Abstract
Bone morphogenic protein (BMP) signaling is essential for the coordinated assembly of the synapse, but we know little about how BMP signaling is modulated in neurons. Our findings indicate that the Nemo (Nmo) kinase modulates BMP signaling in motor neurons. nmo mutants show synaptic structural defects at the Drosophila melanogaster larval neuromuscular junction, and providing Nmo in motor neurons rescues these defects. We show that Nmo and the BMP transcription factor Mad can be coimmunoprecipitated and find a genetic interaction between nmo and Mad mutants. Moreover, we demonstrate that Nmo is required for normal distribution and accumulation of phosphorylated Mad in motor neurons. Finally, our results indicate that Nmo phosphorylation of Mad at its N terminus, distinct from the BMP phosphorylation site, is required for normal function of Mad. Based on our findings, we propose a model in which phosphorylation of Mad by Nmo ensures normal accumulation and distribution of Mad and thereby fine tunes BMP signaling in motor neurons.
Introduction
The ability of neurons to remodel and modify their synaptic connections is a fundamental requirement for higher brain functions such as learning and memory. Defects in this form of synaptic plasticity have been associated with the pathophysiological manifestations of many neurodegenerative diseases and mental illnesses (Zoghbi, 2003; Levitt et al., 2004). Increasing evidence suggests that signaling cascades initiated by secreted molecules such as bone morphogenic protein (BMP), Wnt/wingless (Wg), and FGF are essential for the coordinated assembly of the synapse (Packard et al., 2002; McCabe et al., 2003; Waites et al., 2005; Hodge et al., 2007). Despite recent progress, we know little about how these signaling cascades are modulated in neurons to ensure appropriate synaptic growth. In this study, we describe a mechanism by which BMP signaling is modulated through interaction between the Nemo (Nmo) kinase and the BMP transcription factor Mad in Drosophila melanogaster larval motor neurons.
In vertebrates, BMP signaling has been shown to control different aspects of neuronal development both in the spinal cord and the brain (Liu and Niswander, 2005). The role for BMP signaling in regulating synaptic growth has been well characterized at the Drosophila larval neuromuscular junction (NMJ). Based on the current evidence, the BMP ligand Glass Bottom Boat provides a retrograde signal from the muscle back to the nerve terminal; this signal is processed by a receptor complex comprised of the type I BMP receptors Thickveins (Tkv) and Saxophone and the type II BMP receptor Wishful thinking. Receptor activation then leads to an increase in the phosphorylation of the R-Smad Mad at the NMJ terminals followed by nuclear translocation of phosphorylated Mad (p-Mad) through its interaction with the co-Smad Medea. Mutations of the members of this cascade lead to a drastic reduction in the number of synaptic boutons and the amount of neurotransmitter release at the NMJ (Aberle et al., 2002; Marques et al., 2002; McCabe et al., 2003, 2004).
A large body of research has provided evidence for Smad regulation by several posttranslational mechanisms such as ubiquitination, phosphorylation, and sumoylation, to name a few (Ross and Hill, 2008); however, we know little about how Smads are regulated in neurons. In a forward genetic suppressor screen, we have identified Nmo kinase as a regulator of synaptic growth at the Drosophila larval NMJ. Our findings provide evidence that the normal function of Mad depends on its phosphorylation by Nmo. Interaction between Nmo and Mad provides a mechanism for the modulation of BMP signaling in motor neurons and thus the structural growth and function of synaptic boutons at the NMJ.
Results
Nmo is required in motor neurons for normal synaptic growth at the larval NMJ
Loss-of-function mutations in the E3 ubiquitin ligase highwire (hiw) lead to extensive synaptic expansion at the NMJ (Wan et al., 2000). We reasoned that if a candidate gene could dominantly suppress the synaptic expansion in hiw mutants, it would likely be involved in the regulation of synaptic growth. To search for such candidate genes, we tested whether inclusion of a chromosome containing deletions (deficiency chromosome) could dominantly suppress the synaptic expansion in hiw mutants. We focused on chromosomal deficiency lines available on the left arm of the third chromosome and found that hiw larvae with one copy of Df(3L)pbl-X1 showed significantly less synaptic expansion compared with hiw larvae. We tested genetic interaction with smaller overlapping deficiencies and were able to identify the gene nmo (Choi and Benzer, 1994) as the locus responsible (see Materials and methods; Fig. S1).
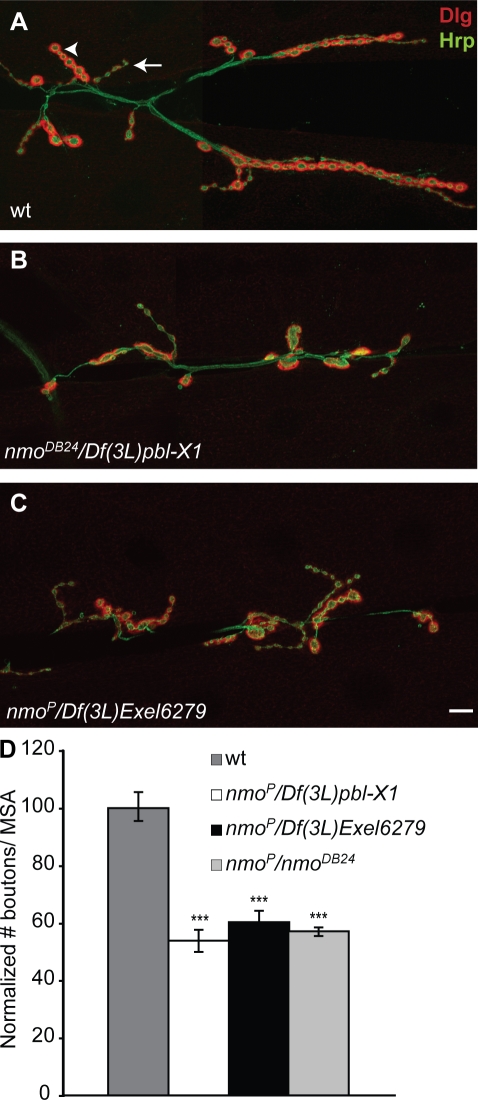
Nmo is the founding member of the evolutionarily conserved Nmo-like kinase family of serine/threonine kinases (Choi and Benzer, 1994). Nmo was first identified as a regulator of epithelial planar cell polarity during eye development (Choi and Benzer, 1994). Moreover, Nmo has been implicated in several developmental processes, including the regulation of patterning and imaginal disc development (Zheng et al., 1995; Verheyen et al., 2001), but its function in the nervous system has not been characterized. To study the role of Nmo in synaptic growth, we examined NMJs from wandering third instar larvae using antibodies against pre- and postsynaptic markers. NMJs undergo rapid structural and functional growth in the few days of larval development; during this time, the number of synaptic boutons increases several fold to keep up with the growing muscle (Schuster et al., 1996; Keshishian, 2002; Davis, 2006). This coordinated growth appeared defective in nmo mutants (Fig. 1); however, we did not observe any detectable abnormalities in the pattern of innervation of the body wall muscles in nmo mutant larvae (not depicted), suggesting that motor neuronal pathfinding and initial target recognition are not affected. A feature of wild-type NMJs is the presence of chains of synaptic boutons organized like beads on a string (Budnik et al., 1996; Zito et al., 1999) forming synaptic branches that contain as many as 20 boutons (Fig. 1 A). However, in nmo mutants, this feature was severely affected, as most NMJs lacked branches containing more than five boutons (Fig. 1, B and C). Different genetically null alleles of nmo in trans-allelic and homozygous combinations showed a strong reduction in the number of type I synaptic boutons (86.5 ± 3.0 and 54.5 ± 3.6 for wild type and nmoP/Df(3L)Exel6279, respectively; n = 20; P < 0.001) at muscles 6 and 7. Type I glutamatergic boutons have been divided into two categories, big (type Ib) and small (type Is), based on their size as well as on the intensity of their Discs large (Dlg) signal (Fig. 1 A, arrowhead and arrow; Budnik et al., 1996). We found that both Ib and Is boutons were reduced in nmo mutants. We observed a similar reduction in the number of boutons at muscle 4 NMJs (58.9 ± 2.6 and 35.3 ± 3.3 for wild type and nmoP/Df(3L)Exel6279, respectively; n = 20; P < 0.001). To accurately reflect the role of Nmo in coordinated synaptic growth, we normalized the total number of boutons at every NMJ to the corresponding muscle surface area (MSA). This approach avoids any biases that might arise as the result of variations in the muscle size among different larvae. Our quantifications showed that in nmo mutants, the number of boutons per MSA was reduced by ∼40% (Fig. 1 D).
