Abstract
Proper epithelial structure requires adherens junction (AJ) assembly. In the early Drosophila embryo, AJ assembly depends on Bazooka (Baz; PAR-3), but it is unclear how Baz affects AJ assembly and what precursors are involved. To understand this process at the molecular level, we counted the number of core AJ proteins and Baz proteins at an average spot AJ (SAJ) and determined their dynamics with fluorescence recovery after photobleaching experiments. These data reveal that SAJs are subdivided into Baz clusters and cadherin–catenin clusters with independent protein numbers and dynamics. This independence suggests that precursory cadherin–catenin clusters might form before SAJ assembly. We identify cadherin–catenin clusters forming between apical microvilli. Further analyses show that they form independently of Baz and that Baz functions in repositioning them to apicolateral sites for full SAJ assembly. Our data implicate cell protrusions in initial cadherin–catenin clustering in the Drosophila melanogaster embryo. Then, independent Baz clusters appear to engage the cadherin–catenin clusters to assemble SAJs.
Introduction
Epithelial tissue structure requires adherens junction (AJ) assembly from E-cadherin (Drosophila melanogaster E-cadherin [DE-cad]) in flies, β-catenin (Armadillo [Arm]), and α-catenin. AJ assembly typically begins with homophilic cis- and trans-cadherin clustering. This breaks the symmetric distribution of cadherin–catenin complexes on the plasma membrane (Tepass et al., 2001; Perez-Moreno et al., 2003; Gumbiner, 2005; Hartsock and Nelson, 2008). In cell culture, cell protrusions break this symmetry by sweeping E-cadherin and β-catenin into puncta at cell–cell contacts (Adams et al., 1996, 1998; Vasioukhin et al., 2000). Recruitment of actin regulators then reorganizes actin to form more stable contacts with expanded belt-like AJs (Drees et al., 2005; Yamada et al., 2005; Yamada and Nelson, 2007).
In Drosophila embryos, the first epithelium forms uniquely. Cells are forced into contact by cellularization, the simultaneous compartmentalization of ∼6,000 nuclei by plasma membranes invaginating from the embryo surface. Two types of junctions form: spot AJs (SAJs) in the apicolateral region and transient basal junctions (BJs) at the base of early invaginating membranes (Tepass et al., 2001; Lecuit, 2004). The polarity regulator Bazooka (Baz; PAR-3) is required for SAJ assembly next to centrosomes in a process involving dynein and microtubules (MTs; Harris and Peifer, 2004, 2005). Thus, Baz acts as part of an early AJ assembly landmark, but does Baz break a symmetric distribution of DE-cad/Arm, or are earlier clustering events involved? We used protein counting and FRAP experiments to define SAJ structure and live imaging and mutant analyses to dissect SAJ assembly.
Results and discussion
Baz and core AJ proteins have different numbers at SAJs
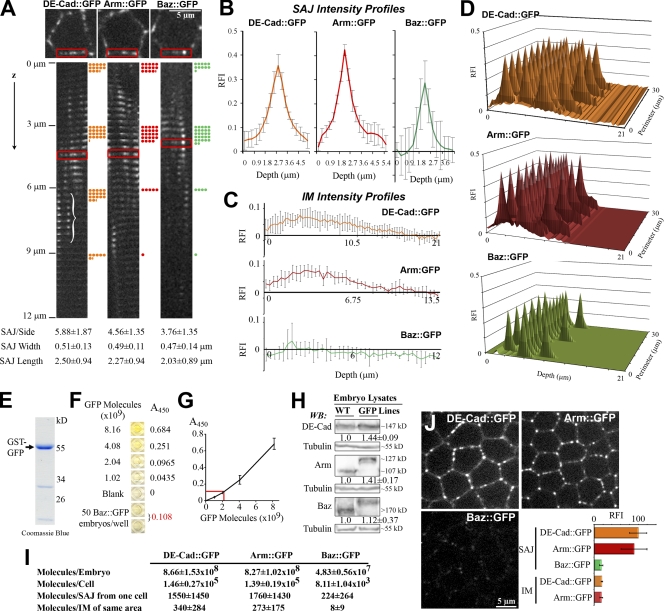
To count protein numbers at a late-cellularization SAJ, we analyzed stocks containing only GFP-tagged forms of DE-cad, Arm, and Baz. Two steps were taken. First, cortical localization maps were made for an average cell. Because the cells are simple hexagonal columns, we first made montages of x-y planes of single cell sides stacked in z (this imaged all SAJs even with membrane bending; Fig. 1 A, bracket). The montages provided SAJ apical–basal position (Fig. 1 A, dots), number, width, length (Fig. 1 A, bottom), and mean fluorescence intensity profiles (Fig. 1 B; see Materials and methods). The montages also provided whole cell height and perimeter plus mean intensity profiles for the intervening membrane (IM) surrounding SAJs (Fig. 1 C). These data were combined into 3D maps of SAJ and IM fluorescence intensities over an average cell cortex (Fig. 1 D). Each protein localized to similar numbers of SAJs in the top 9 µm of the cell.
Figure 1.
