMA upon oz-apraA treatment. (A) Sequence alignment of the EGFR family members from human and mouse. The αC helix contains a 756NKEIL760 pentapeptide sequence (yellow box), which is known as a KFERQ motif recognized by CMA. Pink regions indicate the homogeneity of the amino acid between EGFR family proteins, and purple and red regions indicate the discrepancy between ErbB3 and other members. N, Asn; −, basic amino acid; +, acidic amino acid; Φ, hydrophobic amino acid; *, Hsp90-binding sites. (B) HEK293 cells were transfected with wild-type (WT) EGFR or EGFR(AA) for 24 h and stimulated with 100 ng/ml EGF for 5 min. The levels of selected EGFR downstream protein phosphorylation were detected using specific anti–p-STAT1, anti–p-STAT3, and anti–p-EGFR(Y1068) antibodies. Anti-GAPDH was used as a loading control. Black lines indicate that intervening lanes have been spliced out. (C) The treatment of oz-apraA induces the degradation of EGFR(AA) through MG132-sensitive proteasome pathway. HEK293 cells were transfected with wild-type EGFR or EGFR(AA) for 24 h and treated with 100 nM oz-apraA, 5 µM MG132, or 20 mM NH4Cl for an additional 24 h. The cells were lysed, and the degradation of EGFR was analyzed using anti-EGFR by Western blotting. Anti–α-tubulin was used as a loading control.
Because Hsc70 has been recognized as an important chaperone in mediating CMA (Bandyopadhyay and Cuervo, 2008), the requirement of Hsc70 suggests the possibility that oz-apraA might promote the degradation of Hsp90 client proteins through CMA. Because LAMP-2A recognition is a rate-limiting step in CMA (Kiffin et al., 2004), we further determined whether inhibition of LAMP-2A receptor expression could abrogate the oz-apraA–induced degradation of Hsp90 client proteins. Knockdown of LAMP-2A by vector-mediated RNAi significantly inhibited oz-apraA–induced EGFR and/or ErbB2 degradation in different cell lines, whereas GA-induced ErbB2 and/or EGFR degradation was not affected (Fig. 6 E and Fig. S5 B). These findings suggest that oz-apraA modulates Hsp90 chaperone complexes and enhances the interaction of Hsp90 client proteins and Hsc70, which can then be recognized by LAMP-2A for degradation via CMA.
CMA substrate KFERQ motif is required for oz-apraA–induced EGFR degradation
Cytosolic Hsc70 recognizes a peptide sequence that includes the KFERQ-like motif in its substrate proteins (Chiang et al., 1989). We noted that the EGFR sequence contains a pentapeptide sequence (756NKEIL760) that is consistent with a CMA recognition motif (Dice, 2007), and this pentapeptide sequence is highly conserved between EGFR and ErbB2 in human and mouse, located in the αC helix of the kinase domain (Fig. 7 A). We asked whether this motif was required for oz-apraA–induced EGFR degradation through the lysosomal pathway. We generated a mutant EGFR construct expressing a mutated EGFR with the sequence 756NKEIL760 replaced by 756AAEIL760, termed EGFR(AA). Although endogenous EGFR in HEK293 cells was barely detectable, transiently transfected wild-type and EGFR(AA) expression constructs were expressed at comparable levels (Fig. 7 B). However, the mutant of EGFR does not affect EGF-induced downstream signaling transduction (Fig. 7 B, EGFR and STAT [signal transducer and activator of transcription] phosphorylation) and/or EGF-stimulated EGFR endocytotic degradation (Fig. S4 D).
Mutation of the KFERQ motif impairs the association of mutant proteins with lysosomal membrane and reduces their translocation into the lysosomal lumen (Cuervo et al., 2004). Interestingly, we found that in oz-apraA–treated cells, although the levels of wild-type EGFR were resistant to the effect of MG132 and sensitive to NH4Cl, the levels of EGFR(AA) became sensitive to MG132 but resistant to NH4Cl (Fig. 7 C). From these results, we conclude that the pentapeptide sequence (756NKEIL760) of EGFR is required for the oz-apraA–induced EGFR degradation through CMA. The requirement for the NKEIL polypeptide sequence also supports the involvement of CMA in oz-apraA–induced EGFR degradation rather than other lysosomal-mediated processes such as microautophagy.