Figure 1.
Nmo is required for normal synaptic growth at the NMJ. (A) Wild-type (wt) third instar larval NMJ synapses at muscles 6 and 7 in the third abdominal segment double stained with anti-Dlg (red) and anti-HRP (green). The arrow indicates a type Is bouton; the arrowhead indicates a type Ib bouton. (B and C) NMJs at muscles 6 and 7 in two different nmo mutant combinations. (D) Bar graph showing the normalized total number of boutons/MSA in wild type and in three nmo mutant combinations (n ≥ 20; mean ± SEM; ***, P < 0.001). Bar, 10 µm.
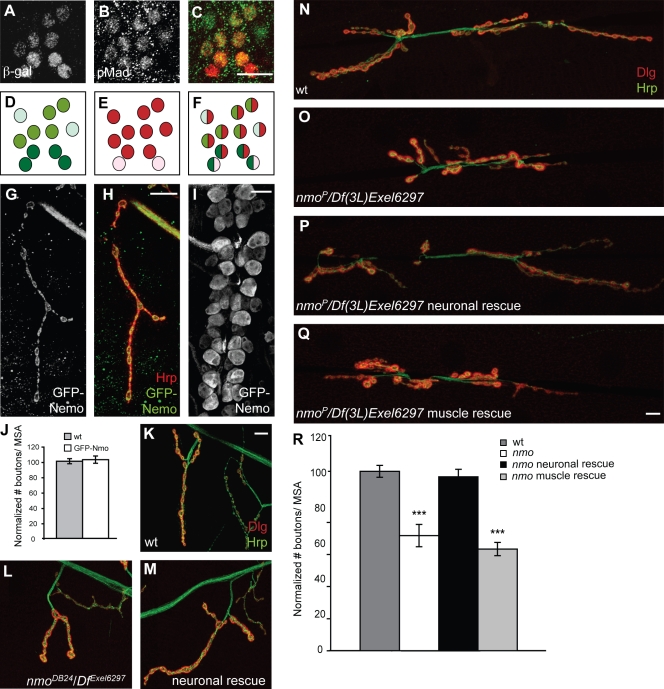
In situ hybridization experiments have previously demonstrated an enrichment of nmo transcript in the ventral nerve cord and in the brain at late embryonic stages (Verheyen et al., 2001). To further test whether the nmo transcript is present in motor neurons, we took advantage of a nmo LacZ enhancer trap fly (nmoP); expression of β-galactosidase (β-gal) from nmoP closely resembles the endogenous expression pattern of nmo (Choi and Benzer, 1994; Zheng et al., 1995; Zeng and Verheyen, 2004). We were able to detect a specific anti–β-gal signal in a subset of neurons, including all motor neurons, in the ventral nerve cord. To confirm the identity of the β-gal–positive neurons in nmoP, we costained the ventral nerve cord with an antibody against p-Mad; p-Mad normally accumulates in the nuclei of motor neurons (Marques et al., 2002; McCabe et al., 2003). We found that all p-Mad–positive motor neurons were also positive for β-gal (Fig. 2, A–F). To further assess the expression pattern of the nmo transcript, we generated larvae that contained one copy of nmoP enhancer trap while expressing upstream activating sequence (UAS)–mCD8-GFP in all motor neurons using motor neuronal Gal4 driver BG380; double staining of ventral nerve cords with anti-GFP and anti–β-gal antibodies revealed that the nmo transcript is not exclusively expressed in motor neurons (unpublished data). Next, we wished to examine the pattern of expression of Nmo protein; however, the available Nmo antibodies failed to detect a specific signal. Thus, we turned to a GFP-Nmo fusion transgene (UAS-GFP-Nmo; Fiehler and Wolff, 2008) to investigate whether Nmo can localize at synaptic sites. We detected GFP signal at all NMJ synapses after overexpression of UAS-GFP-Nmo using BG380-Gal4 (Fig. 2, G and H), suggesting that Nmo protein can localize to the NMJ. In addition, we found that GFP-Nmo was evenly distributed in motor neuronal cell bodies and nuclei without any apparent concentration in any compartments (Fig. 2 I). We did not find a significant change in the number of boutons per MSA in response to Nmo overexpression (Fig. 2 J).
Figure 2.
Nmo is required in motor neurons for normal synaptic growth at the NMJ. (A) β-gal staining in the ventral nerve cord of larvae heterozygous for nmoP. (B) β-gal–positive neurons are also p-Mad positive. (C) Merged image of A and B; p-Mad is shown in green, and β-gal is shown in red. (D–F) Graphic representation of signal intensity in individual motor neurons corresponding to A–C. (G) The transgenic GFP-Nmo fusion protein localizes to the presynaptic domain of the NMJ when expressed in motor neurons. (H) The same NMJ is stained with an anti-HRP antibody, and the merged image is shown. (I) Overexpressed GFP-Nmo also localizes to motor neuronal cell bodies. (J) Bar graph showing the normalized total number of type I boutons per MSA in wild type (wt) and larvae overexpressing GFP-Nmo in motor neurons (n ≥ 20; mean ± SEM). (K–M) Larval NMJ synapses at muscle 4 double stained with anti-Dlg (red) and anti-HRP (green) antibodies in wild type (K), nmo mutant (L), and nmo mutant overexpressing GFP-Nmo in motor neurons (M). (N–Q) Larval NMJ synapses at muscles 6 and 7 in wild type (N), nmo mutant (O), nmo mutant overexpressing Nmo in motor neurons (P), or nmo mutant overexpressing Nmo in muscles (Q). (R) Bar graph showing the normalized total number of type I boutons per MSA in the genotypes shown in N–Q (n = 20; mean ± SEM; ***, P < 0.001). Bars, 10 µm.
To determine whether Nmo is required pre- or postsynaptically, we performed genetic rescue experiments in nmo mutant larvae. We found that motor neuronal overexpression of UAS-Nmo or UAS-GFP-Nmo but not muscle overexpression was able to rescue the reduction in the number of boutons per MSA in nmo mutants (NMJs at muscles 6 and 7, Fig. 2, N–R; NMJs at muscle 4, Fig. 2 K–M). The presence of nmo transcription in motor neurons, the ability of Nmo protein to localize to presynaptic structures, and the rescue of synaptic defects in nmo mutants by presynaptic Nmo together led us to conclude that Nmo is required in presynaptic motor neurons for normal growth of NMJ synaptic structures.