1:7:7 stoichiometry of Baz, DE-cad, and Arm at SAJs. (A–D) Mapping protein distributions. (A, top) X-y planes are shown with single-cell sides boxed. (middle) Montages of single-cell sides shown from apical surface to 12 µm basal (0.3-µm z steps). SAJ is shown in a bracket. Dots indicate five SAJs and are shown at 3-µm intervals. (bottom) SAJ parameters are shown (50 sides each). (B) SAJ RFI profiles are shown of DE-cad∷GFP (n = 288), Arm∷GFP (n = 229), and Baz∷GFP (n = 188). (C) IM RFI profiles (n = 50 each) are shown. (D) Maps of SAJ and IM RFIs are shown over cell cortex. (E–G) Embryo protein counts are shown. (E) Purified GST-GFP is shown (12% SDS-PAGE; coomassie). (F) Example ELISA assay is shown with GST-GFP standard curve, and Baz∷GFP late cellularization embryos are shown. (G) Graph of the example standard curve is shown (done in duplicate), and the example Baz sample is indicated in red. (H) Levels of GFP-tagged proteins versus endogenous proteins in WT embryos (3–7-h embryo lysates; 6% SDS-PAGE). Blots were probed with DE-cad, Arm, Baz, and β-tubulin antibodies (non–GFP-tagged portion of DE-cad∷GFP detected; Oda and Tsukita, 1999). (I) Protein counts are shown. (J) Single-plane images are shown with the same coverslips and settings. Late cellularization is shown. RFIs were measured at the 10 brightest SAJs and equal IM areas. Normalized means ± SD are shown for five embryos each. WB, Western blot.
Next, we counted protein numbers per cell to assign them to the maps. We compared known numbers of late cellularization embryos to GFP standard curves by ELISA assays (Fig. 1, E–G). DE-cad∷GFP and Arm∷GFP had similar numbers per embryo, but Baz∷GFP had 17–18-fold fewer numbers (P < 0.01; Fig. 1 I). The GFP-tagged proteins had similar levels to corresponding endogenous proteins in wild-type (WT) embryos (Fig. 1 H). Based on cellularizing embryos having 5,952 ± 329 cells (Fowlkes et al., 2008), we calculated protein numbers per cell (Fig. 1 I). Applying this to our localization maps revealed similar DE-cad∷GFP and Arm∷GFP protein numbers per membrane at an SAJ but about sevenfold fewer Baz∷GFP proteins (Fig. 1 I). In an equal IM area, we calculated ∼4.5-fold fewer DE-cad∷GFP proteins, ∼6.5-fold fewer Arm∷GFP proteins, and 28-fold fewer Baz∷GFP proteins than at an SAJ. We assumed zero cytoplasmic protein.
A limitation of assigning whole embryo protein counts to localization maps was the high final SDs created by combining many parameters (see Materials and methods). To confirm the DE-cad/Arm to Baz ratio, we directly compared fluorescence levels at late cellularization SAJs under the same coverslip and settings. DE-cad∷GFP and Arm∷GFP fluorescence overlapped, but Baz∷GFP fluorescence was about sixfold lower (Fig. 1 J). IM DE-cad∷GFP and Arm∷GFP were also about sixfold lower. Thus, DE-cad and Arm have a 1:1 ratio in SAJs, as expected, but Baz is at 6–7-fold lower levels. More specifically, we calculated densities of 1,236 DE-cad∷GFP, 1,565 Arm∷GFP, and 220 Baz∷GFP proteins/µm2 per membrane at an SAJ (based on SAJ length and width; Fig. 1 A). These DE-cad/Arm densities are ∼10-fold lower than fully packed desmosomal cadherins detected at 17,500 proteins/µm2 per membrane by EM (Al-Amoudi et al., 2007) and more similar to chicken retinal epithelial AJs, which, by EM, showed 700 cadherins/µm2 per membrane arranged as subclusters (Miyaguchi, 2000).
Baz and core AJ proteins have different dynamics at SAJs
How does Baz organize DE-cad/Arm into SAJs with a 1:7 ratio? We tested three models with distinct dynamics. (1) Baz recruiting DE-cad/Arm to SAJ assembly sites in a 1:1 ratio but most Baz exiting for the 1:7 SAJ ratio; each would have equal entry rates, but Baz would have an approximately sevenfold lower SAJ immobile fraction. (2) DE-cad/Arm diffusing to SAJ assembly sites where Baz seeds their clustering in a 1:7 ratio; entry rates could be independent, but Baz would have an equal or higher immobile fraction. (3) Independently formed Baz and DE-cad/Arm clusters engaging at 1:7 molecular ratios; entry rates and immobile fractions could both be independent.
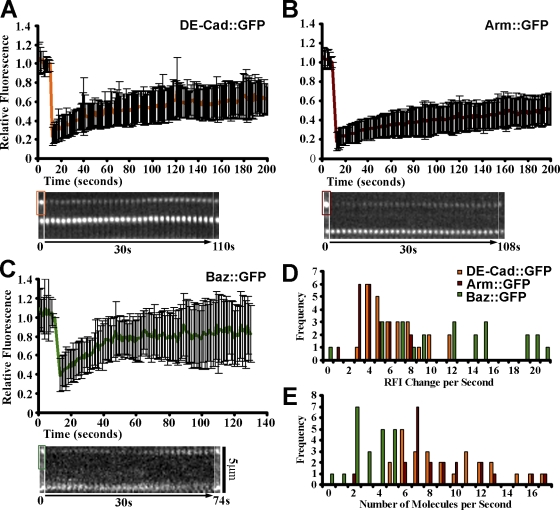
To test the models, we used FRAP to probe late cellularization SAJs. We calculated recovery rates for the first 30 s after bleaching (Fig. 2, A–C), first relative to initial SAJ fluorescence (Fig. 2 D) and then relative to protein numbers in an SAJ (Fig. 2 E). Baz∷GFP net entry rates (mean of 2.54 proteins/s) covered a lower range than those of DE-cad∷GFP and Arm∷GFP, which overlapped (means of 8.87 and 6.96 proteins/s, respectively; Fig. 2 E), arguing against the 1:1 entry model. Baz∷GFP immobile fractions (33 ± 24%) were lower than those of DE-cad∷GFP (48 ± 16%, P < 0.05) and Arm∷GFP (63 ± 11%, P < 0.01; Fig. 2, A–C), arguing against the seeding model. Baz∷GFP t1/2 values (14 ± 4 s) were also lower than those of DE-cad∷GFP (37 ± 12 s, P < 0.01) and Arm∷GFP (44 ± 14 s, P < 0.01). Arm∷GFP immobile fractions were higher than DE-cad∷GFP values, but their t1/2 values were indistinguishable. Overall, the lower Baz entry rates and immobile fractions argued against SAJ assembly via 1:1 entry or seeding alone, suggesting interactions between independent Baz and DE-cad/Arm clusters.