Discussion
Hsp90 has emerged as an exciting molecular target for cancer therapy. Promoting the degradation of Hsp90 client proteins, many of which are important positive regulators of cell growth and proliferation, provides the key underlying mechanism for the success of anti-Hsp90 therapy. We demonstrate in this study that oz-apraA also specifically and potently induces the degradation of at least a subset of Hsp90 client proteins. Although most of the known inhibitors of Hsp90 function by interrupting the intrinsic ATPase activity of Hsp90 and promoting the degradation of the client proteins via the ubiquitin-proteasome pathway, we show that oz-apraA induces the degradation of Hsp90 client proteins through CMA, a lysosomal degradation mechanism distinct from proteasome-dependent mechanisms. Our study provides the first example for the ability of CMA to mediate the degradation of membrane receptors and the cross talk of CMA and proteasomal degradation mechanisms.
The Hsp90 chaperone–client protein cycle involves successive association and dissociation of several cochaperones to form various multimeric protein complexes and is dictated by the ATP-binding state of Hsp90. A client protein initially associates with an Hsp70–Hsp40 complex and is then bound to Hsp90 via HOP (Hsp90/Hsp70-organizing protein). A conformational change in Hsp90, induced by ATP hydrolysis, leads to the release of Hsp70–Hsp40 and HOP and association of other cochaperones to form a mature complex (Sharp and Workman, 2006). Increased association of Hsp70 with Hsp90 client proteins in cells treated with GA might reflect an inability to release the cochaperones from Hsp90 client proteins. By the same analogy, the increased association of not only Hsp70 but also Hsc70 with Hsp90 client proteins in cells treated with oz-apraA suggests that at least a subpopulation of Hsp90 client proteins might regularly interact with Hsc70.
Although Hsc70 and Hsp70 are highly homologous (86% identity in amino acid sequence), only Hsc70 but not Hsp70 has been implicated in mediating CMA as a molecular chaperone that stimulates cargo protein translocation across membrane (Bandyopadhyay and Cuervo, 2008). Hsc70 is constitutively expressed and has also been found to be a part of Hsp90 complex and interact with HOP (Gebauer et al., 1997). Interestingly, Hsp90 megacomplexes, including Hsp90, Hsp40, HOP, Hsp70-interacting protein, and BAG-1 (Bcl-2–associated athanogene 1 protein), were found to be associated with the cytosolic side of the lysosomal membrane in cells under prolonged starvation condition (Agarraberes and Dice, 2001). Thus, the cells may target the Hsp90 client proteins for degradation through lysosomal pathway under nutrient-limiting conditions.
A previous study has reported that the increase in total proteolysis induced by GA is reduced by the treatment of NH4Cl but not by 3-methyladenine, which inhibits macroautophagy but not CMA, which has led Finn et al. (2005) to suggest that GA also stimulates CMA. However, because a relatively high concentration of GA (2 µM) was used by Finn et al. (2005), the apparent induction of CMA might be a result of toxicity rather than a direct result of inhibiting Hsp90. Because the degradation of a vast majority of Hsp90 client proteins induced by GA depends on the proteasomal pathway (Zhang and Burrows, 2004) and because we found that GA does not induce the association of Hsc70 and Hsp90 client proteins, we propose that GA does not directly promote CMA.
Another recent study has reported that the GA-induced degradation of IKK, which is also an Hsp90 client protein, is mediated by the macroautophagic pathway (Qing et al., 2006). In this case, the degradation of IKK was found to require Atg5, a gene required for macroautophagy but not for CMA. However, because a high concentration of 10 µM GA was used in this study, it is also possible that the toxicity of GA activates macroautophagy, which in turn promotes the degradation of IKK.