Nmo binds Mad and is required for normal accumulation of p-Mad in motor neurons
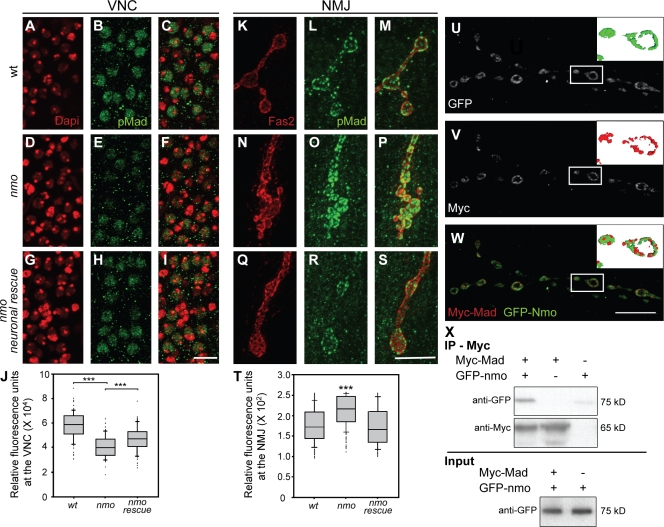
Our findings indicated that genetic reduction of nmo in motor neurons can significantly suppress NMJ overexpansion in hiw mutants (Fig. S1, A–D). Previously, it has been shown that loss of Wallenda (Wnd), a MAPK kinase kinase, can lead to suppression of the hiw loss-of-function phenotype (Collins et al., 2006). Wnd levels are up-regulated in the central nervous system of hiw mutant larvae (Collins et al., 2006). Therefore, we tested whether Wnd protein levels in the central nervous system are affected as a consequence of loss of nmo. We found no detectable changes in the levels of anti-Wnd antibody signal associated with loss of nmo (Fig. S1, E–I). Synaptic overexpansion in hiw mutants is also dependent on normal BMP signaling in motor neurons (McCabe et al., 2004). Moreover, it has recently been demonstrated that Nmo can interact with and phosphorylate the BMP transcription factor Mad in vitro (Zeng et al., 2007). Therefore, we examined whether Nmo participates in the regulation of BMP signaling in motor neurons. As a first step, we set out to test whether the normal accumulation of p-Mad is affected in nmo mutants. p-Mad accumulation at the terminals and in the nuclei of motor neurons is an indicator of retrograde BMP signaling at the NMJ (Marques et al., 2002; McCabe et al., 2003). Using an anti–p-Mad antibody, we were able to detect p-Mad in the nuclei of motor neurons in wild-type and nmo mutants (Fig. 3, A–F); however, our quantification of the fluorescence intensity associated with the p-Mad signal in the ventral nerve cord showed an ∼30% (P < 0.001) decrease in nmo mutants compared with wild type (Fig. 3, A–F and J). In contrast, we found a strong increase in the p-Mad signal at the NMJs. We quantified this increase in two ways. First, we measured the intensity of the signal per individual p-Mad punctum between the two groups and found that nmo mutants show a 25% increase compared with wild type (P < 0.001; Fig. 3 K–P and T). Second, we measured the total amount of fluorescence per synaptic area for p-Mad and normalized it to the amount of fluorescence for Fas2 in the same synaptic area between the two groups; using this measurement, we found that p-Mad signal was increased by 35% in nmo mutants compared with wild type (n = 16; P < 0.001). The p-Mad signal in nmo mutants was restored to normal levels when we provided Nmo in motor neurons (Fig. 3, G–J and Q–T). To rule out the possibility that the abnormal accumulation of p-Mad is merely caused by a general defect in axonal transport in nmo mutants, we examined motor axons in nmo and wild-type larvae after overexpression of UAS-Mito-GFP (Cox and Spradling, 2003). UAS-Mito-GFP is a marker for mitochondria and can be used in live larvae to assess retrograde and anterograde axonal transport (Pilling et al., 2006). We did not find any abnormalities or apparent differences in the accumulation of Mito-GFP puncta in nmo mutant larvae (Fig. S2, A–E). Similarly, we did not observe any apparent differences in the accumulation of synaptotagmin (Syt; Fig. S2, F–N), Bruchpilot, or Fas2 in axons of nmo mutant larvae (not depicted). These results suggest that Nmo specifically influences p-Mad distribution and accumulation in motor neurons.
Figure 3.
Nmo is required for the normal accumulation of p-Mad in motor neurons, and it colocalizes and coimmunoprecipitates with Mad. (A–I) Motor neuron nuclei in third instar larval ventral nerve cords double stained with DAPI (red) and p-Mad (green) in wild-type (wt; A–C), nmo mutant (D–F), and nmo mutant larva overexpressing UAS-Nmo in motor neurons (G–I). (K–S) Terminal boutons at muscle 4 NMJs double stained with anti-Fas2 (red) and anti–p-Mad (green) antibodies in wild-type (K–M), nmo mutant (N–P), and nmo mutant larva overexpressing the Nmo transgene in motor neurons (Q–S). (J and T) Box plots showing the total fluorescence corresponding to the p-Mad signal in motor neuron nuclei (J) or the mean intensity of p-Mad puncta at the NMJ (T) for the aforementioned genotypes. n ≥ 80 motor neurons in J, and n ≥ 175 synaptic boutons in T from at least 10 larvae (mean ± SEM; ***, P < 0.001). (U–W) Synaptic boutons stained with anti-GFP (green; U) and anti-Myc (red; V) and the merged image (W) are shown. Insets show partial 3D reconstructions of a synaptic bouton. (X) Western blots showing the presence of GFP-Nmo after immunoprecipitation (IP) of Myc-Mad using an anti-Myc antibody. VNC, ventral nerve cord. Bars, 10 µm.
Nmo and Mad have been shown to interact in vitro (Zeng et al., 2007); therefore, we wished to verify whether Nmo and Mad also interact in larval tissue. In the absence of reliable anti-Mad and anti-Nmo antibodies, we decided to use tagged transgenes of Mad and Nmo. For this, we generated an Myc-tagged Mad transgene and coexpressed it together with GFP-Nmo in all motor neurons. We found a significant overlap between the signals associated with Myc-Mad and GFP-Nmo at the NMJ (Fig. 3, U–W). In addition, we used Myc-Mad together with GFP-Nmo in coimmunoprecipitation experiments. We found that, in preparations which were immunoprecipitated with the anti-Myc antibody, we could also detect the presence of Nmo using an anti-GFP antibody (Fig. 3 X), suggesting that Mad and Nmo can be present in the same protein complex.
Phosphorylation of Mad by Nmo is required for the normal function of Mad
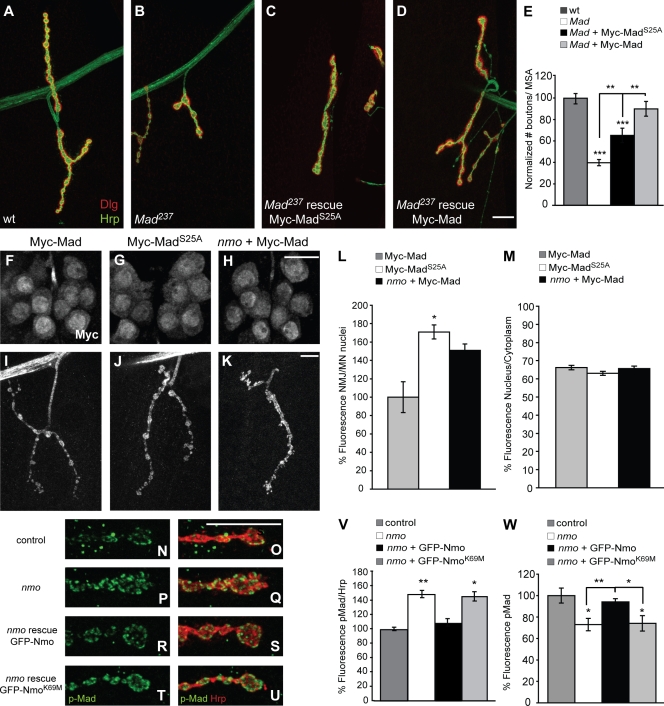
Next, we investigated whether phosphorylation of Mad by Nmo is required for Mad's normal physiological function in motor neurons. Nmo can specifically phosphorylate Mad in vitro at serine 25, a site distinct from the phosphorylation site by type I BMP receptors at the C terminus of Mad (ten Dijke and Hill, 2004; Zeng et al., 2007). If Nmo indeed phosphorylates Mad in vivo, and if this phosphorylation is physiologically relevant, its loss should affect the ability of Mad to function normally in the nervous system. We tested this hypothesis by performing rescue experiments in Mad mutants using a Mad transgene that cannot be phosphorylated by Nmo (UAS-Myc-MadS25A). Neuronal overexpression of UAS-Myc-Mad was able to restore the number of boutons per MSA in Mad mutants; however, UAS-Myc-MadS25A rescued Mad mutants only partially (Fig. 4, A–E). We verified that Myc-Mad and Myc-MadS25A showed similar levels of expression in motor neurons by Western blotting (Fig. S3). Protein levels were quantified by densitometry, and comparable protein levels were found.