Figure 2.
Distinct Baz and DE-cad/Arm dynamics at SAJs. (A–C) Late cellularization SAJ bleach/recovery plots for DE-cad∷GFP (26 SAJs), Arm∷GFP (21 SAJs), and Baz∷GFP (22 SAJs). Example data show prebleach (0 s; the bleached areas are boxed, and the unbleached SAJs are shown below), recovery (30-s kymograph), and recovery plateau (later point). Normalized means ± SD are shown for five embryos each. (D) First 30-s recovery rates are shown relative to prior SAJ fluorescence. (E) Absolute recovery rates are shown.
DE-cad and Arm form puncta at the apical surface during early cellularization
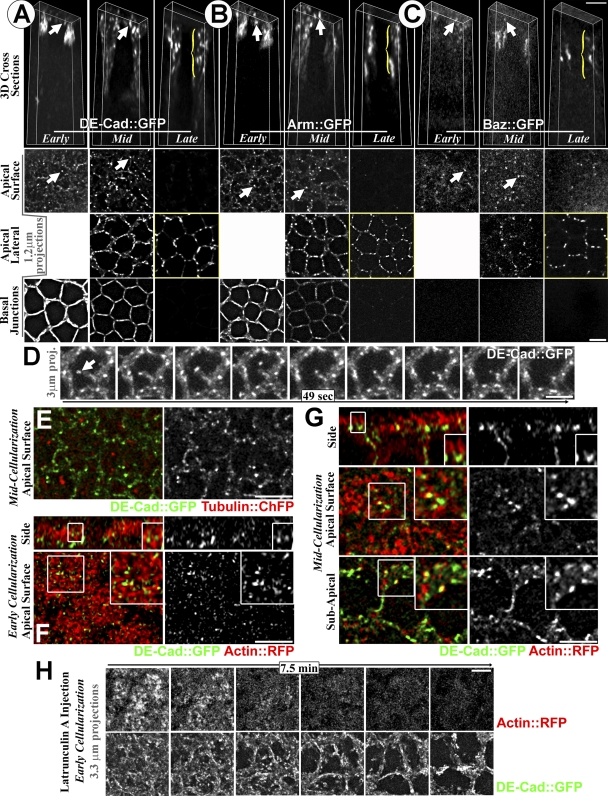
To look for DE-cad/Arm clustering before SAJ assembly, we imaged DE-cad∷GFP and Arm∷GFP by live 3D microscopy over cellularization. With fixation, DE-cad and Arm are in relatively smooth BJs and apicolateral SAJs at early cellularization with BJ loss by late cellularization (Muller and Wieschaus, 1996; Hunter and Wieschaus, 2000). DE-cad∷GFP and Arm∷GFP showed both patterns live (Fig. 3, A and B, bottom two rows). Surprisingly, we also saw previously undetected DE-cad∷GFP and Arm∷GFP puncta at peripheral and central regions of the apical cell surface (Fig. 3, A and B, arrows). DE-cad∷GFP apical surface puncta were lost with fixation (unpublished data), suggesting endogenous puncta are also sensitive to fixation. By late cellularization, apical surface puncta were lost, and DE-cad∷GFP and Arm∷GFP localized primarily to apicolateral SAJs (Fig. 3, A and B, brackets). As with past fixed imaging (Harris and Peifer, 2004), Baz∷GFP was absent from BJs (Fig. 3 C). At early and mid cellularization, Baz∷GFP was detected weakly near the apical surface (Fig. 3 C, arrows), but fewer Baz∷GFP puncta were detected in the central region of the apical surface (0.2 ± 0.57 per cell, n = 50 cells) versus DE-cad∷GFP and Arm∷GFP (3.18 ± 1.49 and 3.56 ± 1.37 per cell, respectively, n = 50 cells, P < 0.01). By late cellularization, Baz∷GFP localized primarily to apicolateral SAJs (Fig. 3 C, bracket), similar to DE-cad/Arm. We hypothesized that the DE-cad/Arm apical surface puncta might be SAJ precursors formed without Baz.
Figure 3.
DE-cad and Arm form puncta between apical surface microvilli. (A–C) Deconvolved images over cellularization are shown. (top) 3D views of single cells shown with front and back removed. (bottom) Projections of apical surface, apicolateral, and BJs. Arrows indicate surface puncta, and brackets indicate SAJs. (D) Mobile DE-cad∷GFP apical surface puncta (arrow). (E) DE-cad∷GFP (green) and tubulin∷mCherry (red) mid cellularization is shown. (F–G) DE-cad∷GFP (green) and actin∷RFP (red) are shown. Images are deconvolved. Early (F) and mid (G) cellularization are shown. Single plane side and surface views are shown. Insets show DE-cad∷GFP puncta between microvilli. (H) Latrunculin A effect on apical surface DE-cad∷GFP/actin∷RFP shown with dual imaging. Images are deconvolved. Experiment was performed at early cellularization. Bars, 5 µm.
