Abstract
The kinetochore-bound protein kinase Bub1 performs two crucial functions during mitosis: it is essential for spindle checkpoint signaling and for correct chromosome alignment. Interestingly, Bub1 mutations are found in cancer tissues and cancer cell lines. Using an isogenic RNA interference complementation system in transformed HeLa cells and untransformed RPE1 cells, we investigate the effect of structural Bub1 mutants on chromosome segregation. We demonstrate that Bub1 regulates mitosis through the same mechanisms in both cell lines, suggesting a common regulatory network. Surprisingly, Bub1 can regulate chromosome segregation in a kinetochore-independent manner, albeit at lower efficiency. Its kinase activity is crucial for chromosome alignment but plays only a minor role in spindle checkpoint signaling. We also identify a novel conserved motif within Bub1 (amino acids 458–476) that is essential for spindle checkpoint signaling but does not regulate chromosome alignment, and we show that several cancer-related Bub1 mutants impair chromosome segregation, suggesting a possible link to tumorigenesis.
Introduction
Chromosome segregation is controlled by the mitotic spindle, which attaches sister chromatids via kinetochores, which are multiprotein complexes located on centromeric DNA (Musacchio and Salmon, 2007; Cheeseman and Desai, 2008). Correct chromosome segregation requires that kinetochore pairs bind to microtubules (MTs) emanating from opposite spindle poles in a bipolar manner and that they control the forces that align the chromosomes on a metaphase plate. Kinetochores also monitor bipolar MT attachment and control mitotic progression through the spindle checkpoint, which arrests cells before anaphase by inhibiting the anaphase-promoting complex in the presence of incorrectly attached chromosomes (Musacchio and Salmon, 2007).
The spindle checkpoint requires the conserved proteins Mad1, Mad2, Bub1, Bub3, Mad3/BubR1, and Mps1, which accumulate on unattached kinetochores during mitosis (Musacchio and Salmon, 2007). The protein kinase Bub1 is not only essential for the spindle checkpoint, it is also required for correct kinetochore–MT attachments (Williams et al., 2007). Bub1 loss delays the formation of stable end-on attachments, causing an accumulation of lateral kinetochore–MT attachments (Gillett et al., 2004; Meraldi and Sorger, 2005). The two functions of Bub1 are conserved, as its inactivation causes loss of spindle checkpoint and severe chromosome segregation defects in all tested eukaryotes (Bernard et al., 1998; Warren et al., 2002; Meraldi and Sorger, 2005; Perera et al., 2007). Studies in yeast and vertebrates have identified several downstream targets that require Bub1 for kinetochore binding, including Mad1, Mad2, and BubR1, the MT–depolymerase mitotic centromere-associated kinesin (MCAK), and the outer kinetochore protein, centromere protein F (CENP-F; Sharp-Baker and Chen, 2001; Warren et al., 2002; Johnson et al., 2004; Meraldi et al., 2004; Boyarchuk et al., 2007; Huang et al., 2007; Kiyomitsu et al., 2007). Moreover, Bub1 regulates the targeting of cohesion protein Sgo1 (shugoshin) to the centromere through PP2A (Tang et al., 2004b, 2006; Kitajima et al., 2005).
Bub1 deregulation is also linked to apoptosis and tumorigenesis. Reduction of Bub1 levels can lead to tumorigenesis, senescence, and p53-dependent and -independent apoptosis (Gjoerup et al., 2007; Jeganathan et al., 2007; Niikura et al., 2007), whereas a Bub1 knockout causes early embryonic lethality (Perera et al., 2007). Primary Bub1−/− mouse embryo fibroblasts lack a functional spindle checkpoint, show aberrant chromosome segregation, and fail to proliferate (Perera et al., 2007). In humans, aberrant Bub1 gene expression is detected in esophageal, gastric, and colon tumors, melanoma, and breast cancer cell lines (Shigeishi et al., 2001; Shichiri et al., 2002; Doak et al., 2004). Moreover, point mutations and deletions were found in leukemia, lymphoma, and thyroid cancer as well as colon and lung cancer cell lines (Cahill et al., 1998; Ohshima et al., 2000; Ru et al., 2002; Shichiri et al., 2002). However, the functional consequence of these mutations is unknown.
Bub1 contains several domains, including an N-terminal domain, which binds to the kinetochore protein KNL1/Blinkin (Kiyomitsu et al., 2007), a Bub3-binding domain (Taylor et al., 1998), and a C-terminal kinase domain. The role of these domains and interactions has been studied in Saccharomyces cerevisiae and Schizosaccharomyces pombe, revealing conserved features of ScBub1 and SpBub1 but also species-dependent differences: Bub3 is essential for Bub1 function in S. cerevisiae but dispensable for checkpoint control in S. pombe, implying that SpBub1 regulates the spindle checkpoint without SpBub3 (Roberts et al., 1994; Warren et al., 2002; Tange and Niwa, 2008). The role of the kinase domain is also different; in both yeasts, it regulates kinetochore–MT attachment, but inactivation of the kinase does not perturb the checkpoint in S. cerevisiae, although partially compromising the checkpoint in S. pombe (Warren et al., 2002; Yamaguchi et al., 2003; Fernius and Hardwick, 2007).
Given these phenotypical variations in fungal systems, the role of the Bub1 domains and of its interaction partners remains unclear or has been studied only in a punctual manner in human cells. It has been reported that, in contrast to yeast, the kinase domain is required for the spindle checkpoint (Kang et al., 2008) and that the KNL1-binding domain is important for chromosome congression, raising the question as to which extent the functions are conserved (Warren et al., 2002; Vanoosthuyse et al., 2004; Kiyomitsu et al., 2007). An equally important question, which could not be studied in yeast, is whether cancer-related Bub1 mutations impair chromosome segregation. Until now, the lack of robust genetic tools prevented the analysis of multiple human Bub1 mutants. We therefore established a genetic system in human cells, i.e., expressing stable Bub1 mutants in an isogenic RNAi complementation system, and combined it with a cell biological analysis. Our results indicate that Bub1 can regulate chromosome segregation in a kinetochore-independent manner; we identify a novel conserved motif, which is essential for spindle checkpoint signaling, and we demonstrate that the kinase activity is essential for chromosome alignment but not for the spindle checkpoint. Finally, we find that cancer-related Bub1 mutations deregulate chromosome segregation, suggesting a functional link to tumorigenesis.
Results
Construction of HeLa Flp-In cell lines
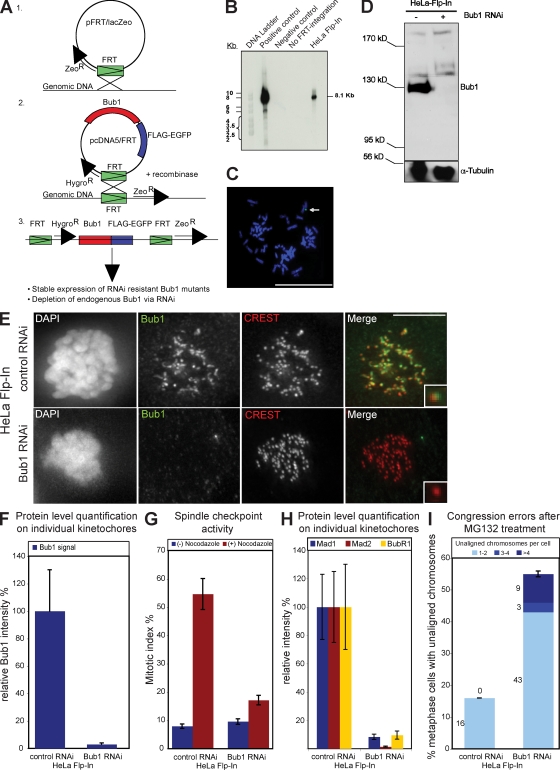
To investigate how human Bub1 regulates chromosome segregation at the molecular level, we generated a Bub1 RNAi complementation system based on stable cell lines expressing different Bub1 mutants. This system is built on the integration of a single Flp recombination target (FRT) site into the genome and the subsequent integration of Bub1 expression constructs via Flp-mediated intermolecular DNA recombination. This guarantees that phenotypical differences observed in different Bub1 mutant cell lines are not a result of the varying genetic background of the integration site. Moreover, all stable cell lines express RNAi-resistant Bub1 mutants, allowing the exclusive depletion of endogenous Bub1 (Fig. 1 A).
Figure 1.