Figure 4.
Nmo phosphorylation of Mad is required for its normal function and distribution in motor neurons. (A–D) Larval NMJs at muscle 4 double stained with anti-Dlg (red) and anti-HRP (green) in wild type (wt; A), homozygous Mad237 (B), and Mad237 rescued by UAS-Myc-MadS25A (C) or UAS-Myc-Mad (D). (E) Bar graph showing the normalized total number of boutons per MSA in the genotypes shown in A–D (n ≥ 14; mean ± SEM; **, P < 0.01; ***, P < 0.001). (F–K) Ventral nerve cords (F–H) and NMJs (I–K) stained with anti-Myc in wild-type larvae overexpressing UAS-Myc-Mad (F and I) or UAS-Myc-MadS25A (G and J) and nmo mutant larvae overexpressing UAS-Myc-Mad (H and K). (L) Bar graph showing the normalized index of mean fluorescence intensity for the Myc signal at the NMJ versus motor neuron nuclei for Myc-Mad, Myc-MadS25A, and Myc-Mad in nmo mutants (n = 16 larvae and two NMJs per larva; mean ± SEM; *, P < 0.05). (M) Bar graph showing the nuclear versus cytoplasmic Myc signal from wild-type larvae overexpressing Myc-Mad or Myc-MadS25A and from nmo mutants overexpressing Myc-Mad using BG380-Gal4 (n ≥ 40 from five larvae; mean ± SEM). (N–U) Terminal boutons at muscle 4 NMJs double stained with anti–p-Mad (green) and anti-HRP (red) in wild type (N and O), nmo mutant (nmoDB24/Df(3L)Exel6279; P and Q), nmo mutant larva overexpressing GFP-Nmo transgene in motor neurons (R and S), and nmo mutant larva overexpressing GFP-NmoK69M in motor neurons (T and U). (V) Bar graph showing p-Mad fluorescence relative to HRP for the genotypes in N–U (n = 8 larvae; mean ± SEM; *, P < 0.05; **, P < 0.01). (W) Bar graph showing p-Mad fluorescence intensity in the nuclei of dorsal motor neurons for the genotypes in N–U (n > 200 motor neurons from 8–10 larvae; mean ± SEM; *, P < 0.05; **, P < 0.01). Bars, 10 µm.
We next analyzed the localization of these two transgenes using immunohistochemistry in larval preparations. We found that both Myc-Mad and Myc-MadS25A were detectable in motor neuronal nuclei and at synaptic terminals (Fig. 4, F–K). However, closer analysis of the Myc signal revealed a proportional increase at the NMJ versus motor neuronal nuclei for Myc-MadS25A compared with Myc-Mad. For this, we generated an index representing the Myc signal at the NMJ in relation to the Myc signal in the nuclei of motor neurons for each larva. Using confocal projections, we measured the amount of fluorescence signal at the NMJ per area and divided that by the amount of fluorescence per area obtained from motor neuronal nuclei for each larva (see Materials and methods). On average, larvae expressing Myc-MadS25A showed a >50% increase in this index compared with larvae that expressed Myc-Mad (P < 0.05; n = 12; Fig. 4 L), which is reminiscent of the distribution of p-Mad in nmo mutants (Fig. 3). Then, we tested the effect of loss of nmo on this distribution index and found that loss of nmo led to an ∼45% increase in the index with a p-value of 0.0504 (n = 12; Fig. 4 L).
Because Nmo has been shown to induce nuclear export of Mad in heterologous cells, we wished to examine the role of Nmo in inducing nuclear export of Mad in motor neurons. For this, we quantified the amount of fluorescence in the nuclei of motor neurons expressing Myc-Mad and divided that by the amount of fluorescence obtained from the entire cell body in each motor neuron in the presence or absence of nmo; we found no significant difference between the nuclear fraction of Myc signal between wild-type and nmo larvae (Fig. 4 M). Similarly, the nuclear fraction of Myc signal in larvae overexpressing UAS-Myc-MadS25A was not different from that of wild-type UAS-Myc-Mad (Fig. 4 M). These findings are consistent with our earlier observations of p-Mad distribution in nmo mutant larvae and suggest that phosphorylation of Mad at serine 25 by Nmo is not essential for normal nuclear export of Mad in motor neurons. Nevertheless, these results do not rule out the possibility that Nmo can induce nuclear export of Mad when overexpressed, as observed previously by Zeng et al. (2007).
Finally, we tested the importance of Nmo's kinase activity in the regulation of p-Mad accumulation and distribution in motor neurons. We generated a transgenic Nmo carrying a point mutation in a critical residue in the catalytic domain of Nmo, UAS-GFP-NmoK69M. When overexpressed in motor neurons, GFP-NmoK69M localized to NMJ terminals in a manner similar to GFP-Nmo but caused a mild reduction in the number of boutons (Fig. S4, A–E), suggesting that it perhaps acts as a dominant negative. We tested whether NmoK69M can rescue the abnormal p-Mad distribution in motor neurons in nmo mutants, and, not surprisingly, we found that it failed to restore p-Mad to normal levels (Fig. 4, N–W; and Fig. S4, F–I). These findings are consistent with a model in which Nmo phosphorylation of Mad at serine 25 influences the distribution and accumulation of Mad in motor neurons and is required for Mad's normal function in promoting synaptic growth.
nmo and Mad genetically interact
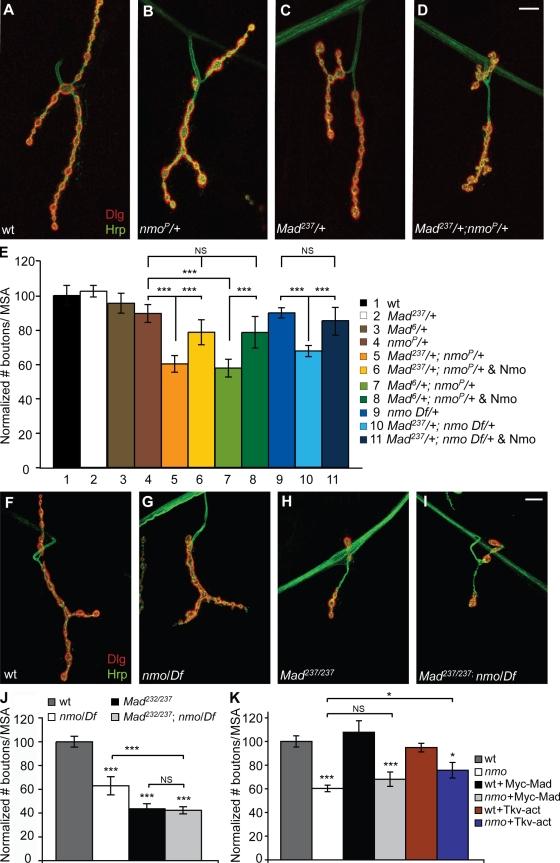
The effect of nmo loss-of-function on p-Mad distribution as well as the biochemical interaction between Mad and Nmo prompted us to test whether nmo and Mad could genetically interact. To test this possibility, we asked whether Mad and nmo would show trans-heterozygous genetic interaction. We found that synaptic structures showed significant abnormalities when one copy of nmo and one copy of Mad were genetically removed (Fig. 5, A–E), whereas loss of one copy of either was not significantly different from wild type. The number of type I boutons per MSA in Mad+/−;nmo+/− trans-heterozygous larvae was reduced by >30% compared with heterozygous control larvae (Fig. 5 E). This trans-heterozygous interaction was rescued by neuronal overexpression of Nmo in different trans-allelic combinations (Fig. 5 E). To further examine the functional interaction between Mad and Nmo, we tested synaptic structures in Mad;nmo double mutants. We found that the number of boutons per MSA in these larvae was statistically similar to that in Mad mutants alone but significantly lower from that in nmo mutants (Fig. 5, F–J). These results suggest that Mad and Nmo most likely function together in the same pathway to control synaptic growth at the NMJ. These results also raise the possibility that an increase in BMP signaling may be capable of restoring synaptic defects in nmo mutants. We tested this hypothesis by overexpressing either UAS-Myc-Mad or a constitutively active form of the type I BMP receptor Tkv (UAS-Tkv-act; Hoodless et al., 1996) in motor neurons in nmo mutant larvae. UAS-Tkv-act has been shown to increase the amount of p-Mad in the nuclei of motor neurons (Collins et al., 2006) without affecting the number of boutons at the NMJ (McCabe et al., 2004). Overexpression of either UAS-Myc-Mad or UAS-Tkv-act did not affect the number of boutons per MSA (Fig. 5 K). Although overexpression of Mad did not significantly increase the number of boutons in nmo mutant larvae, overexpression of UAS-Tkv-act did so significantly (60.77 ± 2.6 for BG380;nmo−/− compared with 75.67 ± 6.6 for BG380;UAS-Tkv-act/+;nmo−/−; n = 12; P = 0.018; Fig. 5 K and Fig. S5, A–D). These results further support a model in which Nmo regulates synaptic structural growth at the NMJ via its interaction with BMP signaling in motor neurons.