Apical surface DE-cad puncta form between apical microvilli
To assess how the puncta form, we compared them with other proteins. A control transmembrane protein (mouse CD8∷GFP) had a smooth membrane distribution (unpublished data), arguing against DE-cad/Arm clustering via nonspecific effects on receptor diffusion. In fact, DE-cad/Arm puncta were quite dynamic, often moving to and from the center of the apical surface (Fig. 3 D, arrow). To test cytoskeletal associations, we dual imaged DE-cad∷GFP with tubulin∷mCherry or actin∷RFP live. Basolateral MT bundles were seen (not depicted), but few MTs were at the apical surface, and these colocalized minimally with DE-cad∷GFP puncta (Fig. 3 E). Actin-RFP labeled apical microvilli that cover the apical surface during cellularization (Turner and Mahowald, 1976; Grevengoed et al., 2003). At early to mid cellularization, surface views showed DE-cad∷GFP puncta between the microvilli (Fig. 3, F and G). Side views showed the puncta at the base of microvilli (Fig. 3, F and G). By mid cellularization, the puncta also associated with microvilli at apical cell–cell contacts (Fig. 3 G).
To test how actin affects the DE-cad/Arm puncta, we dual imaged DE-cad∷GFP and actin∷RFP live after injecting latrunculin A at early cellularization. This first eliminated apical microvilli (Fig. 3 H) and then led to general cell shape loss. DE-cad∷GFP puncta cleared from the apical surface center with the same timing as the microvilli loss, apparently moving to the periphery (Fig. 3 H). Carrier controls had no effect (unpublished data). Thus, actin-based microvilli appear to position DE-cad/Arm puncta at the apical surface. Because contacts between actin-based protrusions promote cadherin–catenin clustering in mammalian cells (Adams et al., 1996, 1998; Vasioukhin et al., 2000), contacts between apical microvilli may similarly promote trans-cadherin interactions to form DE-cad/Arm puncta. Puncta formed at cell–cell contacts could be direct precursors to SAJs.
The apical puncta enlarge, move basally, and join Baz to form SAJs
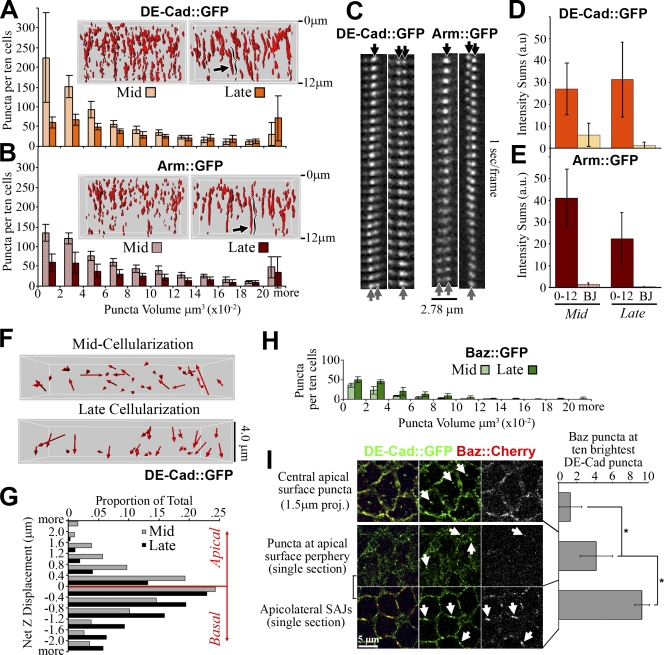
How do the apical puncta contribute to SAJs? 3D quantification of DE-cad∷GFP and Arm∷GFP puncta in the apical 12 µm of cells from mid to late cellularization showed a significant decrease in puncta number per cell (n = 5 embryos each, P < 0.01; Fig. 4, A and B) and a loss of smaller volume puncta (n = 5 embryos each, P < 0.01; Fig. 4, A and B). Many puncta became elongated and lined up along the z axis by late cellularization (Fig. 4, A and B, arrows and brackets), suggesting puncta clustering. Analyzing individual puncta revealed mergers and fissions (Fig. 4 C, arrows). In 2-min periods between mid and late cellularization, 20/50 DE-cad∷GFP and 8/50 Arm∷GFP puncta merged, and 6/50 DE-cad∷GFP and 7/50 Arm∷GFP puncta split. Thus, the puncta interact dynamically, and a net tendency to merge may produce SAJs. Total BJ intensity was relatively low at mid cellularization (Fig. 4, D and E) before the apical changes began, indicating that reorganization of mid cellularization apical puncta may be sufficient for forming late cellularization SAJs. Also, the total intensity of all puncta in the apical 12 µm did not increase from mid to late cellularization (an apparent drop for Arm∷GFP was statistically insignificant; Fig. 4, D and E).
Figure 4.
DE-cad/Arm puncta cluster, reposition, and join Baz to form SAJs. (A and B) DE-cad∷GFP and Arm∷GFP puncta volumes (n = 5 embryos; apical, 12 µm). Mid and late cellularization are shown. Insets show example puncta in 3D. Arrows indicate elongated SAJs. (C) SAJ mergers/fissions are indicated by arrows. (D and E) Fluorescence intensity sums are shown. All apical 12-µm puncta versus BJs. Mid and late cellularization (n = 5 embryos) are shown. (F) Net displacement arrows of DE-cad∷GFP puncta (apical, 4 µm). Rows of two to three cells. Puncta tracked for 1 min. Mid and late cellularization are shown. (G) Apical versus basal movement (n = 878 mid and 1,000 late DE-cad∷GFP puncta, 8 embryos each). (H) Baz∷GFP puncta volumes (n = 5 embryos; apical, 12 µm). Mid and late cellularization are shown. (I) DE-cad∷GFP versus Baz∷Cherry. (top) Apical surface projection is shown. Arrows indicate central apical surface puncta. Bottom rows show single sections. Apical surface periphery and apicolateral SAJs are shown. Arrows indicate puncta. (right) Quantification is shown as the number of Baz∷mCherry puncta overlapping the 10 brightest DE-Cad∷GFP puncta per field (n = 10 embryos). Means ± SD are shown. *, P < 0.01.