Integration of the FRT site into the genome of HeLa cells does not affect the Bub1 RNAi phenotype. (A) Schematic diagram illustrating the generation of stable Flp-In and Bub1 mutant cell lines. (B) Southern blot against the DNA of two HeLa Flp-In clones probed for lacZ. The clone in the right lane was selected. The transfected vector pFRT/lacZeo was used as a positive control, and DNA from untransfected HeLa Kyoto H2B-mRED cells were used as negative control. (C) FISH analysis of selected HeLa Flp-In cells with a probe against lacZ. The arrow indicates positive FISH signal. Bar, 5 µm. (D–I) Characterization of HeLa Flp-In cells treated with control or Bub1 RNAi. (D) Immunoblot of whole cell lysates probed with Bub1 and α-tubulin antibodies. Black line indicates that intervening lanes have been spliced out. (E) Immunofluorescence images of mitotic cells stained with DAPI (DNA), Bub1 antisera (green), and CREST antisera (red, kinetochores). Bar,10 µm. (F) Quantification by immunofluorescence of Bub1 levels at kinetochores. (G) Mitotic index of HeLa Flp-In cells treated for 16 h with or without nocodazole. (H) Quantification by immunofluorescence of Mad1 (blue), Mad2 (red), and BubR1 (yellow) levels at kinetochores. (I) Cumulative plot of the percentage of metaphase cells with unaligned chromosomes after a 1-h MG132 treatment. Insets show a higher magnification view of a single kinetochore. Error bars represent standard deviation.
The Flp-In system was integrated into a HeLa cell line expressing histone H2B–monomeric RED protein (mRED) in 50% of the cells to allow both immunofluorescence measurements and the monitoring of chromosome segregation by time-lapse imaging. Southern blot analysis with a probe directed against the lacZ gene localized on the pFRT/lacZeo vector identified one HeLa Flp-In clone with a single FRT integration site, and FISH analysis with a hybridization sample directed against lacZ confirmed the uniqueness of the integration site (Fig. 1, B and C).
Depletion of Bub1 by RNAi abrogates the spindle checkpoint and impairs chromosome congression (Meraldi and Sorger, 2005). We tested the efficiency of Bub1 RNAi in HeLa Flp-In cells to confirm that FRT integration did not affect the Bub1 RNAi phenotype. Immunoblotting with Bub1 antibodies showed that Bub1 RNAi depleted Bub1, and quantification by immunofluorescence indicated that Bub1 protein levels at kinetochores were reduced to 3% when compared with control-treated cells (Fig. 1, D–F). To test for spindle checkpoint abrogation, cells were treated for 16 h with the MT-depolymerizing drug nocodazole, which abrogates kinetochore−MT attachment, resulting in spindle checkpoint–dependent mitotic arrest and a strong enrichment of Mad1, Mad2, and BubR1 at kinetochores (Hoffman et al., 2001). Depletion of Bub1 in HeLa Flp-In cells abrogated both the mitotic arrest, as visualized by phase-contrast microscopy, and the recruitment of Mad1, Mad2, and BubR1 to kinetochores, as quantified by immunofluorescence (Fig. 1, G and H). We next measured whether Bub1 depletion also impaired chromosome congression. We treated our cells for 1 h with the proteasome inhibitor MG132, which arrests cells in metaphase in a spindle checkpoint–independent manner (Rock et al., 1994), fixed, and stained them for DNA, kinetochores, and the mitotic spindle. We identified cells with a metaphase plate, indicative of a metaphase arrest, and quantified the percentage of cells with unaligned chromosomes (chromosomes were counted as unaligned when located outside of the central 30% of the mitotic spindle). Consistent with previous studies, Bub1 depletion strongly increased the percentage of HeLa Flp-In cells with uncongressed chromosomes when compared with control depletion (55 vs. 16%; Fig. 1 I; Johnson et al., 2004; Meraldi and Sorger, 2005) but did not affect chromosome cohesion (not depicted; Perera et al., 2007). We conclude that FRT integration did not affect the Bub1 RNAi phenotype.
Expression of Bub1–wild type (wt) complements Bub1 RNAi
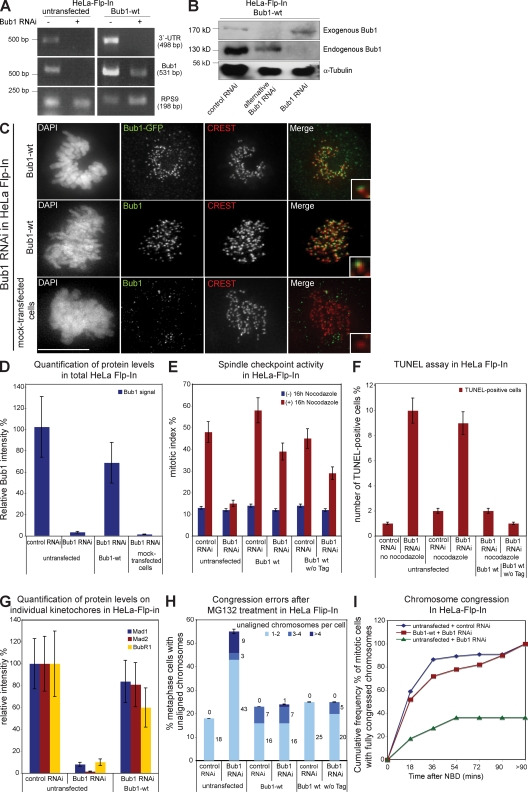
We next tested the ability of exogenous Bub1-wt to complement Bub1 RNAi when stably expressed in HeLa Flp-In cells. Bub1-wt encodes for wt Bub1 protein fused at its C terminus with a Flag-EGFP tag and carries four silent mutations in the target RNAi site to guarantee Bub1-wt expression in a Bub1 RNAi background.
We quantified the Bub1 mRNA levels in HeLa Flp-In and stable Bub1-wt cells treated with Bub1 or control RNAi by RT-PCR to confirm that Bub1-wt is resistant to Bub1 RNAi. We amplified a fragment of the coding Bub1 mRNA to measure total Bub1 expression and a fragment of the Bub1 3′ untranslated region (UTR) to measure expression of endogenous Bub1. Bub1 RNAi abolished expression of the Bub1 3′ UTR in both cell lines when compared with control RNAi–treated cells, confirming the degradation of endogenous Bub1 mRNA (Fig. 2 A). Bub1 RNAi also abolished expression of total Bub1 in Flp-In cells but only reduced total Bub1 mRNA levels in Bub1-wt cells, indicating the presence of an RNAi-resistant exogenous Bub1-wt mRNA (Fig. 2 A). These results were confirmed at the protein level, as anti-Bub1 immunoblotting indicated that the endogenous Bub1 protein band of 130 kD disappeared after Bub1 RNAi, whereas a 170-kD Bub1-Flag-EGFP protein band remained in Bub1-wt cells (Fig. 2 B). In contrast, treatment with an alternative Bub1 siRNA, which targets a different sequence within Bub1, depleted both endogenous and exogenous Bub1 (Fig. 2 B). Quantitative anti-Bub1 immunofluorescence indicated that after Bub1 RNAi, total Bub1-wt was expressed at roughly 70% of endogenous Bub1 and localized to kinetochores (Fig. 2, C and D). In contrast, stable cell lines having incorporated an empty vector contained only 1% of endogenous Bub1 after Bub1 RNAi (Fig. 2, C and D).
Figure 2.
Bub1-Flag-EGFP wt complements Bub1 RNAi. (A) RT-PCR of a region within the 3′ UTR of Bub1, a region within the ORF of Bub1, and the housekeeping gene RPS9. (B) Immunoblot of lysates of Bub1-wt cells treated with RNAi as indicated and probed with Bub1 and α-tubulin antibodies. (C) Immunofluorescence images of mitotic cells stained with DAPI (DNA), GFP, or Bub1 antisera (green) and CREST antisera (red, kinetochores). Bar, 10 µm. (D) Quantification by immunofluorescence of Bub1 levels in total cells. (E) Mitotic index of the indicated cells treated with or without nocodazole for 16 h. (F) Quantification of TUNEL-positive cells after nocodazole and Bub1 RNAi. (G) Quantification by immunofluorescence of Mad1, Mad2, and BubR1 levels on kinetochores. (H) Cumulative plot of percentage of metaphase cells with unaligned chromosomes after a 1-h MG132 treatment. (I) Cumulative frequency plots of chromosome alignment time using nuclear breakdown (NBD) as t = 0. Insets show a higher magnification view of a single kinetochore. Error bars represent standard deviation.
We next analyzed the efficiency at which Bub1-wt rescued the Bub1 RNAi phenotype. HeLa Flp-In cells treated with Bub1 RNAi did not arrest in mitosis when treated with nocodazole (mitotic index of 15%), whereas control-depleted cells had a high mitotic index (48%). In contrast, both control- and Bub1-depleted Bub1-wt cells arrested in mitosis after a nocodazole treatment (39 vs. 58%; Fig. 2 E). Identical results were obtained with an untagged Bub1-wt, indicating that the weaker spindle checkpoint response was not a consequence of the C-terminal Flag-EGFP tag (Fig. 2 E). As Bub1 depletion also induces mitotic cell death, we used a TUNEL assay to quantify the extent to which a nocodazole treatment induced apoptosis (Jeganathan et al., 2007; Niikura et al., 2007). Although Bub1 siRNA treatment increased the number of TUNEL-positive Flp-In cells (9 vs. 2% in control-depleted cells), stable expression of Bub1-wt with or without tag suppressed apoptosis (Fig. 2 F and Fig. S1 A). Immunofluorescence showed that Bub1-wt expression also restored recruitment of Mad1, Mad2, and BubR1 to kinetochores (Fig. 2 G; and Fig. S1, B–D). This indicated that Bub1-wt complements Bub1 RNAi in terms of spindle checkpoint and apoptosis.