Figure 5.
nmo and Mad genetically interact. (A–D) NMJs at muscle 4 double stained with anti-Dlg (red) and anti-HRP (green) in wild type (wt; A), heterozygous nmo mutants (B), heterozygous Mad mutants (C), and trans-heterozygous Mad−/+;nmo−/+ mutants (D). (E) Bar graph showing the normalized total number of boutons/MSA in wild type, heterozygous nmo and Mad mutants, trans-heterozygous Mad;nmo mutants, and trans-heterozygous Mad;nmo mutants overexpressing Nmo in motor neurons (n ≥ 12; mean ± SEM; ***, P < 0.001). (F–I) NMJs at muscle 4 double stained with anti-Dlg (red) and anti-HRP (green) in wild type (F), homozygous nmo mutants (G), homozygous Mad mutants (H), and Mad;nmo double mutants (I). (J) Bar graph showing the normalized total number of boutons/MSA for the genotypes in F–I (n ≥ 20; mean ± SEM; ***, P < 0.001). (K) Bar graph showing the normalized total number of boutons/MSA in wild-type and nmo mutant larvae as well as in wild-type or nmo mutant larvae overexpressing UAS-Myc-Mad or UAS-Tkv-act using BG380-Gal4 (n ≥ 10; mean ± SEM; *, P < 0.05; ***, P < 0.001). Df corresponds to Df(3L)Exel6279. Bars, 10 µm.
Nmo overexpression negatively regulates synaptic release
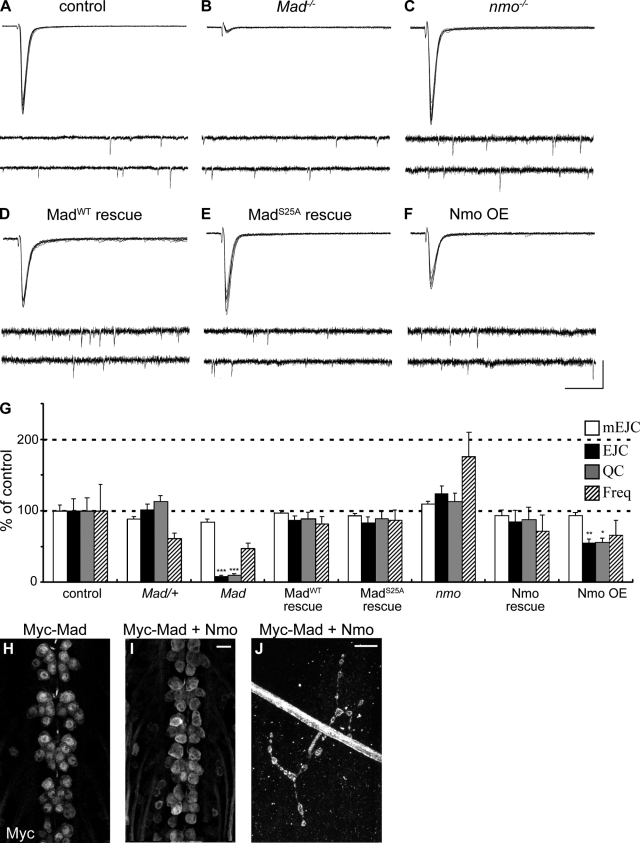
Loss of BMP signaling has a profound effect on neurotransmitter release at the NMJ (Aberle et al., 2002; McCabe et al., 2004). Based on our results, we predicted that loss of nmo would mimic partial loss of BMP signaling, leading to a decrease in quantal release. Using a standard two-electrode voltage-clamp technique, we evaluated electrophysiological properties of nmo mutant larvae by measuring postsynaptic evoked junctional currents (EJCs) as well as miniature EJCs (mEJCs). To our surprise, quantal content in nmo mutants appeared normal. Although EJCs, quantal content, and mEJC frequency showed an increase on average compared with wild type, we did not find any statistically significant difference between the two groups (quantal content, 25.92 ± 4.7 for control vs. 29.23 ± 3.25 for nmo; n = 12; P = 0.56; EJC, P = 0.25; frequency of mEJC, P = 0.16; Fig. 6, A, C, and G). In contrast, Mad mutants, as reported previously (McCabe et al., 2004), showed a severe reduction in quantal release compared with wild type (Fig. 6, B and G). These results suggested that the normal function of Mad in the regulation of synaptic strength is not dependent on Nmo. In support of this conclusion, both UAS-Myc-Mad and UAS-Myc-MadS25A were capable of restoring normal synaptic function in Mad mutant larvae (Fig. 6, D, E, and G). These results suggest that the regulation of synaptic structural growth and tuning of synaptic strength have differential requirements for Mad signaling in motor neurons.
Figure 6.
Loss of nmo does not lead to electrophysiological defects at the NMJ. (A–F) Representative traces of EJCs and mEJCs from control (A), Mad mutant (B), or nmo mutant larvae (C) or from Mad mutant larvae presynaptically expressing UAS-Myc-Mad (D) or UAS-Myc-MadS25A (E) and from larvae overexpressing UAS-GFP-Nmo (F). In each panel, the top traces show 10 superimposed EJCs, and the bottom traces show the continuous recording of spontaneous mEJCs without nerve stimulation. The horizontal scale indicates time (40 ms for EJCs and 400 ms for mEJCs), and the vertical scale indicates current (8 nA for EJCs and 2 nA for mEJCs). (G) Graph of mEJC amplitude, EJC amplitude, quantal content (QC), and frequency of mEJCs for the following genotypes: BG380/+ (control), BG380/+;Mad237/+ (Mad/+), BG380/+;Mad237/237 (Mad), BG380/+;Mad237/237;UAS-Myc-Mad (MadWT rescue), BG380/+;Mad237/237;UAS-Myc-MadS25A/+ (MadS25A rescue), BG380/+;nmoDB24/Df(3L)Exel6279 (nmo), BG380/+;nmoDB24, UAS-GFP-Nmo/Df(3L)Exel6279 (Nmo rescue), BG380/+;UAS-GFP-Nmo (Nmo OE; n ≥ 10 for all genotypes; mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (H and I) Dorsal motor neurons in the ventral nerve cords of larvae expressing UAS-Myc-Mad alone or UAS-Myc-Mad together with UAS-GFP-Nmo are shown. Note the change in the distribution of Myc signal. (J) An NMJ at muscle 4 for the same genotype as in I is shown; Myc-Mad accumulates normally at the NMJ. OE, overexpression. Bars, 10 µm.