The EGFR family of receptors (EGFR [ErbB1], ErbB2, ErbB3, and ErbB4) are important regulators of normal growth and differentiation. Antagonizing the functions of EGFR family of receptors has been an important therapeutic strategy for the treatment of cancers. Oz-apraA–induced EGFR degradation requires the 756NKEIL760 peptide for degradation through CMA. Interestingly, in a genomic screen for somatic cancer mutations from a subset of 58 nonsmall cell lung cancer samples that included 41 lung adenocarcinomas, Paez et al. (2004) identified 10 tumor samples that carried small deletions in EGFR for codons 746–750 (Del-1) and one tumor sample that carried a deletion of 752–759 (Del-2; Table S3), which deleted most of the CMA recognition sequence identified by us. Although the potential significance of such deletions for the degradation of EGFR through CMA remains to be examined in future studies, we propose that degradation of EGFR through CMA might serve as a tumor suppressor mechanism by down-regulating EGFR levels.
Both GA and oz-apraA can induce the down-regulation of EGFR protein levels but through distinct mechanisms. Such down-regulation of EGFR may occur during the folding and ER-Golgi transport process before localization on the plasma membrane. Misfolded membrane proteins are known to be retro transported from the ER to the cytosol through ER-associated degradation and degraded through proteasomal pathway (Strous and Gent, 2002). The ability of oz-apraA to induce the appearance of aggresomes without impairing the function of proteasomes is consistent with a significant increase in the retro transport of misfolded proteins from the ER to the cytosol through ER-associated degradation. Alternatively, oz-apraA–induced down-regulation of EGFR may occur after ligand binding and receptor activation during receptor recycling and degradation process, which is mediated through clathrin-coated endocytic vesicles and subsequent fusion with early endosomes (Roepstorff et al., 2008). The ability of oz-apraA to dramatically stimulate the polyubiquitination of EGFR in an NH4Cl−-sensitive manner is also consistent with the possibility that oz-apraA may be able to promote the polyubiquitination and degradation of ligand-bound EGFR. The exact form of EGFR that is degraded after oz-apraA treatment needs to be further characterized.
The existence of cross talks between different forms of autophagy pathways has been proposed by Massey et al. (2006a). It was reported that the cells with impaired CMA function were able to maintain homeostatic protein degradation through up-regulation of macroautophagy (Massey et al., 2006b). Thus, inhibition of macroautophagy by oz-apraA at later time points may also contribute to further induction of CMA. Although the molecular mechanism that mediates the cross talk of macroautophagy and CMA is not yet clear, Hsc70, a cytosolic chaperone essential for translocation of substrates via CMA, has been proposed as a good candidate for acting as a cross-talking molecule. Because experimental loading of CMA-incompetent lysosomes with Hsc70 is sufficient to activate CMA capability, increased expression or localization of Hsc70 within a particular subcellular area may activate CMA. We propose that an increased interaction of EGFR with Hsc70 induced by oz-apraA is sufficient to promote the degradation of EGFR through the CMA.
Materials and methods
Cells and reagents
A549, MDA-MB-453, HEK293, SKoV3, and H4 cells were cultured in DME containing 10% FCS (Hyclone). The sources of chemicals, peptides, and antibodies were as follows: GA and MG132 (EMD), rapamycin and nigericin (BIOMOL International L.P.), ammonium chloride (Shanghai Reagents, Inc.), recombinant human EGF (Invitrogen), DAPI (Sigma-Aldrich), lysotracker (Invitrogen), secondary HRP-conjugated antibodies (Jackson ImmunoResearch Laboratories), secondary FITC- and TRITC-conjugated antibodies (Shanghai Kangcheng), mouse anti–α-tubulin (Santa Cruz Biotechnology, Inc.), rabbit anti-EGFR (Cell Signaling Technology), mouse anti-Hsp70/Hsc70 (Santa Cruz Biotechnology, Inc.), rabbit anti-GP130 (Santa Cruz Biotechnology, Inc.), mouse antiubiquitin (Cell Signaling Technology), rabbit anti-LC3 (Novus Biologicals), mouse anti-HA (Tiangen), rabbit anti-ErbB2 (Cell Signaling Technology), mouse anti-Hsp90 (BD), mouse anti-RIP1 (BD), rabbit antiphospho-EGFR (Y1068; Invitrogen), rabbit antiphospho-STAT1 (Y701; Cell Signaling Technology), mouse antiphospho-STAT3 (Y705; Cell Signaling Technology), rabbit antiphospho-ERK1/2 (T202/Y204; Cell Signaling Technology), and mouse anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Shanghai Kangcheng).