To test whether Bub1-wt also rescued chromosome congression, we first detected the amount of uncongressed chromosomes in the MG132 assay and found that Bub1 RNAi did not increase the proportion of cells with unaligned chromosomes in Bub1-wt cells (Fig. 2 H). Second, we monitored chromosome congression by live cell imaging using the H2B-mRED signal. The time point of nuclear breakdown was set at t = 0, and the time until all chromosomes congressed on a metaphase plate was recorded. Bub1 RNAi did not affect chromosome congression in Bub1-wt cells compared with control RNAi–treated cells, as 50% of the cells aligned all their chromosomes after 17 min in both depletions (Fig. 2 I). In contrast, in Flp-In HeLa cells, Bub1 depletion caused a severe delay of congression (after 8 h, only 36% of the cells had aligned all their chromosomes; Fig. 2 I). This indicated that Bub1-wt also complemented Bub1 RNAi in terms of chromosome congression. Overall, we conclude that we have established a system that allows the functional comparison of Bub1 mutants. We further note that exogenous Bub1 is expressed at roughly 70% of endogenous Bub1 and that it complements the spindle checkpoint only to 80%, suggesting that we are working with a sensitized system.
Differential effects of Bub1 mutants on spindle checkpoint and chromosome congression
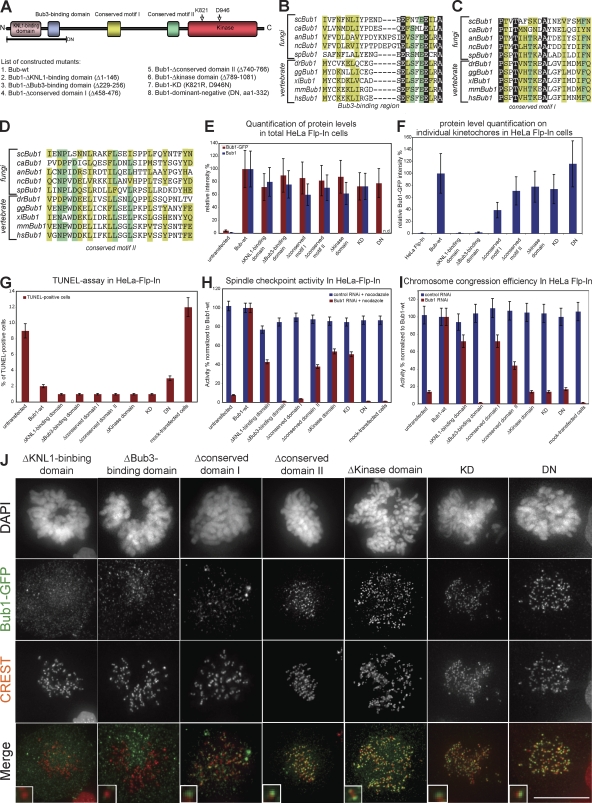
Alignments of fungal and vertebrate Bub1 sequences reveal several conserved domains, including two previously uncharacterized, conserved motifs. To investigate the different roles of Bub1, we generated mutants lacking one of the following domains: (a) the KNL1-binding domain, which is required for kinetochore localization, (b) the Bub3-binding domain, (c) the conserved motif I (aa 458–476), (d) the conserved motif II (aa 740–766), (e and f) two different catalytically inactive mutants (one lacking the whole kinase domain and the other harboring a lysine to arginine mutation in the catalytic motif and an aspartate to asparagine mutation in the DFG motif, which both abolish kinase activity; van den Heuvel and Harlow, 1993), and (g) a truncated Bub1 fragment (dominant negative [DN]; aa 1–332), which acts in a DN manner when overexpressed (Fig. 3, A–D; Taylor and McKeon, 1997).
Figure 3.
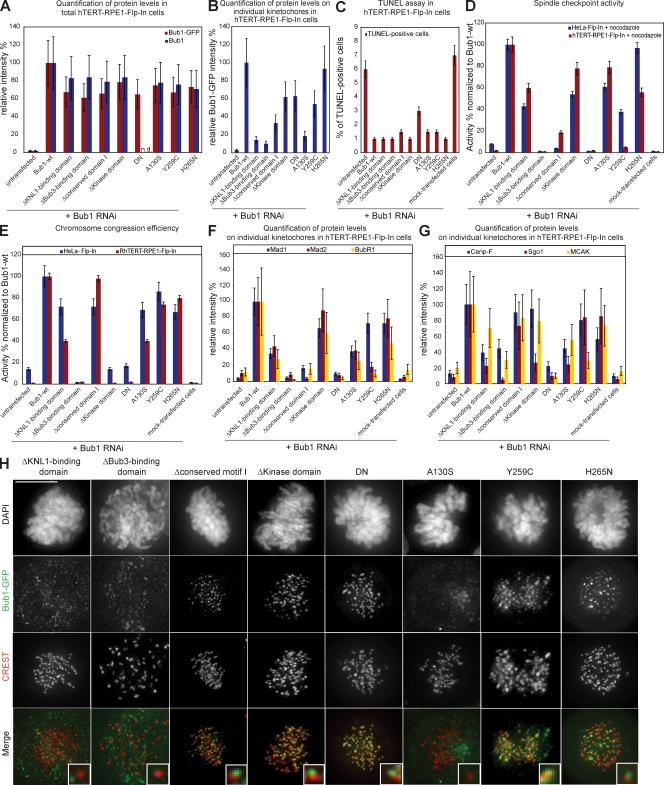
Differential ability of Bub1 mutants to complement apoptosis, spindle checkpoint signaling, and chromosome congression. (A) Schematic structure of Bub1 protein indicating conserved domains and a set of constructed Bub1 mutants. (B–D) Multiple sequence alignment of the Bub3-binding domain (B), the conserved domain I (C), and the conserved domain II (D) in five fungi and five vertebrates. Identical residues are in a black, residues conserved in ≥80% of the species are in a dark green, and similar residues in ≥80% of the species are in a light green background. Sc, S. cerevisiae; ca, Candida albicans; an, Aspergillus nidulans; nc, Neurospora crassa; sp, S. pombe; dr, Danio rerio; gg, Gallus gallus; xl, X. laevis; mm, Mus musculus; hs, Homo sapiens. (E) Immunofluorescence quantification of relative total cellular Bub1 levels with Bub1 (blue) and GFP antisera (red). Note that Bub1 antibodies do not recognize Bub1-DN. (F) Immunofluorescence quantification of relative Bub1 levels on kinetochores with GFP antisera. (G) Quantification of TUNEL-positive cells after nocodazole and Bub1 RNAi. (H) Mitotic index after 16 h nocodazole treatment normalized to Bub1-wt. (I) Ability to complement congression errors in cells treated with MG132 normalized to Bub1-wt. Error bars show standard deviation. (J) Immunofluorescence images of mitotic cells stained with DAPI (DNA), GFP antisera (green), and CREST sera (red, kinetochores). KD, kinase dead. Insets show a higher magnification view of a single kinetochore. Bar, 10 µm.
All of these mutants were expressed in stable HeLa Flp-In cells as Flag-EGFP fusion proteins. We quantified by immunofluorescence their total cellular levels (Fig. 3 E) and their specific abundance on unattached kinetochores (Fig. 3, F and J) in Bub1 RNAi–treated cells using GFP and Bub1 antibodies. All mutants were expressed at equal levels but showed distinct abilities to bind unattached kinetochores; Bub1–Δkinase domain, Bub1–kinase dead, and Bub1-DN levels on kinetochores were similar to Bub1-wt (75–120% of Bub1-wt), whereas Bub1–Δconserved motif I and II had a decreased ability to bind to kinetochores (40–60%). Finally, Bub1-ΔKNL1–binding domain and Bub1-ΔBub3–binding domain did not localize to unattached kinetochores.
Using the nocodazole and the MG132-based congression assays, we found that none of the tested mutants affected the spindle checkpoint or chromosome alignment, indicating that they did not act in a DN manner (Fig. 3, H and I). Interestingly, in the presence of endogenous Bub1, most of the mutants did not localize to kinetochores, suggesting, as previously reported, a low exchange rate of endogenous Bub1 at kinetochores (Fig. S1 E and not depicted; Shah et al., 2004).