To further explore the action of Nmo on the regulation of neurotransmitter release, we conducted quantal analysis of larvae overexpressing Nmo (Fig. 6, F and G). Our analysis of synaptic structures showed no significant change in the number of synaptic boutons in response to Nmo overexpression (Fig. 2 J); however, unexpectedly, we found a drastic reduction in both EJCs and quantal content in response to Nmo overexpression in motor neurons (quantal content, 25.92 ± 4.7 for control vs. 14.38 ± 1.49 for UAS-GFP-Nmo; n = 12; P = 0.012; EJC, P = 0.007; Fig. 6, F and G). To learn about the relationship between this negative regulation and Nmo's interaction with Mad, we evaluated the distribution of Myc-Mad in motor neurons in response to Nmo gain-of-function. We found that in these larvae, Myc-Mad was present in both motor neuronal cell bodies and nuclei as well as at the NMJ (Fig. 6, H–J); however, the nuclear distribution of Myc-Mad appeared severely affected. Although Myc-Mad normally shows a strong concentration in the nuclei of motor neurons, when overexpressed together with GFP-Nmo, the Myc signal appeared evenly distributed in the cell body to a point that most nuclei were not easily detectable (Fig. 6 I). These results reveal a potential role for Nmo in the negative regulation of Mad, a role which was not revealed through our analysis of nmo loss-of-function, and highlight the ability of Nmo to regulate synaptic structural growth and neurotransmitter release differentially.
Discussion
Our findings point to a model in which Nmo phosphorylation of Mad promotes its accumulation in the nuclei of motor neurons and thereby ensures effective BMP signaling at the NMJ. nmo mutant larvae show a significant aberration in the accumulation and/or distribution of p-Mad in motor neurons, with elevated levels of p-Mad at the NMJ and decreased levels of p-Mad in the nuclei of motor neurons. In addition, when Mad is mutated at its phosphorylation site for Nmo (MadS25A), it shows an expression pattern that qualitatively resembles that of p-Mad in nmo mutants, with more accumulation at the NMJ and less accumulation in the nucleus compared with wild-type Mad. Consistent with the importance of this phosphorylation, MadS25A fails to rescue synaptic structural defects in Mad mutants effectively. These observations suggest that phosphorylation of Mad by Nmo most likely modulates Mad's function by regulating its distribution and accumulation in motor neurons. Based on our findings, it is tempting to conclude that the reduction in the number of NMJ synaptic boutons in nmo mutants is, to a large extent, caused by the failure of p-Mad to signal to the nucleus effectively. In support of this, we reveal a strong trans-heterozygous interaction between nmo and Mad and show that synaptic defects in nmo mutants can be partially rescued by overexpression of a constitutively active form of Tkv (Tkv-act). Although our current findings provide strong support for this model, we cannot rule out the possibility that Nmo is involved in other processes that contribute to the growth of synaptic boutons at the NMJ.
We find that, in contrast to its critical role in the regulation of synaptic structure, Nmo does not play an important role in the regulation of synaptic function; in the absence of nmo, quantal content remains at normal levels. Consistently, we find that the MadS25A transgene is capable of rescuing the severe electrophysiological defects of Mad mutants as efficiently as a wild-type Mad transgene. Previously, it has been suggested that structural growth and the homeostasis of neurotransmitter release at the NMJ have different requirements for BMP signaling (Goold and Davis, 2007). Similarly, our findings highlight the differential requirements for the regulation of synaptic structure and synaptic strength via BMP signaling. Interestingly, although overexpression of Nmo in motor neurons does not influence synaptic structural growth, it does cause a significant reduction in neurotransmitter release. This observation is consistent with those made by Zeng et al. (2007), showing an antagonistic effect of Nmo gain-of-function on Mad in the wing imaginal discs. Nevertheless, although this observation shows a potential for Nmo to act as a negative regulator of Mad, our findings argue against a significant negative regulatory role for Nmo in motor neurons under normal physiological conditions.
We propose that Nmo exerts its action primarily by modulating Mad's retrograde movement from the NMJ to the nucleus. Nmo has been implicated in the regulation of Mad nuclear export in heterologous cells (Zeng et al., 2007); however, we find no evidence for changes in Mad nuclear export as a consequence of loss of nmo. In contrast, we find that Nmo is required for accumulation of p-Mad in the nuclei of motor neurons. In the absence of nmo, p-Mad levels increase at the NMJ and decrease in the nuclei of motor neurons, suggesting that Nmo is required for normal translocation/trafficking of Mad from the NMJ to the nucleus.
Finally, we show that, consistent with previous findings (Zeng et al., 2007), overexpression of Nmo can reduce the proportion of Mad concentration in nuclei versus that in cell bodies of motor neurons. Based on the phenotypic consequences of nmo loss- and gain-of-function, it appears that normal growth of synaptic structures at the NMJ depends on continuous and efficient BMP signaling from the NMJ to the nuclei of motor neurons and is less sensitive to the residence time of Mad in the nucleus. However, it appears that regulation of neurotransmitter release is more sensitive to the residence time of Mad in the nucleus and less dependent on the continuous retrograde signaling from the NMJ. Our findings highlight the importance of Nmo phosphorylation of Mad at serine 25 in this process; however, a comprehensive understanding of the regulation of Mad trafficking and movement dynamics in different cellular compartments will require future studies.
Finally, an intriguing possibility would be the involvement of the Wg pathway in the regulation of Mad dynamics through Nmo. Nmo has been implicated in the Wg pathway during wing development and has been shown to be a transcriptional target of Wg (Zeng and Verheyen, 2004). As Wg has been shown to participate in the regulation of synaptic growth at the NMJ (Packard et al., 2002), it would be tempting to envisage a role for Wg in the regulation of Nmo transcription in motor neurons and thus a link between the Wg and BMP pathways in the regulation of synaptic growth and function at the NMJ.
Materials and methods
Fly stocks
Flies were cultured on standard medium at 25°C. The following fly stocks were used: UAS-GFP-Nmo (gift from R. Fiehler and T. Wolff, Washington University, St. Louis, MO; Fiehler and Wolff, 2008), UAS-Mito-GFP (Cox and Spradling, 2003), UAS-mCD8-GFP (Lee and Luo, 1999), UAS-TkvQ199D (UAS-Tkv-act in this study; Hoodless et al., 1996), Df(3L)pbl-X1, Df(3L)pbl-NR, Df(3L)Exel6279 (Parks et al., 2004), MadK00237 (Mad237 in this study; Bach et al., 2003), Mad6 (Sekelsky et al., 1995), nmoP (Choi and Benzer, 1994), nmoDB24 (Zeng and Verheyen, 2004), and hiwEMS (Wan et al., 2000). Wild type refers to w1 or w1118 (Bloomington Stock Center). The neuronal Gal4 driver BG380 (Budnik et al., 1996) or OK6 and the muscle-specific Gal4 driver G14 (Aberle et al., 2002) were used.
hiw suppressor screen
To identify dominant suppressors of synaptic overgrowth in hiw mutants, we crossed hiwEMS recombined with Shaker-mCD8-GFP (Wan et al., 2000; McCabe et al., 2004) females to chromosomal deficiency lines available on the left arm of the third chromosome from the Bloomington Deficiency kit (Bloomington Stock Center). We examined the hemizygous male hiwEMS/Y;;Df/+ offsprings for reductions in NMJ boutons. We identified Df(3L)pbl-X1 as a dominant suppressor and subsequently found that the smaller deficiency's Df(3L)pbl-NR and Df(3L)Exel6279 also suppressed hiw. Among candidate genes uncovered by these deficiencies was nmo. We found that nmo mutants also suppressed hiw overgrowth; thus, we initiated our analysis of nmo.