Immunofluorescence analysis
Cells were fixed with 3.8% formaldehyde for 10 min at RT. After washing with PBS, cells were permeabilized with 0.1% Triton X-100 for 10 min at RT and rinsed with PBS three times. Samples were treated with 5% BSA-containing PBS for 1 h, incubated first with primary antibodies and then with FITC- or TRITC-conjugated secondary antibodies, and images of immunolabeled samples were obtained at RT with a laser-scanning confocal microscope (TCS SP2; Leica) using a Plan Apo 100× 1.40 or 63× NA 1.40 objective lens and argon ion (488 nm) and HeNe (543 nm) lasers. Images were acquired sequentially using confocal software (Leica); for double labeling experiments, images of the same confocal plane were generated and superimposed. Tagged image file format images were processed using Photoshop software (version 7.0; Adobe).
Gene expression signature analysis
Apratoxin A gene expression raw data were obtained from the National Center for Biotechnology Information’s Gene Expression Omnibus (accession no. GSE2742; http://www.ncbi.nlm.nih.gov/geo/). Raw data were processed by robust multiarray analysis and normalized using quantile normalization. For marker gene selection, we used the signal to noise statistic to rank the genes that correlated with apratoxin A–treated versus vehicle-treated cell distinction. Signal to noise = (µ0 − µ1)/(σ0 + σ1), where µ and σ represent the mean and standard deviation of the expression, respectively, for each class with performing 1,000 permutation (Golub et al., 1999). The top 30 class neighbor marker genes were used as the signatures of apratoxin A. The enrichment of these signatures in the gene expression profiles of the Connectivity Map database was assessed using a gene-enrichment metric, the connectivity score, based on the Kolmogorov-Smirnov statistic (Lamb et al., 2006).
Plasmids, RNAi, and transfection
The expression construct of full-length human EGFR in pcDNA3 was provided by X. Fu (Indiana University, Bloomington, IN). EGFR αC helix 756N and 757K point mutation (NK→AA) were constructed by the QuickChange PCR amplification method using appropriately designed oligonucleotide primers (Agilent Technologies). The site-directed mutagenesis primer sequence was 5′-AGAAGCAACATCTCCGAAAGCCGCCGCGGAAATCCTCGATGAAGCCTAC-3′. pSuper–LAMP-2A RNAi plasmid was provided by A.M. Cuervo (Albert Einstein College of Medicine, New York, NY). Chemically synthetic siRNA sequences were listed as follows (Dickey et al., 2007): Hsc70 siRNA, 5′-CAGCACGGAAAAGUCGAGA-3′; Hsp70 siRNA, 5′-UCCUGUGUUUGCAAUGUUGAA-3′; Hsp90 siRNA, 5′-UCCCGACGAUAUUACUAAUGA-3′.
Coimmunoprecipitation
Cells were harvested on ice for 30 min using the MNX buffer containing 25 mM MES, pH 6.5, 1% Triton X-100, 150 mM NaCl, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, and a complete protease inhibitor cocktail (Sigma-Aldrich). For the transient transfection experiments, cells were first transfected with pcDNA-HA-Hsp70 or pcDNA-HA-Hsc70 (provided by G. Kudla, University of Edinburgh, Edinburgh, Scotland, UK) for 24 h, and cell lysates were precleared with protein A/G plus agarose (Santa Cruz Biotechnology, Inc.) for 1 h at 4°C followed by incubating with each primary antibody at 4°C overnight with gentle agitation and with protein A/G plus agarose for anther 2 h. After agarose pellets had been washed five times with MNX buffer, bound proteins were extracted with Laemmli 2× sample buffer (Sigma-Aldrich) by heating at 100°C for 10 min and subjected to Western blotting.