We next evaluated the importance of the Bub1 domains with regard to apoptosis, spindle checkpoint, and chromosome congression in cells lacking endogenous Bub1. All Bub1 mutants with the exception of the short Bub1-DN fully rescued the apoptotic response associated with Bub1 RNAi, suggesting that cell death associated with Bub1 loss is not associated with a particular domain (Fig. 3 G). This also indicated that all of our mutants are biologically active, excluding misfolding artifacts. We also found that both Bub1-ΔBub3–binding domain and Bub1-DN cells could not activate the spindle checkpoint or align chromosomes after Bub1 RNAi (Fig. 3, H and I). Neither of these two mutants rescued kinetochore binding of Mad1, Mad2, BubR1, MCAK, Sgo1, or CENP-F, indicating that Bub3 binding is essential for Bub1 chromosome segregation functions in human cells and that expression of the DN domain does not rescue any Bub1 function (Fig. S2, A and B). Expression of Bub1–Δconserved motif II only weakly complemented the spindle checkpoint (42%) and chromosome congression (42%), suggesting that it plays an important role for Bub1 function (Fig. 3, H and I). This was also reflected at the level of the Bub1 downstream targets, as loss of the conserved domain II resulted in a reduced kinetochore binding of Mad1, Mad2, BubR1, MCAK, CENP-F, and Sgo1 (Fig. S2, A and B).
Complementing roles of Bub1–conserved domain I and kinase domain
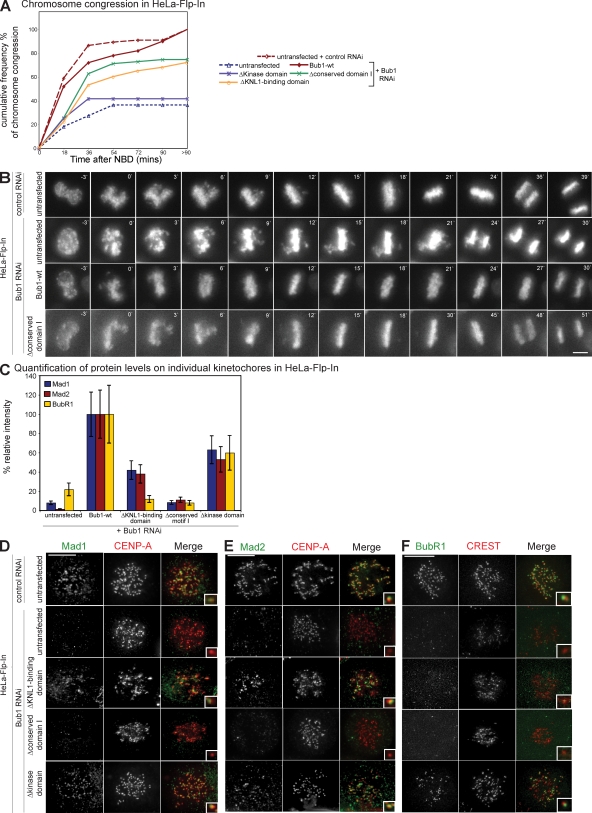
We next focused on the domains that were specifically required for one function of Bub1: the two catalytically inactive mutants, Bub1–Δkinase domain and Bub1–kinase dead, did not rescue chromosome alignment in Bub1 RNAi–treated cells (14% of Bub1-wt activity) but partially complemented the spindle checkpoint response (51–54% of Bub1-wt; Fig. 3, H and I). In contrast, cells expressing Bub1–Δconserved motif I did not activate the spindle checkpoint (4% of Bub1-wt) but efficiently congressed their chromosomes (72% of Bub1-wt; Fig. 3, H and I). Additional experiments confirmed the separation of function for both types of Bub1 mutants. Bub1–Δkinase domain cells treated with Bub1 RNAi failed to align chromosomes on a metaphase plate when measured by live cell imaging, whereas expression of Bub1–Δconserved motif I rescued the timing of chromosome alignment in cells treated with Bub1 RNAi (Fig. 4, A and B). Conversely, Bub1–Δconserved motif I failed to recruit Mad1, Mad2, and BubR1 to kinetochores in the absence of endogenous Bub1, whereas expression of a catalytically inactive Bub1 rescued the ability of kinetochores to bind the checkpoint proteins (Fig. 4, C–F). We conclude that the kinase activity is primarily required for chromosome alignment, whereas the conserved domain I is only required for the spindle checkpoint, indicating that these two Bub1 functions can be separated.
Figure 4.
Loss of Bub1 kinase activity, conserved domain I, or kinetochore binding differentially affect chromosome congression and the spindle checkpoint. (A) Cumulative frequency plots of chromosome alignment time from live cell videos using nuclear breakdown (NBD) as t = 0. (B) Successive frames every 3 min from live cell videos of HeLa Flp-In, Bub1-wt, and Bub1–Δconserved motif I cells transfected with indicated RNAi. (C) Quantification by immunofluorescence of Mad1, Mad2, and BubR1 levels on kinetochores of indicated stable cell lines treated with Bub1 RNAi. Error bars represent standard deviation. (D–F) Immunofluorescence images of the indicated control or Bub1 RNAi–treated prometaphase cell lines stained with Mad1 (D), Mad2 (E), or BubR1 (F) antisera (green) and CENP-A, or CREST antisera (red, kinetochores). Insets show a higher magnification view of a single kinetochore. Bars,10 µm.
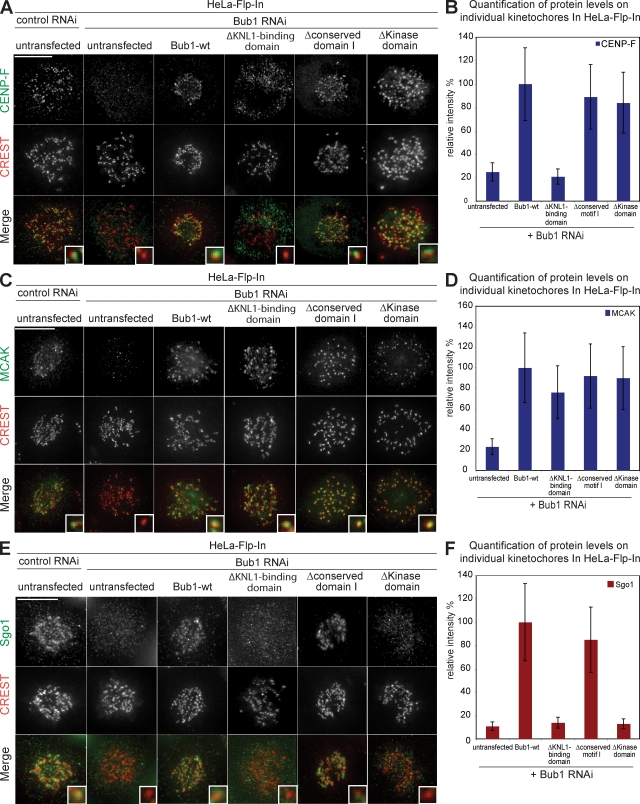
To obtain a better understanding at the molecular level of these two mutants, we tested their ability to rescue the recruitment at kinetochores of three downstream targets of Bub1 that are implicated in chromosome congression: CENP-F, Sgo1, and MCAK (Johnson et al., 2004; Tang et al., 2004b; Huang et al., 2007). We detected normal levels of CENP-F and MCAK at kinetochores in cells lacking the conserved motif I or cells lacking Bub1-kinase activity, indicating that both domains are dispensable for the loading of MCAK and CENP-F (Fig. 5, A–D). In contrast, loss of Bub1 kinase activity abrogated Sgo1 targeting to centromeres, whereas deletion of the conserved motif I did not impair the recruitment of Sgo1 (Fig. 5, E and F). We conclude that in contrast to CENP-F and MCAK, Sgo1 recruitment is a crucial target of human Bub1 kinase activity.
Figure 5.
Loss of Bub1 kinase activity, conserved domain I, or kinetochore binding differentially affect the recruitment of CENP-F, MCAK, and Sgo1 to kinetochores. (A, C, and E) Immunofluorescence images of mitotic control or Bub1 RNAi–treated Bub1 mutant cell lines stained with CENP-F (A), MCAK (C), Sgo1 (E, green), and CREST antisera (red, kinetochores). Bar, 10 µm. (B, D, and F) Quantification of CENP-F (B), MCAK (D), and Sgo1 (F) levels on kinetochores in indicated cell lines relative to Bub1-wt. Insets show a higher magnification view of a single kinetochore. Errors bars represent standard deviation.
Bub1 can also function when not bound to kinetochores
In a third step, we investigated the importance of kinetochore binding for Bub1 function by analyzing cells expressing Bub1-ΔKNL1–binding domain, which does not bind to kinetochores (Fig. 3, F and J). Interestingly, Bub1-ΔKNL1–binding domain partially rescued both the spindle checkpoint (42% of Bub1-wt activity) and chromosome congression (72% of Bub1-wt activity; Fig. 3, H and I; and Fig. 4 A). The Bub1-ΔKNL1–binding domain mutant also rescued the ability to recruit substantial amounts of Mad1, Mad2, and MCAK to unattached kinetochores (Fig. 4, C–E; and Fig. 5, C and D). In contrast, recruitment of CENP-F, Sgo1, and BubR1 was impaired, indicating that the presence of Bub1 at kinetochores is essential for the binding of these proteins to kinetochores (Fig. 4, C and F; and Fig. 5, A, B, E, and F). We conclude that Bub1 can exert some of its function when not bound to kinetochores and that loss of kinetochore binding only affects a subset of downstream Bub1 targets.