To test how directly the puncta could contribute to SAJ assembly, we tracked single DE-cad∷GFP puncta in the apical 4 µm of the cells (Fig. 4 F). At early mid cellularization, there was relatively equal apical and basal movement (55.7 ± 14.3% basal vs. 43.8 ± 14.4% apical, n = 8 embryos). At late cellularization, the displacements became mainly basal (77.1 ± 11.9% basal vs. 22.3 ± 12.0% apical, n = 8 embryos; Fig. 4 G), which is a significant change (P < 0.01) directed toward SAJ assembly sites.
Contrasting DE-cad∷GFP and Arm∷GFP, low numbers of Baz∷GFP puncta existed per cell at mid cellularization, and puncta numbers and volumes significantly increased by late cellularization (n = 5 embryos each, P < 0.01; Fig. 4 H). Late Baz∷GFP puncta numbers were similar to those for DE-cad∷GFP and Arm∷GFP, likely a result of colocalization at SAJs. The fewer earlier Baz∷GFP puncta correlated with the fewer Baz∷GFP puncta counted at the apical surface at these stages (Fig. 3 C). To further test whether Baz has low overlap with apical surface DE-cad puncta, we generated and coexpressed Baz∷mCherry with DE-cad∷GFP. At mid cellularization, apical surface DE-cad∷GFP puncta rarely colocalized with Baz∷mCherry puncta (Fig. 4 I, arrows), which were mainly absent from the domain. DE-cad∷GFP puncta at peripheral, protrusive edges of the apical surface showed more overlap with Baz∷mCherry (Fig. 4 I, arrows), which was still relatively sparse at this position. In contrast, apicolateral SAJ assembly sites showed almost full colocalization of DE-cad∷GFP and Baz∷mCherry (Fig. 4 I, arrows). Thus, DE-cad/Arm puncta appear to form at the apical surface with minimal Baz association and then associate with Baz at SAJs.
Baz recruits the apical puncta into apicolateral SAJs
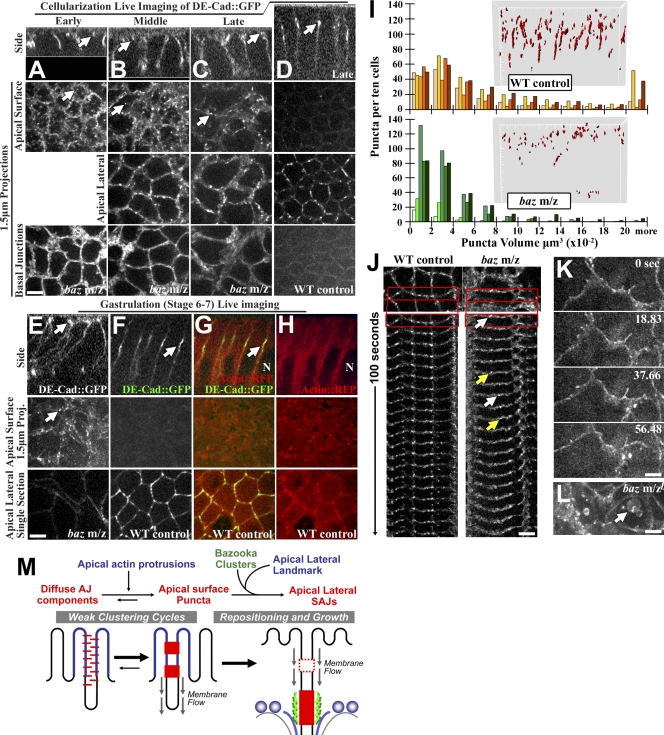
To test how Baz affects the DE-cad/Arm puncta, we imaged DE-cad∷GFP in baz maternal zygotic mutants live. At early and mid cellularization, baz mutants had apical surface puncta (Fig. 5, A and B, arrows) and BJs (Fig. 5, A and B, bottom). At late cellularization, apical surface puncta persisted abnormally versus WT controls (Fig. 5, C vs. D, arrows), and basal DE-cad∷GFP was detected (Fig. 5, C vs. D, bottom). As gastrulation began, baz mutants continued to have apical surface puncta (Fig. 5, E vs. F, arrows) plus abnormal cell shapes and basal DE-cad∷GFP (Muller and Wieschaus, 1996). 7/7 mutant embryos had this developmental progression. Dual DE-cad∷GFP/actin∷RFP imaging at WT gastrulation showed the apical surface devoid of DE-cad∷GFP puncta and DE-cad∷GFP at apicolateral SAJs next to the top of nuclei (Fig. 5, F–H, arrows) where centrosomes localize (Harris and Peifer, 2005). Thus, apical surface DE-cad∷GFP puncta can form without Baz, but normally, Baz repositions them to the apicolateral region next to centrosomes.
Figure 5.
Baz is required for puncta repositioning and SAJ growth. (A–F) Live DE-cad∷GFP. (A–C and E) baz maternal and zygotic mutants shown over cellularization to gastrulation. Arrows indicate apical surface puncta. (D and F) WT controls with equal DE-cad∷GFP transgenes at late cellularization and gastrulation. Arrows indicate apicolateral SAJs. (G and H) WT gastrulation and DE-cad∷GFP/actin∷RFP dual imaging are shown. Arrow indicates SAJ. N, nucleus. (I) Puncta volumes, WT controls versus baz mutants (n = 5 embryos) at gastrulation onset are shown. Insets show example puncta in 3D. (J) Puncta mobility is shown. WT control versus baz mutants are shown at gastrulation onset as 1.5-µm projections. White and yellow arrows show puncta appearance and disappearance, respectively. Boxes indicate regions shown in the kymographs below. (K and L) Later gastrulation is shown. (K) baz mutant cell protrusions and DE-cad∷GFP are shown. (L) baz mutant DE-cad∷GFP in vesicles (arrow) is shown. (M) Model of early AJ assembly in Drosophila. Bars, 5 µm.