In vitro Hsp90-binding assay
1 × 107 SKoV3 cells were harvested and lysed by MNX buffer on ice for 30 min. After centrifugation at 4°C (12,000 g) for 10 min, supernant was aliquoted into three tubes. Lysates were treated with or without 100 nM oz-apraA and1 µM GA at 4°C for 2 h followed by immunoprecipitation of ErbB2. ErbB2-bound Hsp90 were detected using Western blotting.
Bio–oz-apraA pull-down assay
1 × 108 HeLa cells were harvested and lysed by hypotonic buffer containing 20 mM Hepes, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 0.01% TritonX-100, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, and a complete protease inhibitor cocktail (Sigma-Aldrich; Wang et al., 2007). After centrifugation at 4°C (12,000 g) for 10 min, supernatant was aliquoted into three tubes. Lysates were incubated with 2 µM bio–oz-aprA at 4°C with agitation overnight, and streptavidin-agarose was added to pull-down bio–oz-apraA protein complex by incubating at 4°C for 3 h. After brief centrifugation, pellets were boiled in 2× SDS loading buffer for 5 min, samples were separated by SDS-PAGE, and pulled-down proteins were identified by silver staining.
Chemical data
The following data describe the results of nuclear magnetic resonance of bio–oz-apraA: 1H nuclear magnetic resonance (500 MHz, CDCl3) δ 7.13 (m, 2H), 6.78 (m, 2H), 6.49 (d, J = 7.8 Hz, 1H), 6.19 (d, J = 9.5 Hz, 1H), 5.82 (brs, 1H), 5.30 (m, 1H), 5.21 (d, J = 11.7 Hz, 1H), 5.07 (m, 1H), 4.96 (d, J = 12.6 Hz, 1H), 4.88 (m, 1H), 4.77 (m, 1H), 4.65 (d, J = 10.9 Hz, 1H), 4.52 (m, 1H), 4.36–4.26 (m, 4H), 4.25–4.10 (m, 3H), 3.91 (t, J = 6.6 Hz, 2H), 3.66 (m, 2H), 3.29 (m, 1H), 3.25–3.14 (m, 4H), 3.12 (t, J = 11.7 Hz, 1H), 3.04 (m, 1H), 2.93 (dd, J = 4.9, 12.8 Hz, 1H), 2.84–2.81 (m, 3H), 2.81–2.67 (m, 6H), 2.63 (s, 1H), 2.42 (m, 1H), 2.33 (m, 1H), 2.28–2.11 (m, 4H), 2.11–1.99 (m, 2H), 1.96–1.84 (m, 6H), 1.74 (m, 6H), 1.66 (m, 6H), 1.46 (m, 6H), 1.39–1.29 (m, 8H), 1.29–1.24 (m, 4H), 1.22 (m, 4H), 1.11–1.03 (m, 5H), 1.02–0.91 (m, 8H), 0.91–0.79 (m, 10H); ESIMS m/z 1329 [M+H]+. High-resolution mass spectrometry calculated for C71H114N11O11S+ [M+H]+, which requires 1328.8415, found 1328.8457.