Bub1 mutants harboring cancer mutations
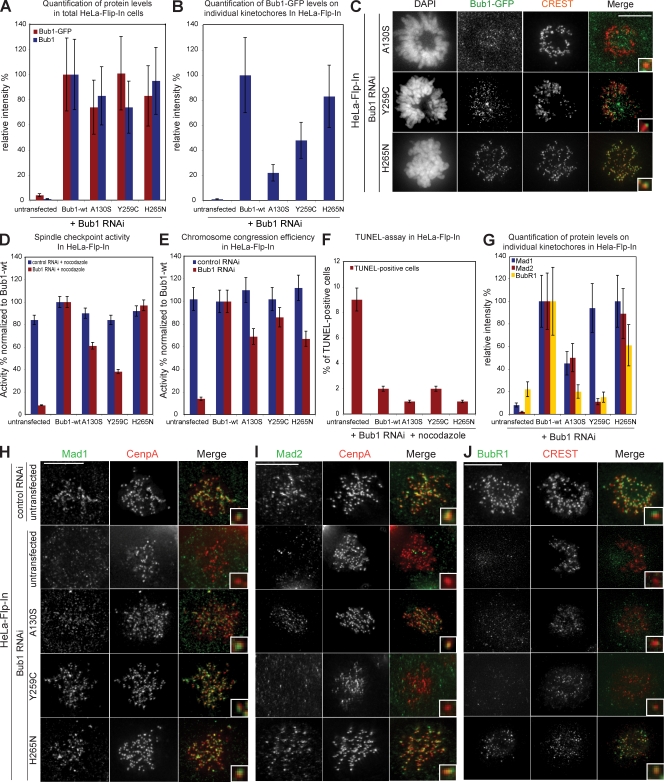
Because Bub1 mutations have been implicated in carcinogenesis, we also investigated whether three Bub1 point mutants found in cancer cell lines affect chromosome segregation: A130S is a conserved aa located in the KNL1-binding domain, and Y259C and H265N are located close to the Bub3-binding domain (Shichiri et al., 2002; Hempen et al., 2003). Immunofluorescence quantification revealed that all three mutants were expressed at levels comparable to Bub1-wt (Fig. 6 A). Bub1-Y259C and Bub1-H265N localized to kinetochores, whereas Bub1-A130S localization at kinetochores was impaired, suggesting a disruption of the KNL1-binding domain (22% of Bub1-wt levels; Fig. 6, B and C).
Figure 6.
Expression of cancer-related Bub1 mutants differentially affects checkpoint efficiency and chromosome congression. (A) Immunofluorescence quantification of total cellular Bub1 mutant levels using Bub1 and GFP antisera. (B) Immunofluorescence quantification of Bub1 mutant levels on kinetochores using GFP antisera. (C) Immunofluorescence images of indicated mitotic cells stained with DAPI (DNA), GFP antisera (green), and CREST antisera (red, kinetochores). (D) Mitotic index of Bub1 mutant cells treated for 16 h with nocodazole normalized to Bub1-wt. (E) Ability to complement congression errors in Bub1 mutant cells treated for 1 h with MG132 normalized to Bub1-wt. (F) Quantification of TUNEL-positive cells after nocodazole and Bub1 RNAi. (G) Immunofluorescence quantification of Mad1, Mad2, and BubR1 levels on kinetochores in indicated cell lines treated with Bub1 RNAi. Error bars represent standard deviation. (H–J) Immunofluorescence images of the indicated control or Bub1 RNAi–treated prometaphase cells stained with Mad1 (H), Mad2 (I), or BubR1 (J) antisera (green) and CENP-A or CREST antisera (red, kinetochores). Insets show a higher magnification view of a single kinetochore. Bars,10 µm.
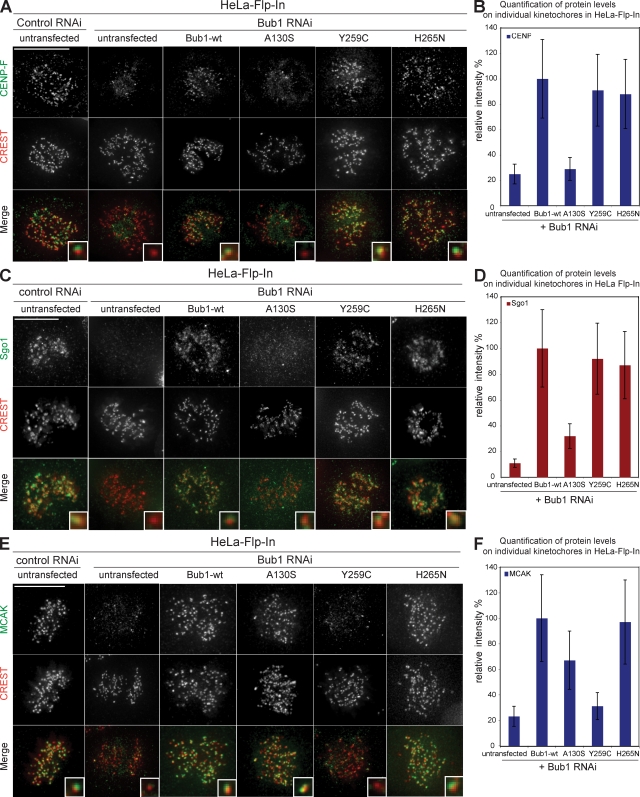
None of the point mutants acted in a DN manner (Fig. 6, D and E), and all three mutants rescued the cell death phenotype (Fig. 6 F). In the absence of endogenous Bub1, Bub1-H265N rescued the spindle checkpoint, chromosome alignment, and the loading of all tested downstream factors (Fig. 6, D, E, G–J; and Fig. 7). Similar to the loss of the KNL1-binding domain, the Bub1-A130S mutation weakened the checkpoint, increased the rate of congression errors, and caused the loss of kinetochore binding of CENP-F, Sgo1, and BubR1 but not of MCAK, Mad1, or Mad2 (Fig. 6, D, E, G–J; and Fig. 7).
Figure 7.
Effect of cancer-related Bub1 mutants on the recruitment of CENP-F, Sgo1, and MCAK to kinetochores. (A, C, and E) Immunofluorescence images of the indicated control or Bub1 RNAi–treated mitotic cells stained with CENP-F (A), Sgo1 (C), and MCAK (E) antisera (green) and CREST antisera (red, kinetochores). Bars, 10 µm. (B, D, and F) Quantification of CENP-F (B), Sgo1 (D), and MCAK (F) levels on kinetochores in the indicated cell lines treated with Bub1 RNAi. Insets show a higher magnification view of a single kinetochore. Error bars represent standard deviation.
The most interesting mutant was Bub1-Y259C, which did not rescue the spindle checkpoint (38% of Bub1-wt), although efficiently restoring chromosome congression (86% of Bub1-wt; Fig. 6, D and E). Bub1-Y259C cells treated with Bub1 RNAi had normal levels of Mad1 but low levels of Mad2 and BubR1 (Fig. 6, G–J). Given the usual tight binding of Mad1 to Mad2, we tested whether the low levels of Mad2 on kinetochores were caused by reduced Mad2 levels or the inability to recruit Mad2 on kinetochores by immunoblotting with Mad2 antibodies and found the latter to be the case (Fig. S2 C). Thus, we conclude that Bub1-Y259C fails to rescue the spindle checkpoint as the result of a deficient recruitment of Mad2 and BubR1 to kinetochores. Expression of Bub1-Y259C also rescued the ability of kinetochores to bind Sgo1 and CENP-F but not MCAK (Fig. 7). Interestingly, Bub1-Y259C was, apart from the null mutants Bub1-ΔBub3–binding domain and Bub1-DN, the only Bub1 mutant that did not rescue MCAK localization. This suggests that this region of Bub1 must be critical for MCAK regulation and indicates that Bub1 does not regulate chromosome alignment through MCAK. Given the proximity of the Y259C mutation to the Bub3-binding domain, we further tested the ability of Bub1-Y259C and other Bub1 mutants to bind to Bub3 in an in vitro assay. Our analysis revealed that all mutants except the ΔBub3-binding domain and Bub1-DN bind to Bub3 even though the binding of Bub1-Y259C was reduced compared with Bub1-wt (Fig. S2, D and E). However, this was also the case for catalytically inactive Bub1 mutants, which do not have strong spindle checkpoint defects and still load MCAK, indicating that the defects observed in cells expressing Bub1-Y259C cannot result just from reduced Bub3 binding.