Transgenic flies
For UAS-Myc-Mad, the full-length ORF of the Mad gene (FBgn0011648) was subcloned into a pUAST vector (Brand and Perrimon, 1993) containing six N-terminal Myc repeats (pUAST-Myc). For UAS-Myc-MadS25A, serine 25 was mutated to alanine by PCR using the primers 5′-TCACAGCGCCGGCGGTGAAGAAGCTGCTGGGCTGG-3′ and 5′-CCGGCGCTGTGAAGGAGAATAGCGAGCCCAGTGTG-3′, respectively, in combination with the XbaI-linked primer 5′-GGGGATCCTAATGGACACCGACGAT-3′ and the BamHI-linked primer 5′-GCTCTAGAGCTTAGGATACCGAACTAATTGCAT-3′. After an overlapping PCR, the assembled product was subcloned into pUAST-Myc. For UAS-GFP-NmoK69M, site-directed mutagenesis was performed on UAS-GFP-Nmo wild type using the primers 5′-GGACGACGAGTGGCCTTGATGAAGCTGCCAAATGTGTTCC-3′ and 5′-GGAACACATTTGGCAGCTTCATCAAGGCCACTCGTCGTCC-3′. Transgenic Mad lines were generated according to standard protocols (Spradling and Rubin, 1982), whereas the GFP-NmoK69M construct was injected by Best Gene, Inc.
Western blots and coimmunoprecipitation
Dissected third instar larvae were used for Western blotting as well as for immunoprecipitation experiments. Protein extraction was performed according to standard protocols. The following antibodies were used: mouse anti-GFP (1:500; Clontech Laboratories, Inc.), mouse anti–α-actin (1:2,000; Millipore), and mouse anti-Myc (1:200; Developmental Studies Hybridoma Bank). Peroxidase-conjugated secondary antibodies (1:5,000; Invitrogen) and ECL reagents (GE Healthcare) were used to detect the antigens. Densitometric analysis was performed using MetaMorph software (MDS Analytical Technologies). For immunoprecipitation experiments, protein from larvae overexpressing Myc-Mad was immunoprecipitated using an Anti–c-Myc Immunoprecipitation kit (Sigma-Aldrich). Immunoprecipitation was followed by Western blot analysis.
Immunochemistry
Wandering third instar larvae were dissected in cold HL3 (Stewart et al., 1994), fixed in 4% PFA (Sigma-Aldrich), incubated overnight with primary antibodies at 4°C, and incubated with fluorescence-conjugated secondary antibodies at room temperature. The following antibodies were used: rabbit polyclonal against GFP (1:500; A6455; Invitrogen), rabbit polyclonal against PS1 (1:500) and PS3 (1:200; p-Mad; provided by P. ten Dijke [Leiden University, Leiden, Netherlands] and C.H. Heldin [Ludwig Institute for Cancer Research, Stockholm, Sweden]; Tanimoto et al., 2000; Sun et al., 2007), mouse monoclonal against Dlg (1:500; provided by C. Goodman, University of California, San Francisco, San Francisco, CA; Parnas et al., 2001), rabbit anti-Syt (1:2,000; provided by H. Bellen, Baylor College of Medicine, Houston, TX; Littleton et al., 1993), rabbit anti-Wnd (1:300; provided by A. DiAntonio [Washington University, St. Louis, MO] and C. Collins [University of Michigan, Ann Arbor, MI]; Collins et al., 2006), mouse monoclonal against Fas2 (1:20; Schuster et al., 1996), and Cy5-conjugated goat against HRP (1:250; Jackson ImmunoResearch Laboratories). Antibodies obtained from the Developmental Studies Hybridoma Bank were mouse monoclonal against Myc (1:20; 9E10) and mouse monoclonal against β-gal (1:500; 40-1A). Secondary antibodies used were Alexa Fluor 488–conjugated goat against rabbit (1:500; Invitrogen) and Cy3-conjugated goat against mouse (1:500; GE Healthcare). DAPI was used at 1× (Sigma-Aldrich) together with the secondary antibodies. Samples were mounted in Vectashield (Vector Laboratories).
Confocal imaging and bouton counts
Synapses and ventral nerve cords were imaged using a ConfoCor LSM 510 META on an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.). Settings were optimized for detection without saturating the signal. Images were obtained at room temperature using a Plan-Apochromat 63× NA 1.4 immersion oil objective lens (Carl Zeiss, Inc.) and Immersol oil 518N (Carl Zeiss, Inc.). Images were cropped using Photoshop CS2 (Adobe).
Synaptic boutons were counted as previously described (Schuster et al., 1996) using the muscles of segment 3, with the following modifications: NMJs were costained with anti-Dlg and anti-HRP antibodies, and the counts were performed with a 63× NA 1.4 oil immersion objective using an epifluorescence microscope (Imager Z1; Carl Zeiss, Inc.).
Fluorescence quantification
Maximum projection images of NMJs and ventral nerve cords were generated from stacked confocal images (0.32 and 0.5 µm thickness, respectively). To compare intensities of p-Mad puncta at NMJs, synaptic boutons were manually traced using Fas2 staining. p-Mad immunofluorescence levels were quantified by measuring the intensity using the LSM5 META MK4 software (Carl Zeiss, Inc.). The peak signal intensities of p-Mad puncta, from ∼150 boutons per genotype, were then plotted and compared across genotypes. Alternatively, we quantified p-Mad at the NMJ by measuring the integrated fluorescence signal associated with p-Mad and normalizing it to the Fas2 signal at the same NMJ using MetaMorph software.
For Myc quantification at the NMJ, the integrated fluorescence signal per synaptic area was measured using the HRP signal to delineate synaptic boutons. To assess the fluorescence signals associated with p-Mad, β-gal, and Myc in the nuclei of motor neurons, a standard area of 25 µm2 per motor neuron nucleus was used for larval segments A5, A6, and A7. Signals were quantified with MetaMorph software.
To generate the relative fluorescence intensity between the NMJ and motor neuronal nuclei for Myc-Mad versus Myc-MadS25A, we divided the mean intensity per area, measured for at least 50 boutons, by the mean intensity per area for at least 30 motor neurons for each larva. To quantify Syt accumulation in axons, maximum projections of larval axons stained with anti-Syt were generated from stacked confocal images. MetaMorph software was used to determine the number of large Syt puncta (≥0.75 µm in diameter) present in each projection.
Electrophysiology
Wandering third instar larvae were dissected in cold HL3 solution according to standard protocol (Stewart et al., 1994). A standard two-electrode voltage-clamp technique was used to measure mEJC and EJC at muscle 6 in segment A3 in wandering third instar larvae using an amplifier (AxoClamp 2B; MDS Analytical Technologies). All recordings were performed at room temperature in HL3 with 0.5 mM external Ca2+, and muscles were held at −80 mV. The holding current was <3 nA.
Statistical analysis
Data are presented as mean values ± SEM (n = number of NMJs unless otherwise indicated), and Student's t tests were used to calculate statistical significance (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
Online supplemental material
Fig. S1 shows dominant suppression of hiw mutants by loss of one copy of nmo as well as staining for Wnd in the ventral nerve cord. Fig. S2 shows accumulation of Mito-GFP or Syt in axons. Fig. S3 shows a Western blot for Myc-Mad and Myc-MadS25A. Fig. S4 shows expression of NmoKD and its failure to rescue p-Mad abnormalities in nmo mutants. Fig. S5 shows the partial rescue of nmo by overexpression of Tkv-act. Onine supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200809127/DC1.
Acknowledgments
We would like to thank H. Bellen for the anti-Syt antibody, P. ten Dijke and C.H. Heldin for anti–p-Mad antibody, and A. DiAntonio and C. Collins for the anti-Wnd antibody. We also thank the Bloomington Stock Center for fly stocks and the Developmental Hybridoma Bank for monoclonal antibodies. We are grateful to R. Fiehler and T. Wolff for providing us with UAS-GFP-Nmo before publication. We also thank M. Rahnama for the generation of the UAS-GFP-NmoK69M transgenic fly, M. Warren-Paquin for her invaluable technical help with confocal microscopy, and R. Ball and E. Liao for their comments on the manuscript. We thank J. Young for technical advice. We would also like to thank all of the members of the Haghighi laboratory for their support, especially C. Schuetze for technical support. We are grateful to C. Goodman for his continuous support of our research program and for providing us with numerous reagents.