Online supplemental material
Fig. S1 shows that apratoxin A and its analogue oz-apraA induce degradation of Hsp90 client proteins. Fig. S2 shows that oz-apraA disrupts the interaction of Hsp90 and Hsp70/Hsc70 at a later time point (8 h). Fig. S3 shows that oz-apraA induces aggresome formation in HeLa cells. Fig. S4 shows that oz-apraA–mediated Hsp90 client degradation is not through endocytosis and/or through inducing lysosomes. Fig. S5 shows the efficiency of siRNA knockdown on different proteins. Table S1 shows class neighbor gene signatures of apratoxin A for 3-h treatment. Table S2 shows class neighbor gene signatures of apratoxin A for 6-h treatment. Table S3 shows the Del-2 somatic mutation in human nonsmall cell lung cancers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810183/DC1.
Acknowledgments
We thank Profs. Ana Maria Cuervo, Xinyuan Fu, Takashi Nonaka (Tokyo Institute of Psychiatry, Tokyo, Japan), and Grzegorz Kudla for shRNA vector for LAMP-2A, expression vectors of EGFR, GFP-CL1, and pcDNA-HA-Hsp70/pcDNA-HA-Hsc70, respectively.
This work was supported in part by the China National Science and Technology 973 grant (2004CB518903), China National Natural Science Foundation grants (30672481 and 30771097 to Q. Yu), China National Science Foundation grants (20632050 and 20621062 to D. Ma), a merit award from the U.S. National Institute on Aging (to J. Yuan), the National Basic Research Program of China grant (2006CB910700), and the PROTEOMAGE Project (FP6) of the European Union (to R. Zeng).
Footnotes
Abbreviations used in this paper: bio–oz-apraA, biotin derivative of oz-apraA; CHX, cycloheximide; CMA, chaperone-mediated autophagy; EGFR, EGF receptor; GA, geldanamycin; MEF, mouse embryonic fibroblast; oz-apraA, oxazoline analogue of apratoxin A.
References
- Agarraberes F.A., Dice J.F. 2001. A molecular chaperone complex at the lysosomal membrane is required for protein translocation.J. Cell Sci. 114:2491–2499 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U., Cuervo A.M. 2008. Entering the lysosome through a transient gate by chaperone-mediated autophagy.Autophagy. 4:1101–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation.Science. 292:1552–1555 [DOI] [PubMed] [Google Scholar]
- Chiang H.L., Terlecky S.R., Plant C.P., Dice J.F. 1989. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins.Science. 246:382–385 [DOI] [PubMed] [Google Scholar]
- Citri A., Harari D., Shohat G., Ramakrishnan P., Gan J., Lavi S., Eisenstein M., Kimchi A., Wallach D., Pietrokovski S., Yarden Y. 2006. Hsp90 recognizes a common surface on client kinases.J. Biol. Chem. 281:14361–14369 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. 2004. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy.Science. 305:1292–1295 [DOI] [PubMed] [Google Scholar]
- Dice J.F. 2007. Chaperone-mediated autophagy.Autophagy. 3:295–299 [DOI] [PubMed] [Google Scholar]
- Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C.B., et al. 2007. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins.J. Clin. Invest. 117:648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong H., Rizzo K., Fang S., Kulpa V., Weissman A.M., Kohn E.C. 2003. CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins.J. Biol. Chem. 278:28490–28500 [DOI] [PubMed] [Google Scholar]
- Finn P.F., Mesires N.T., Vine M., Dice J.F. 2005. Effects of small molecules on chaperone-mediated autophagy.Autophagy. 1:141–145 [DOI] [PubMed] [Google Scholar]
- Gebauer M., Zeiner M., Gehring U. 1997. Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity.FEBS Lett. 417:109–113 [DOI] [PubMed] [Google Scholar]
- Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., et al. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring.Science. 286:531–537 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing.EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Kim T., Ko Y.G., Rho S.B., Park S.G., Kim M.J., Kwon H.J., Kim S. 2000. Heat shock protein 90 mediates protein-protein interactions between human aminoacyl-tRNA synthetases.J. Biol. Chem. 275:31682–31688 [DOI] [PubMed] [Google Scholar]