Bub1 mutants in nontransformed hTERT-RPE1 cells
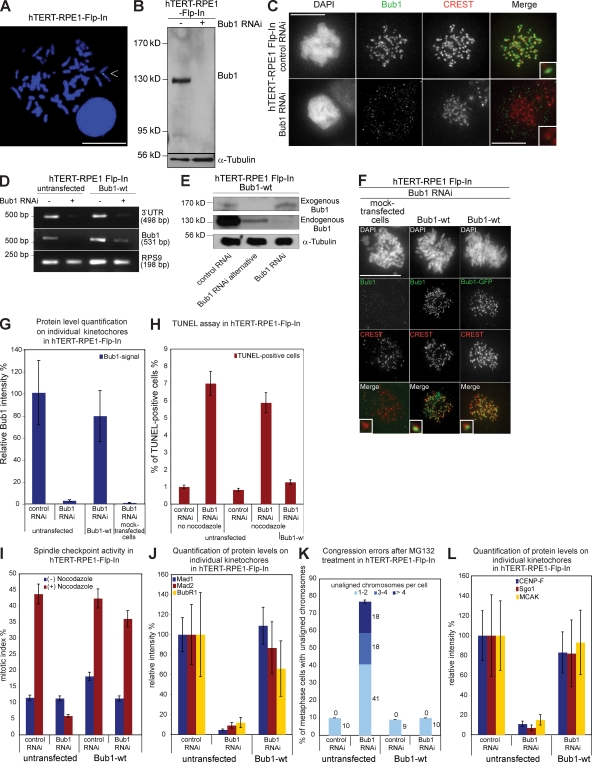
To investigate whether Bub1 function depends on its genetic background, we compared our Bub1 mutant HeLa cell lines with the same set of mutants expressed in nontransformed hTERT-RPE1 (human telomerase-immortalized retinal pigment epithelial) Flp-In cells. We generated hTERT-RPE1 cells with a single Flp-In recombination site (Fig. 8 A) in which we stably incorporated Bub1-wt, Bub1-ΔKNL1–binding domain, Bub1-ΔBub3–binding domain, Bub1–Δconserved domain I, Bub1–Δkinase domain, Bub1-DN, Bub1-A130S, Bub1-Y259C, and Bub1-H265N. We first confirmed that Bub1 RNAi depleted endogenous Bub1, abolished the spindle checkpoint, disrupted chromosome segregation, and impaired the recruitment of Mad1, Mad2, BubR1, CENP-F, MCAK, and Sgo1 to kinetochores in hTERT-RPE1 Flp-In cells (Fig. 8, B–L; Fig. S3; and Fig. S4). Stable expression of Bub1-wt-Flag-EGFP suppressed apoptosis and completely rescued the spindle checkpoint, chromosome congression, and the recruitment of all tested downstream kinetochore proteins, indicating that hTERT-RPE1 Flp-In cells are suitable for the characterization of Bub1 function in nontransformed cells (Fig. 8, D–L; Fig. S3; and Fig. S4).
Figure 8.
Integration of the FRT site into the genome of hTERT-RPE1 cells does not affect the Bub1 RNAi phenotype, and expression of Bub1-wt-Flag-EGFP rescues Bub1 RNAi. (A) FISH analysis of the selected hTERT-RPE1 Flp-In clone with a probe against lacZ. The arrowhead indicates a positive FISH signal. Bar, 5 µm. (B) Immunoblot of hTERT-RPE1 Flp-In cell lysates probed with Bub1 and α-tubulin antibodies. Black line indicates that intervening lanes have been spliced out. (C) Immunofluorescence images of mitotic cells stained with DAPI (DNA), Bub1 (green), and CREST antisera (red, kinetochores). Bar, 10 µm. (D) RT-PCR of a region within the 3′ UTR of Bub1, the ORF of Bub1, and the housekeeping gene RPS9. (E) Immunoblot of cell lysates of Bub1-wt cells treated with RNAi as indicated and probed with Bub1 and α-tubulin antibodies. (F) Immunofluorescence images of mitotic cells stained with DAPI (DNA), GFP, or Bub1 antisera (green) and CREST antisera (red, kinetochores). Bar, 10 µm. (G) Quantification by immunofluorescence of total Bub1 levels using Bub1 antibodies. (H) Quantification of TUNEL-positive cells after nocodazole and Bub1 RNAi. (I) Mitotic index of the indicated cells treated with or without nocodazole for 16 h. (J) Quantification by immunofluorescence of Mad1, Mad2, and BubR1 levels on kinetochores. (K) Cumulative plot of percentage of metaphase cells with unaligned chromosomes after a 1-h MG132 treatment. (L) Quantification by immunofluorescence of CENPF, Sgo1, and MCAK levels on kinetochores. Insets show a higher magnification view of a single kinetochore. Error bars represent standard deviation.
In a second step, we verified the expression and localization of all tested Bub1 mutants and quantified to which extent they rescued apoptosis, spindle checkpoint, chromosome congression, and target protein recruitment (Figs. 9, S3, and S4) when compared with HeLa Flp-In cells (Fig. 9, D and E). Qualitatively, we found that all the Bub1 mutants regulated the spindle checkpoint, chromosome congression, and the recruitment of all tested downstream factors in RPE1 cells in a manner that was identical to HeLa cells, indicating that our results were independent of the genetic background (Figs. 9, S3, and S4). However, we also noted a small number of significant quantitative differences. First, the functional separation of spindle checkpoint and chromosome congression defects was more consistent in RPE1 than in HeLa cells; Bub1–Δkinase domain did not rescue chromosome congression, but in contrast to HeLa cells, efficiently complemented spindle checkpoint activity (Fig. 9, D and E), strengthening our conclusion that Bub1 kinase activity only plays a minor role in the spindle checkpoint. Bub1–Δconserved domain I still impaired the spindle checkpoint while fully rescuing chromosome alignment, which is in contrast to HeLa cells where it led to weak alignment defects (Fig. 9, D and E). Second, RPE1 cells expressing two cancer-related Bub1 mutants, Y259C and H265N, showed more severe spindle checkpoint defects than the corresponding HeLa cells (Fig. 9, D and E). This was particularly true for Bub1-H265N, which fully rescued the spindle checkpoint in HeLa cells but only showed a partial complementation in RPE1 cells.
Figure 9.
The hTERT-RPE1 Bub1 mutant cells behave in a manner similar to HeLa cells. (A) Quantification by immunofluorescence of total Bub1 mutant levels using Bub1 and GFP antibodies. (B) Quantification by immunofluorescence of Bub1 mutant levels on kinetochores using GFP antibodies. (C) Quantification of TUNEL-positive cells after nocodazole treatment and Bub1 RNAi. (D) Comparison of mitotic index of Bub1 mutant HeLa and hTERT-RPE1 cells treated for 16 h with nocodazole normalized to Bub1-wt. (E) Comparison of the ability to complement congression errors in Bub1 mutant HeLa and hTERT-RPE1 cell lines treated for 1 h with MG132 normalized to Bub1-wt. (F) Quantification by immunofluorescence of Mad1, Mad2, and BubR1 levels on kinetochores. (G) Quantification by immunofluorescence of CENP-F, Sgo1, and MCAK levels on kinetochores. (H) Immunofluorescence images of mitotic cells stained with DAPI (DNA), GFP antisera (green), and CREST antisera (red, kinetochores). Insets show a higher magnification view of a single kinetochore. Error bars represent standard deviation. Bar, 10 µm.
Discussion
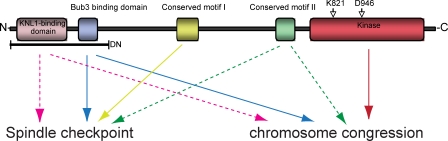
We have established an isogenic expression system in HeLa and RPE1 cells to investigate the function of Bub1 at the molecular level. Even though this system is probably sensitized as a result of mild underexpression of exogenous Bub1, it allowed important molecular insights into the function of human Bub1. First, we conclude that Bub1 can act in a kinetochore-independent manner. Second, our results indicate that the spindle checkpoint and chromosome alignment functions can be, as in yeast, separated in human Bub1; we identify a novel domain within Bub1 that is specifically required for spindle checkpoint signaling and demonstrate that Bub1 kinase activity regulates chromosome alignment but does not play a major role in the spindle checkpoint (Fig. 10). This separation of function is also reflected at the level of downstream effectors: mutants lacking spindle checkpoint control cannot load BubR1, Mad2, and to a lesser extent Mad1, but recruit Sgo1, whereas mutants defective in chromosome congression fail to recruit Sgo1 but load Mad1, Mad2, and BubR1. Finally, analysis of three cancer-related point mutants revealed partial defects in Bub1 function, in particular in their ability to activate the spindle checkpoint, suggesting that hypomorphic Bub1 mutants could contribute to cellular transformation.
Figure 10.
Schematic illustration of the importance of the different Bub1 domains for spindle checkpoint efficiency and chromosome congression. Continuous line, high importance; dashed line, moderate importance.