To assess how Baz affects DE-cad∷GFP puncta size and number, we compared equal 3D volumes of full epithelial monolayers of baz mutant or WT embryos as gastrulation began. After cell number corrections (fewer baz mutant cells were present as a result of their flat morphology), three baz mutant embryos had WT puncta numbers per cell and two had very few puncta (Fig. 5 I). Overall, baz mutants had lower puncta volumes (P < 0.01, n = 5 embryos each) and less puncta elongation in the z axis (Fig. 5 I, insets). As gastrulation began, baz mutant puncta were also more dynamic (Fig. 5 J, arrows). By ∼10 min into gastrulation, baz mutant cells often had large cell protrusions (7/7 embryos; Fig. 5 K). By ∼20 min, large DE-cad–positive vesicles arose (7/7 embryos; Fig. 5 L, arrow). Thus, baz mutants fail at positioning DE-cad puncta and growing SAJs and subsequently lose epithelial structure and display elevated internal DE-cad.
SAJ assembly via higher order interactions between Baz and DE-cad/Arm clusters
Our data indicate that Baz and DE-cad/Arm form independent clusters that engage to form SAJs. Baz and DE-cad/Arm clusters are structurally distinct (with different protein numbers and dynamics). They arise in distinct ways both developmentally and within the cell. They are also genetically separable; apical surface DE-cad puncta can form in baz mutants (Fig. 5, A and B), and apicolateral Baz puncta can form in AJ mutants (Harris and Peifer, 2004). This independent clustering may involve Baz homooligomerization (Benton and St Johnston, 2003) and homophilic cis- and trans-cadherin interactions (Gumbiner, 2005).
How do Baz puncta control the repositioning of DE-cad/Arm puncta? Direct transport is unlikely, as there are few Baz puncta or MTs at the apical surface. However, general basal plasma membrane flow occurs during cellularization (Lecuit and Wieschaus, 2000). Interestingly, we detected apical DE-cad∷GFP puncta moving basally during later cellularization when apical microvilli density decreases (Turner and Mahowald, 1976; Grevengoed et al., 2003). Perhaps the puncta interact more extensively with dense early apical microvilli and thus resist membrane flow. Later, microvilli thinning may release more puncta to flow basally. We propose that Baz clusters act as molecular nets to catch and concentrate these DE-cad/Arm puncta at SAJ assembly sites (Fig. 5 M).
Why reposition apical surface DE-cad/Arm puncta to the apicolateral region? Cell protrusions may have limited clustering ability. They can sweep receptors into clusters, but their movement could also break clusters apart, producing weak clustering cycles (Fig. 5 M). For example, early contacts form and break repeatedly in MDCK cells (McNeill et al., 1993). In these cells, signaling to actin converts the dynamic protrusions into stable contacts for AJ growth (Drees et al., 2005; Yamada et al., 2005; Yamada and Nelson, 2007). In Drosophila, Baz appears to promote AJ growth in a distinct way by repositioning AJ puncta to separate assembly sites next to centrosomes (Fig. 5 M). Without Baz, DE-cad/Arm complexes appear trapped in weak clustering cycles leading to epithelial breakdown.
Materials and methods
Fly stocks
FlyBase describes mutations and constructs (http://flybase.bio.indiana.edu). Flies with DE-cad∷GFP under the ubiquitin promoter in a shotgun-null mutant background were generated with an established protocol (flies with DE-cad∷GFP under the ubiquitin promoter were provided by H. Oda, JT Biohistory Research Hall, Osaka, Japan; Oda and Tsukita, 2001). Flies with arm∷GFP under the arm promoter in an arm-null mutant background were provided by D. McEwen (University of Texas at San Antonio, San Antonio, TX) and M. Peifer (University of North Carolina at Chapel Hill, Chapel Hill, NC). baz∷GFP was a gene trap with GFP inserted into the first intron of the baz locus (Fly Trap). UAS-tubulin∷mCherry was provided by N. Rusan (University of North Carolina at Chapel Hill) and M. Peifer. UAS-mCD8 and UAS-actin∷RFP stocks were obtained from the Bloomington Drosophila Stock Center. UAS-baz∷mCherry was generated with standard molecular methods and inserted into the chromosome 2 attp20 site (Genetic Services). UAS constructs were expressed maternally using maternal-α4-tubulin-GAL4. bazXi106 mutants were provided by A. Wodarz (University of Göttingen, Göttingen, Germany). bazXi106 maternal zygotic mutants were made by the FLP dominant female sterile method as described previously (Harris and Peifer, 2004) but were heterozygous for ubi-DE-cad∷GFP. WT was yellow white.
Time-lapse microscopy
Dechorionated embryos were mounted in halocarbon oil (series 700; Halocarbon Products) on petriPERM dishes (Sigma-Aldrich). Images were collected with a spinning-disk confocal system (Quorum Technologies) at RT with a 63× Plan Apochromat NA 1.4 objective (Carl Zeiss, Inc.), a piezo top plate, an EM charge-coupled device camera (Hamamatsu Photonics), and Volocity software (PerkinElmer). Z stacks were collected with 300-nm step sizes. In all experiments, the autofluorescent vitelline membrane of the egg shell was used as a marker for the apical surface of the cells just below it.