- Kiffin R., Christian C., Knecht E., Cuervo A.M. 2004. Activation of chaperone-mediated autophagy during oxidative stress.Mol. Biol. Cell. 15:4829–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. 2004. The role of autophagy during the early neonatal starvation period.Nature. 432:1032–1036 [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease.Science. 313:1929–1935 [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy.Dev. Cell. 6:463–477 [DOI] [PubMed] [Google Scholar]
- Luesch H., Yoshida W.Y., Moore R.E., Paul V.J., Corbett T.H. 2001. Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula.J. Am. Chem. Soc. 123:5418–5423 [DOI] [PubMed] [Google Scholar]
- Luesch H., Chanda S.K., Raya R.M., DeJesus P.D., Orth A.P., Walker J.R., Izpisua Belmonte J.C., Schultz P.G. 2006. A functional genomics approach to the mode of action of apratoxin A.Nat. Chem. Biol. 2:158–167 [DOI] [PubMed] [Google Scholar]
- Ma D., Zou B., Cai G., Hu X., Liu J.O. 2006. Total synthesis of the cyclodepsipeptide apratoxin A and its analogues and assessment of their biological activities.Chemistry. 12:7615–7626 [DOI] [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Cuervo A.M. 2006a. Lysosomal chat maintains the balance.Autophagy. 2:325–327 [DOI] [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. 2006b. Consequences of the selective blockage of chaperone-mediated autophagy.Proc. Natl. Acad. Sci. USA. 103:5805–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A.J. 2003. Amino acids as regulators and components of nonproteinogenic pathways.J. Nutr. 133:2057S–2062S [DOI] [PubMed] [Google Scholar]
- Meyer U., Schweim P., Fracella F., Rensing L. 1995. Close correlation between heat shock response and cytotoxicity in Neurospora crassa treated with aliphatic alcohols and phenols.Appl. Environ. Microbiol. 61:979–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy.Science. 304:1497–1500 [DOI] [PubMed] [Google Scholar]
- Peng X., Guo X., Borkan S.C., Bharti A., Kuramochi Y., Calderwood S., Sawyer D.B. 2005. Heat shock protein 90 stabilization of ErbB2 expression is disrupted by ATP depletion in myocytes.J. Biol. Chem. 280:13148–13152 [DOI] [PubMed] [Google Scholar]
- Politz J.C., Yarovoi S., Kilroy S.M., Gowda K., Zwieb C., Pederson T. 2000. Signal recognition particle components in the nucleolus.Proc. Natl. Acad. Sci. USA. 97:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G., Yan P., Xiao G. 2006. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IkappaB kinase (IKK).Cell Res. 16:895–901 [DOI] [PubMed] [Google Scholar]
- Roepstorff K., Grovdal L., Grandal M., Lerdrup M., van Deurs B. 2008. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer.Histochem. Cell Biol. 129:563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffonati L., Tiberio L. 1997. Gene expression in liver after toxic injury: analysis of heat shock response and oxidative stress-inducible genes.Liver. 17:183–191 [DOI] [PubMed] [Google Scholar]
- Sharp S., Workman P. 2006. Inhibitors of the HSP90 molecular chaperone: current status.Adv. Cancer Res. 95:323–348 [DOI] [PubMed] [Google Scholar]
- Strous G.J., Gent J. 2002. Dimerization, ubiquitylation and endocytosis go together in growth hormone receptor function.FEBS Lett. 529:102–109 [DOI] [PubMed] [Google Scholar]
- Wang G., Shang L., Burgett A.W., Harran P.G., Wang X. 2007. Diazonamide toxins reveal an unexpected function for ornithine delta-amino transferase in mitotic cell division.Proc. Natl. Acad. Sci. USA. 104:2068–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L. 2005. HSP90 and the chaperoning of cancer.Nat. Rev. Cancer. 5:761–772 [DOI] [PubMed] [Google Scholar]
- Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu.Proc. Natl. Acad. Sci. USA. 99:12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Burrows F. 2004. Targeting multiple signal transduction pathways through inhibition of Hsp90.J. Mol. Med. 82:488–499 [DOI] [PubMed] [Google Scholar]