Regulation of the spindle checkpoint
Bub1 is required for the loading of the checkpoint proteins Mad1, Mad2, and BubR1/Mad3 to kinetochores (Sharp-Baker and Chen, 2001; Gillett et al., 2004; Johnson et al., 2004; Meraldi et al., 2004) and has been proposed to regulate the spindle checkpoint by phosphorylating the anaphase-promoting complex/cyclosome subunit Cdc20 (Tang et al., 2004a; Kang et al., 2008). Our results show that lack of catalytic activity either compromises the spindle checkpoint only partially (HeLa cells) or not at all (hTERT-RPE1 cells). In contrast, we observe a strong correlation between the spindle checkpoint response and the ability to recruit BubR1, Mad2, and to a lesser degree Mad1, to kinetochores. This indicates that BubR1, Mad1, and Mad2 recruitment is the primary mechanism by which Bub1 contributes to spindle checkpoint signaling and that the kinase activity plays only a minor role in checkpoint signaling. Consistent with our findings, we note that phospho-specific antibodies that identify putative Bub1 phosphorylation sites on Cdc20 fail to detect such a phosphorylation event under physiological conditions (Kang et al., 2008). Our data also indicate that the binding to Bub3 is essential for Bub1 function in human cells. This is different for fission yeast Bub1 because SpBub3 is not essential for the spindle checkpoint (Tange and Niwa, 2008).
We further identify a novel conserved 19-aa motif in the central part of Bub1, which is required for the loading of BubR1, Mad1, and Mad2 to kinetochores but does not control chromosome congression. Experiments in yeast reported that the central 240-aa region, which includes the conserved 19-aa motif, can bind Mad1 in cell extracts (Warren et al., 2002). However, when we tested for such an interaction in in vitro coimmunoprecipitation experiments, we failed to detect a direct interaction between human Mad1 and Bub1 (with or without Bub3; unpublished data). Moreover, we find that Mad1 is still recruited when Bub1 fails to bind to kinetochores. This indicates that Bub1 does not act as a Mad1-binding platform on kinetochores but rather that it facilitates the recruitment of Mad1/Mad2. Interestingly, we note that the conserved motif I contains several invariant serines and threonines (Fig. 3 C) and that serine 459 is phosphorylated in the presence of unattached kinetochores in Xenopus laevis egg extracts (Chen, 2004). Thus, it will be important to test in the future whether the conserved motif I is also phosphorylated when the spindle checkpoint is active and whether this phosphorylation contributes to checkpoint signaling in human cells.
Regulation of chromosome alignment
Loss of Bub1 leads to a high number of congression errors in human cells (Johnson et al., 2004; Meraldi and Sorger, 2005). Catalytically inactive Bub1 mutants do not rescue chromosome congression defects in cells depleted of endogenous Bub1. We therefore conclude that the enzymatic activity of Bub1 is essential for the regulation of chromosome alignment. Previous studies reported that Sgo1 is a key target of Bub1 for chromosome congression (Tang et al., 2004b, 2006; Kitajima et al., 2005). Consistent with data from X. laevis or S. pombe, we find that loss of the kinase domain also impairs loading of Sgo1 onto kinetochores (Kitajima et al., 2004; Boyarchuk et al., 2007; Fernius and Hardwick, 2007). However, our data also indicate that loss of Sgo1 on kinetochores does not strictly correlate with a high rate of chromosome congression defects because cells expressing Bub1-ΔKNL1–binding domain have only moderate congression defects even though they fail to recruit Sgo1 onto kinetochores. Therefore, Sgo1 cannot be the only target of Bub1 that is relevant for chromosome congression, and Bub1 must phosphorylate other substrates to ensure chromosome alignment in human cells.
Role of Bub1 kinetochore localization
Photoactivation experiments indicate that Bub1 is tightly associated to unattached kinetochores and has, among the spindle checkpoint proteins, the longest residency time at kinetochores (Howell et al., 2004; Shah et al., 2004). Moreover, experiments in S. pombe show that telomere-tethered Bub1 recruits Mad3 and Bub3 from the cytoplasm, leading to the proposal that Bub1 acts as a spindle checkpoint scaffold protein at kinetochores (Rischitor et al., 2007). Interestingly, loss of kinetochore binding only weakens but does not abolish human Bub1 functions. Bub1-ΔKNL1–binding domain partially rescues the spindle checkpoint, allows the recruitment of Mad1, Mad2, and MCAK, and strongly reduces the rate of chromosome congression errors when compared with a full Bub1 depletion. We therefore conclude that binding of Bub1 at the kinetochore is important but not essential for its checkpoint and chromosome congression function. Although Bub1 might act as a scaffold for certain proteins such as Mad3/BubR1 or CENP-F, we propose that it regulates chromosome segregation primarily through mechanisms independent of kinetochore binding.
Interestingly, our results also suggest a kinetochore-independent role for BubR1, which also regulates the spindle checkpoint and chromosome alignment (Lampson and Kapoor, 2005). Indeed, HeLa cells complemented with Bub1–Δconserved domain I do not load BubR1 onto kinetochores yet efficiently align their chromosomes on a metaphase plate. Although recent studies showed that BubR1 can regulate the spindle checkpoint in the cytoplasm (Kulukian et al., 2009; Malureanu et al., 2009), our data would suggest that it can also control chromosome alignment when not bound to kinetochores.
Bub1 cancer-related mutants
The occurrence of Bub1 mutations and differential Bub1 gene and protein expression in cancer tissues and cell lines and the occurrence of spontaneous cancers in mice expressing low doses of Bub1 indicate a possible role of Bub1 in cancer formation (Gemma et al., 2000; Shigeishi et al., 2001; Ouyang et al., 2002; Ru et al., 2002; Shichiri et al., 2002; Hempen et al., 2003; Doak et al., 2004; Jeganathan et al., 2007). However, given the multiple roles of Bub1, it is unknown which might be the critical function of Bub1 in the context of tumorigenesis. Interestingly, all tested Bub1 mutants fail to fully rescue Bub1 RNAi. This indicates that Bub1 mutants can deregulate chromosome segregation and suggests that cancer formation could be linked to a weakened Bub1 function. A precise analysis of the involvement of these Bub1 mutants in tumor formation should therefore be an important feature in future works. It is interesting to note that the tested cancer-related Bub1 mutants lead to quantitatively different results in HeLa and hTERT-RPE1 cells, suggesting that the regulatory pathways controlling chromosome segregation are very similar yet also distinct between transformed and untransformed cells.
An important question is whether heterozygous Bub1 mutations are sufficient to disrupt chromosome segregation or whether both Bub1 alleles have to be targeted. Although the A130S mutation was present on both alleles, Y259C and H265N were found only on one allele, which is accompanied by a wt Bub1 allele (Hempen et al., 2003). However, in our expression system, none of the Bub1 mutants, including the DN N-terminal Bub1 fragment, induced a DN effect on mitotic progression, suggesting that the DN effect observed by Taylor and McKeon (1997) requires overexpression of the mutant protein. Therefore, we speculate that epigenetic control mechanisms that down-regulate gene expression such as hypermethylation or hypoacetylation of the second allele are required to sensitize untransformed cells for Bub1 mutations.
Materials and methods
Antibody production
A GST-tagged Bub1 fragment (aa 336–489) was purified from Escherichia coli under native conditions and injected into rabbits (NeoMPS) and goats (BioGenes). Rabbit anti-Bub1 antibodies were affinity purified against GST-tagged Bub1 bound to an AminoLink Plus Immobilization column (Thermo Fisher Scientific).
Cell culture, stable cell lines, RNAi, and functional assays
Stable HeLa Flp-In, hTERT-RPE1 Flp-In, and Bub1 mutant cell lines were constructed according to the manufacturer’s protocol (Invitrogen). We amplified the Bub1 cDNA by PCR and inserted it into the pcDNA5/FRT/V5-His-TOPO vector (Invitrogen). Flag-EGFP was subcloned C-terminally of Bub1. The mutants were constructed via site-directed mutagenesis. Cells were grown at 37°C in 5% CO2 in either Dulbecco’s minimum essential medium + 10% FCS (HeLa cells) or 50:50 Ham’s F-12/DME + 10% FCS (hTERT-RPE1 cells). HeLa Kyoto H2B-mRED cells (provided by D. Gerlich, ETH Zurich, Zurich, Switzerland) were supplemented with 500 µg/ml G418, HeLa Flp-In, and hTERT-RPE1 Flp-In cells with 400 µg/ml zeocin, and stable Bub1 mutant cells were supplemented with 300 µg/ml hygromycine (HeLa) or 5 µg/ml puromycine (RPE1). Cells were transfected as described with 30- (HeLa) or 40-nM (RPE1) siRNAs (Bub1 siRNA, 5′-GAGUGAUCACGAUUUCUAA-3′; alternative Bub1 siRNA, 5′-AAGATGCATTTGAAGCCCAGT-3′) and analyzed 48 h after transfection (Elbashir et al., 2001). Cells were treated for 1 h with 1 µM MG132 prior to fixation to measure congression efficiency. To measure spindle checkpoint activity, cells were treated with 1 nM nocodazole for 16 h, and the fraction of rounded-up cells was determined by phase-contrast microscopy. To measure apoptosis, cells were incubated with 1 nM nocodazole for 16 h and fixed with 3.7% formaldehyde in phosphate-buffered saline, pH 7.4. A TUNEL assay was performed using an in situ cell death detection system that contained fluorescein–deoxy UTP (dUTP; Roche).