Latrunculin A injections
Dechorionated embryos were attached to coverslips with tape adhesive dissolved in heptane, dried, and overlaid with halocarbon oil. Latrunculin A (Sigma-Aldrich) was dissolved in DMSO at 500 µM and injected using Femotips (Eppendorf). Embryos were imaged (as described in Time-lapse microscopy) 1–2 min after injection.
FRAP analyses
Samples were photobleached with an argon laser using a mosaic digital diaphragm (Photonic Instruments) attached to the aforementioned spinning-disk confocal system. A 2.2–2.5-µm-wide line typically covering half a cell and spanning four to six cells in length was selected as the area for photobleaching. Two to three of these areas were simultaneously bleached for 1 s per field of view per embryo. The samples were continually imaged at a single z plane with a separate laser before, during, and after the photobleaching. Imaging was stopped after a clear recovery plateau was reached. Intensities of bleached SAJs that remained in the focal plane for the full recovery were measured using ImageJ (National Institutes of Health). These values were first corrected for background by subtracting a mean of three fluorescence values for areas of the same size outside of the embryo. To correct for general bleaching of the embryo from imaging, the values were divided by the total fluorescence of a square containing eight to ten cells outside the FRAP area. The corrected fluorescence intensities for the SAJs were normalized to the time point just before bleaching and were plotted using Excel (Microsoft). Recovery rates were calculated from the slopes of best-fit lines for the first 30 s after photobleaching. Immobile fractions and recovery half-times were calculated based on fluorescence levels at the recovery plateau versus the prebleach level and the level immediately after bleaching. Three to five bleached SAJs were analyzed per embryo (five embryos total).
Postacquisition image analysis and manipulation
Fluorescence intensity measurements in Fig. 1 were performed with ImageJ. Image deconvolution (iterative restoration) and maximum intensity projections were performed with Volocity software where noted. 3D reconstructions were performed with Imaris software (version 6.2; Bitplane). Puncta were quantified in 3D datasets with Imaris software, and puncta selection was standardized for each experiment based on particle intensity versus IM intensity, particle surface area, and particle volume. Data from the full field of view were quantified and normalized to 10 cells per embryo. Particle tracking and standardized particle selection were also performed with Imaris software, but only puncta that could be tracked for 30–60 s were included in the tracking analyses. Particle tracking was performed for 4-µm-deep stacks collected with 2–4-s intervals. 60-s periods showing no movement of the vitelline membrane were analyzed. Calculations and graphing were performed using Excel. For figure preparation, Photoshop (Adobe) was used to adjust input levels so the main range of signals spanned the entire output greyscale, and bicubic interpolation was used for image resizing (minimal changes seen at normal viewing magnifications).
ELISAs and Western blots
For ELISAs, late cellularization DE-cad∷GFP and Arm∷GFP embryos were selected by hand under a dissecting microscope and placed on ice (10 embryos collected each). Because of lower Baz∷GFP protein levels, 50 Baz∷GFP embryos were counted from a 3–4-h collection and placed on ice (separate collections from this period showed that the majority of embryos were at late cellularization). Embryos were dechorionated and transferred to a prechilled mini homogenizer and lysed with 100 µl NP-40 lysis buffer (150 mM sodium chloride, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml PMSF, 1.0% NP-40, and 50 mM Tris, pH 8.0). The lysates were centrifuged for 6 min at 1,050 g, transferred to a new tube, and centrifuged for 1 min at 16,900 g. Then, the full lysate volumes were loaded into ELISA plates coated with goat anti-GFP antibody (Thermo Fisher Scientific).
For the GFP standard curve, GST-GFP was cloned by standard methods, expressed in DL21 cells, purified with glutathione resin (GE Healthcare), and quantified with a BCA protein assay (Thermo Fisher Scientific) versus BSA standard curves. The GFP standard curve was created by serial dilution of the pure GST-GFP in WT embryo lysate (prepared in the same way as the lysates of embryos expressing GFP-tagged proteins) and applied in neighboring wells of the ELISA plate. The ELISA protocol was performed at 4°C following the supplier’s instructions. Rabbit anti-GFP antibodies (ab290; Abcam), goat anti–rabbit-HRP antibodies (Thermo Fisher Scientific), detection reagent (1-Step Ultra TMB-ELISA; Thermo Fisher Scientific), and a plate reader (Spectramax Plus 384; MDS Analytical Technologies) were used for detection. For Western blots, dechorionated embryo pellets of equal volume were mixed 1:5 (vol/vol) with 2× SDS-PAGE sample buffer, homogenized, boiled for 5 min, separated by 6% SDS-PAGE, blotted, probed, and imaged with a FluorChem 8900 (Alpha Innotech). Antibodies used were rat anti–DE-cad (DCAD1; 1:500; Developmental Studies Hybridoma Bank), mouse anti-Arm (N27A1; 1:250; Developmental Studies Hybridoma Bank), rabbit anti-Baz (1:2,000), mouse anti–β-tubulin (E7; 1:200; Developmental Studies Hybridoma Bank), and corresponding HRP-conjugated secondary antibodies (Thermo Fisher Scientific) and HRP detection reagents (Thermo Fisher Scientific).
Protein-counting calculations
To count the number of GFP-tagged proteins in an SAJ, we first calculated the total relative fluorescent intensity (RFI) at an SAJ by multiplying the area under the mean RFI profile at an SAJ (Fig. 1 B) by the width of an SAJ (Fig. 1 A). Mean RFI profiles were created by normalizing maximum intensity values and aligning the z positions of these peaks (note that because the individual curves were not symmetrically distributed around these peaks, the RFI intensity profiles suggested SAJ lengths longer than those measured directly; Fig. 1 A). The total RFI values at an SAJ were multiplied by the mean number of SAJs in a cell (Fig. 1 A) to give the total relative cortical fluorescence from all SAJs.