This work was supported by grants from the Canadian Institutes of Health Research, the EJLB Foundation, and Genome Canada to A.P. Haghighi. M.B. O'Connor is an investigator with Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper: β-gal, β-galactosidase; BMP, bone morphogenic protein; Dlg, Discs large; EJC, evoked junctional current; hiw, highwire; mEJC, miniature EJC; MSA, muscle surface area; NMJ, neuromuscular junction; Nmo, Nemo; p-Mad, phosphorylated Mad; Syt, synaptotagmin; Tkv, Thickveins; UAS, upstream activating sequence; Wg, wingless; Wnd, Wallenda.
References
- Aberle H., Haghighi A.P., Fetter R.D., McCabe B.D., Magalhaes T.R., Goodman C.S. 2002. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila.Neuron. 33:545–558 [DOI] [PubMed] [Google Scholar]
- Bach E.A., Vincent S., Zeidler M.P., Perrimon N. 2003. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway.Genetics. 165:1149–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes.Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Budnik V., Koh Y.H., Guan B., Hartmann B., Hough C., Woods D., Gorczyca M. 1996. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg.Neuron. 17:627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.W., Benzer S. 1994. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene.Cell. 78:125–136 [DOI] [PubMed] [Google Scholar]
- Collins C.A., Wairkar Y.P., Johnson S.L., DiAntonio A. 2006. Highwire restrains synaptic growth by attenuating a MAP kinase signal.Neuron. 51:57–69 [DOI] [PubMed] [Google Scholar]
- Cox R.T., Spradling A.C. 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during oogenesis.Development. 130:1579–1590 [DOI] [PubMed] [Google Scholar]
- Davis G.W. 2006. Homeostatic control of neural activity: from phenomenology to molecular design.Annu. Rev. Neurosci. 29:307–323 [DOI] [PubMed] [Google Scholar]
- Fiehler R.W., Wolff T. 2008. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation.Dev. Biol. 313:533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold C.P., Davis G.W. 2007. The BMP ligand Gbb gates the expression of synaptic homeostasis independent of synaptic growth control.Neuron. 56:109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge L.K., Klassen M.P., Han B.X., Yiu G., Hurrell J., Howell A., Rousseau G., Lemaigre F., Tessier-Lavigne M., Wang F. 2007. Retrograde BMP signaling regulates trigeminal sensory neuron identities and the formation of precise face maps.Neuron. 55:572–586 [DOI] [PubMed] [Google Scholar]
- Hoodless P.A., Haerry T., Abdollah S., Stapleton M., O'Connor M.B., Attisano L., Wrana J.L. 1996. MADR1, a MAD-related protein that functions in BMP2 signaling pathways.Cell. 85:489–500 [DOI] [PubMed] [Google Scholar]
- Keshishian H. 2002. Is synaptic homeostasis just wishful thinking? Neuron. 33:491–492 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis.Neuron. 22:451–461 [DOI] [PubMed] [Google Scholar]
- Levitt P., Eagleson K.L., Powell E.M. 2004. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders.Trends Neurosci. 27:400–406 [DOI] [PubMed] [Google Scholar]
- Littleton J.T., Stern M., Schulze K., Perin M., Bellen H.J. 1993. Mutational analysis of Drosophila synaptotagmin demonstrates its essential role in Ca(2+)-activated neurotransmitter release.Cell. 74:1125–1134 [DOI] [PubMed] [Google Scholar]
- Liu A., Niswander L.A. 2005. Bone morphogenetic protein signalling and vertebrate nervous system development.Nat. Rev. Neurosci. 6:945–954 [DOI] [PubMed] [Google Scholar]
- Marques G., Bao H., Haerry T.E., Shimell M.J., Duchek P., Zhang B., O'Connor M.B. 2002. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function.Neuron. 33:529–543 [DOI] [PubMed] [Google Scholar]
- McCabe B.D., Hom S., Aberle H., Fetter R.D., Marques G., Haerry T.E., Wan H., O'Connor M.B., Goodman C.S., Haghighi A.P. 2004. Highwire regulates presynaptic BMP signaling essential for synaptic growth.Neuron. 41:891–905 [DOI] [PubMed] [Google Scholar]
- McCabe B.D., Marques G., Haghighi A.P., Fetter R.D., Crotty M.L., Haerry T.E., Goodman C.S., O'Connor M.B. 2003. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction.Neuron. 39:241–254 [DOI] [PubMed] [Google Scholar]
- Packard M., Koo E.S., Gorczyca M., Sharpe J., Cumberledge S., Budnik V. 2002. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation.Cell. 111:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome.Nat. Genet. 36:288–292 [DOI] [PubMed] [Google Scholar]
- Parnas D., Haghighi A.P., Fetter R.D., Kim S.W., Goodman C.S. 2001. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix.Neuron. 32:415–424 [DOI] [PubMed] [Google Scholar]
- Pilling A.D., Horiuchi D., Lively C.M., Saxton W.M. 2006. Kinesin-1 and Dynein are primary motors for fast transport of mitochondria in Drosophila motor axons.Mol. Biol. Cell. 17:2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Hill C.S. 2008. How the Smads regulate transcription.Int. J. Biochem. Cell Biol. 40:383–408 [DOI] [PubMed] [Google Scholar]
- Schuster C.M., Davis G.W., Fetter R.D., Goodman C.S. 1996. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth.Neuron. 17:641–654 [DOI] [PubMed] [Google Scholar]
- Sekelsky J.J., Newfeld S.J., Raftery L.A., Chartoff E.H., Gelbart W.M. 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster.Genetics. 139:1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C., Rubin G.M. 1982. Transposition of cloned P-elements into Drosophila germline chromosomes.Science. 218:341–347 [DOI] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wnag J., Wu C.-F. 1994. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solution.J. Comp. Physiol. A: Neuroethol. Sens. Neural. Behav. Physiol. 175:179–191 [DOI] [PubMed] [Google Scholar]
- Sun M., Thomas M.J., Herder R., Bofenkamp M.L., Selleck S.B., O'Connor M.B. 2007. Presynaptic contributions of chordin to hippocampal plasticity and spatial learning.J. Neurosci. 27:7740–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H., Itoh S., ten Dijke P., Tabata T. 2000. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs.Mol. Cell. 5:59–71 [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Hill C.S. 2004. New insights into TGF-beta-Smad signalling.Trends Biochem. Sci. 29:265–273 [DOI] [PubMed] [Google Scholar]
- Verheyen E.M., Mirkovic I., MacLean S.J., Langmann C., Andrews B.C., MacKinnon C. 2001. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development.Mech. Dev. 101:119–132 [DOI] [PubMed] [Google Scholar]
- Waites C.L., Craig A.M., Garner C.C. 2005. Mechanisms of vertebrate synaptogenesis.Annu. Rev. Neurosci. 28:251–274 [DOI] [PubMed] [Google Scholar]
- Wan H.I., DiAntonio A., Fetter R.D., Bergstrom K., Strauss R., Goodman C.S. 2000. Highwire regulates synaptic growth in Drosophila.Neuron. 26:313–329 [DOI] [PubMed] [Google Scholar]
- Zeng Y.A., Verheyen E.M. 2004. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development.Development. 131:2911–2920 [DOI] [PubMed] [Google Scholar]
- Zeng Y.A., Rahnama M., Wang S., Sosu-Sedzorme W., Verheyen E.M. 2007. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation.Development. 134:2061–2071 [DOI] [PubMed] [Google Scholar]
- Zheng L., Zhang J., Carthew R.W. 1995. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye.Development. 121:3045–3055 [DOI] [PubMed] [Google Scholar]
- Zito K., Parnas D., Fetter R.D., Isacoff E.Y., Goodman C.S. 1999. Watching a synapse grow: noninvasive confocal imaging of synaptic growth in Drosophila.Neuron. 22:719–729 [DOI] [PubMed] [Google Scholar]
- Zoghbi H.Y. 2003. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 302:826–830 [DOI] [PubMed] [Google Scholar]