To calculate the total relative cortical fluorescence for IM, we calculated the regions of an average cell cortex not occupied by SAJs (Fig. 1 D) and applied the mean RFI profiles of IM (Fig. 1 C) to these regions. Adding the total relative cortical fluorescence from SAJs and IM gives the overall total relative cortical fluorescence of an average cell.
Next, we applied our counts of proteins per cell to these relative cortical fluorescence measurements. To determine the number of proteins per SAJ, the counts of proteins per cell were multiplied by the fraction of total relative cortical fluorescence caused by SAJs, and this was then divided by the number of SAJs per cell (this gave the number of proteins per SAJ from one cell). To determine the number of proteins in the IM, the counts of proteins per cell were multiplied by the fraction of total relative cortical fluorescence caused by IM. To determine the number of proteins in an area of IM equal to the area of an SAJ, we divided the number of proteins in the IM by the total area of the IM and then multiplied by the area of an SAJ. SDs for each final mean were calculated by the standard procedure of (a) dividing the SD of each parameter by its individual mean, (b) squaring this value, (c) summing these squared values for all parameters used, (d) taking the square root of this sum, and (e) multiplying this value by the final mean.
Statistics
All comparisons with Student’s t tests were performed using Excel.
Acknowledgments
We thank R. Winklbauer and U. Tepass for critiques, C.Q. Yu for technical help, A. Wilde for embryo injection help, and L. Buck for ELISA plate reader use. We thank M. Peifer, D. McEwen, N. Rusan, H. Oda, A. Wodarz, Fly Trap, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for reagents.
This work was supported by a Canadian Institutes of Health Research operating grant. T. Harris holds a Tier 2 Canada Research Chair.
Footnotes
Abbreviations used in this paper: AJ, adherens junction; Arm, Armadillo; Baz, Bazooka; BJ, basal junction; DE-cad, Drosophila E-cadherin; IM, intervening membrane; MT, microtubule; RFI, relative fluorescent intensity; SAJ, spot AJ; WT, wild type.
References
- Adams C.L., Nelson W.J., Smith S.J. 1996. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion.J. Cell Biol. 135:1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C.L., Chen Y.T., Smith S.J., Nelson W.J. 1998. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein.J. Cell Biol. 142:1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A., Diez D.C., Betts M.J., Frangakis A.S. 2007. The molecular architecture of cadherins in native epidermal desmosomes.Nature. 450:832–837 [DOI] [PubMed] [Google Scholar]
- Benton R., St Johnston D. 2003. A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity.Curr. Biol. 13:1330–1334 [DOI] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., Weis W.I. 2005. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly.Cell. 123:903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes C.C., Hendriks C.L., Keranen S.V., Weber G.H., Rubel O., Huang M.Y., Chatoor S., DePace A.H., Simirenko L., Henriquez C., et al. 2008. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm.Cell. 133:364–374 [DOI] [PubMed] [Google Scholar]
- Grevengoed E.E., Fox D.T., Gates J., Peifer M. 2003. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled.J. Cell Biol. 163:1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. 2005. Regulation of cadherin-mediated adhesion in morphogenesis.Nat. Rev. Mol. Cell Biol. 6:622–634 [DOI] [PubMed] [Google Scholar]
- Harris T.J., Peifer M. 2004. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila.J. Cell Biol. 167:135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., Peifer M. 2005. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila.J. Cell Biol. 170:813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock A., Nelson W.J. 2008. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton.Biochim. Biophys. Acta. 1778:660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Wieschaus E. 2000. Regulated expression of nullo is required for the formation of distinct apical and basal adherens junctions in the Drosophila blastoderm.J. Cell Biol. 150:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T. 2004. Junctions and vesicular trafficking during Drosophila cellularization.J. Cell Sci. 117:3427–3433 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Wieschaus E. 2000. Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo.J. Cell Biol. 150:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H., Ryan T.A., Smith S.J., Nelson W.J. 1993. Spatial and temporal dissection of immediate and early events following cadherin-mediated epithelial cell adhesion.J. Cell Biol. 120:1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaguchi K. 2000. Ultrastructure of the zonula adherens revealed by rapid-freeze deep-etching.J. Struct. Biol. 132:169–178 [DOI] [PubMed] [Google Scholar]
- Muller H.A., Wieschaus E. 1996. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila.J. Cell Biol. 134:149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H., Tsukita S. 1999. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins.Dev. Biol. 216:406–422 [DOI] [PubMed] [Google Scholar]
- Oda H., Tsukita S. 2001. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells.J. Cell Sci. 114:493–501 [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M., Jamora C., Fuchs E. 2003. Sticky business: orchestrating cellular signals at adherens junctions.Cell. 112:535–548 [DOI] [PubMed] [Google Scholar]
- Tepass U., Tanentzapf G., Ward R., Fehon R. 2001. Epithelial cell polarity and cell junctions in Drosophila.Annu. Rev. Genet. 35:747–784 [DOI] [PubMed] [Google Scholar]
- Turner F.R., Mahowald A.P. 1976. Scanning electron microscopy of Drosophila embryogenesis. 1. The structure of the egg envelopes and the formation of the cellular blastoderm.Dev. Biol. 50:95–108 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion.Cell. 100:209–219 [DOI] [PubMed] [Google Scholar]
- Yamada S., Nelson W.J. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell–cell adhesion.J. Cell Biol. 178:517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W.I., Nelson W.J. 2005. Deconstructing the cadherin-catenin-actin complex.Cell. 123:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]