FISH analysis and Southern blotting
For FISH analysis, chromosome spreads were prepared according to standard cytogenetic methodology. The sample directed against the LacZ gene was labeled with Atto488-dUTP (Jena Bioscience). Hybridization was performed with 20-ng sample. Images were acquired using a microscope (Axio Imager M1; Carl Zeiss, Inc.) and a fluorescence imaging system (ISIS version 5.2; Metasystems). For Southern blotting, genomic DNA was extracted following standard procedures (proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation). 5 µg DNA was digested with HindIII. The hybridization probes were prepared by labeling PCR products against lacZ with DIG-dUTP (Roche), and the signal was detected by adding CDP-star chemiluminescent substrate (Roche) and exposure to x-ray films.
Immunoblotting and RT-PCR
Whole cell extracts were resolved by SDS-PAGE and transferred to nitrocellulose membranes by semidry blotting. Membranes were blocked in blocking buffer (5% low-fat dried milk, PBS, and 0.1% Tween 20) and incubated with 0.3 µg/ml rabbit anti-Bub1, rabbit anti-Mad2 (1:5,000; Bethyl Laboratories, Inc.), or mouse anti–α-tubulin (1:10,000; Sigma-Aldrich) antibodies in blocking buffer. Anti–mouse and anti–rabbit HRP-conjugated secondary antibodies (Sigma-Aldrich) were applied in blocking buffer and developed by enhanced chemiluminescence. For RT-PCR, total RNA was isolated as described previously (Chen et al., 2005) and subjected to random-primed reverse transcription using SuperScript II Rnase H-Reverse transcription (Invitrogen). PCR reactions were performed with 100 ng template DNA. The optimal cycle number was determined as described previously (for Bub1 and 3′ UTR, 38 cycles; RPS9, 28 cycles; Chen et al., 2003). Controls were performed with primers for ribosomal protein S9. PCR products were resolved by electrophoresis, visualized with ethidium bromide, and their intensities were measured densitometrically.
In vitro translation and immunoprecipitation
Bub1 mutants and Bub3 were in vitro translated using the TNT T7 Coupled Reticulocyte Lysate System (Promega) using 50 µCi [35S]-methionine (Perkin Elmer) for each reaction. Flag-tagged Bub1 mutants were immunoprecipitated with anti-Flag M2 affinity gel (Sigma-Aldrich) and equilibrated with TNES buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 2 mM EDTA, 1% NP-40, and 1 mM DTT). After a 2-h incubation at 4°C, the agarose beads were washed three times with TNES and loaded onto an SDS-PAGE gel. After exposure to x-ray films for autoradiography, the autoradiogram was scanned, and the intensity of the signals was measured using softWoRx (Applied Precision, LLC).
Immunofluorescence microscopy
Cells were fixed, permeabilized, blocked, and imaged as described previously (McClelland et al., 2007) using a 60× oil NA 1.3 objective on a microscope (Deltavision RT; Applied Precision, LLC) equipped with a camera (CoolSnapHQ; Roper Scientific). The following primary antibodies were used for staining: goat anti-Bub1 (1:2,000; this study), human anti-CREST (1:400; Antibodies, Inc.), mouse anti–CENP-A (1:1,000; Abcam), mouse anti–α-tubulin (1:10,000; Sigma-Aldrich), rabbit anti-Sgo1 (1:500; Abcam), rabbit anti–CENP-F (1:1,000; Abcam), rabbit anti-Mad1 (1:2,000; Meraldi et al., 2004), rabbit anti-Mad2 (1:500; Covance), mouse anti-BubR1 (1:1,500; Abcam), rabbit anti-MCAK (1:500; Cytoskeleton, Inc.), and rabbit anti-GFP (1:1,000; Abcam). The levels of kinetochore-bound protein were quantified as described previously (McClelland et al., 2007). For quantification of total Bub1 mutant protein expression levels, cells were stained with GFP or Bub1 antibodies, and the total signal intensity per cell was measured using a low magnification 20× NA 0.45 objective with high optical thickness. We determined the mean intensity and standard deviation of 10 cells for each Bub1 mutant.
Live cell time-lapse imaging and analysis
For live cell imaging, cells were monitored at 37°C in LabTechII (Thermo Fisher Scientific) chambers in Leibovitz’s L-15 medium containing 10% FCS. Images were acquired every 3 min for 8 h using a 20× NA 0.75 objective on a microscope (Life; Carl Zeiss, Inc.) equipped with a DAPI-FITC-Rhod/TR-CY5 (Chroma Technology Corp.) filter set.
Online supplementary material
Fig. S1 shows the TUNEL assay and the immunofluorescence images of Mad2, Mad1, and BubR1 in HeLa Flp-In cells after Bub1 RNAi. Fig. S2 shows the quantification of Mad2, Mad1, BubR1, MCAK, Sgo1, and CENP-F levels on kinetochores in Bub1-ΔBub3–binding domain and Bub1-DN cells, the immunoblotting of Mad2 in Bub1-Y259C cells, and the in vitro Bub3-binding assay. Figs. S3 and S4 show immunofluorescence images of Mad1, Mad2, and BubR1 (Fig. S3) and MCAK, Sgo1, and CENP-F (Fig. S4) in hTERT-RPE1 Flp-In cells. Online supplementary material is available at http://www.jcb.org/cgi/content/full/jcb.200902128/DC1.
Acknowledgments
We thank Daniel Gerlich for the HeLa Kyoto H2B-Red cells, Satyarebala Borusu for generating goat anti-Bub1 antibodies, Michaela Gerber, René Holtackers, and Sandra Maar (all ETH Zurich) for excellent technical assistance, and Ulrike Kutay (ETH Zurich), Silke Hauf (Max Planck Institute, Tübingen, Germany), Kevin Hardwick (University of Endinburgh, Endinburgh, Scotland, UK), Andrew McAinsh (Marie Curie Research Institute, Surrey, England, UK), and the Meraldi group for stimulating discussions. We thank the Light Microscopy Center of ETH Zurich for technical support.
This work was supported by the Deutsche Forschungsgemeinschaft (C. Klebig), Krebsliga Schweiz (P. Meraldi), and the Swiss National Science Foundation (P. Meraldi).
Footnotes
Abbreviations used in this paper: CENP, centromere protein; DN, dominant negative; dUTP, deoxy UTP; FRT, Flp recombination target; MCAK, mitotic centromere-associated kinesin; mRED, monomeric RED protein; MT, microtubule; UTR, untranslated region; wt, wild type.
References
- Bernard P., Hardwick K., Javerzat J.P. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis.J. Cell Biol. 143:1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y., Salic A., Dasso M., Arnaoutov A. 2007. Bub1 is essential for assembly of the functional inner centromere.J. Cell Biol. 176:919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B. 1998. Mutations of mitotic checkpoint genes in human cancers.Nature. 392:300–303 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface.Nat. Rev. Mol. Cell Biol. 9:33–46 [DOI] [PubMed] [Google Scholar]
- Chen R.H. 2004. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint.EMBO J. 23:3113–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Petersen S., Pacyna-Gengelbach M., Pietas A., Petersen I. 2003. Identification of a novel homeobox-containing gene, LAGY, which is downregulated in lung cancer.Oncology. 64:450–458 [DOI] [PubMed] [Google Scholar]
- Chen Y., Huhn D., Knosel T., Pacyna-Gengelbach M., Deutschmann N., Petersen I. 2005. Downregulation of connexin 26 in human lung cancer is related to promoter methylation.Int. J. Cancer. 113:14–21 [DOI] [PubMed] [Google Scholar]
- Doak S.H., Jenkins G.J., Parry E.M., Griffiths A.P., Baxter J.N., Parry J.M. 2004. Differential expression of the MAD2, BUB1 and HSP27 genes in Barrett’s oesophagus-their association with aneuploidy and neoplastic progression.Mutat. Res. 547:133–144 [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells.Nature. 411:494–498 [DOI] [PubMed] [Google Scholar]
- Fernius J., Hardwick K.G. 2007. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis.PLoS Genet. 3:e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemma A., Seike M., Seike Y., Uematsu K., Hibino S., Kurimoto F., Yoshimura A., Shibuya M., Harris C.C., Kudoh S. 2000. Somatic mutation of the hBUB1 mitotic checkpoint gene in primary lung cancer.Genes Chromosomes Cancer. 29:213–218 [DOI] [PubMed] [Google Scholar]
- Gillett E.S., Espelin C.W., Sorger P.K. 2004. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast.J. Cell Biol. 164:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoerup O.V., Wu J., Chandler-Militello D., Williams G.L., Zhao J., Schaffhausen B., Jat P.S., Roberts T.M. 2007. Surveillance mechanism linking Bub1 loss to the p53 pathway.Proc. Natl. Acad. Sci. USA. 104:8334–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempen P.M., Kurpad H., Calhoun E.S., Abraham S., Kern S.E. 2003. A double missense variation of the BUB1 gene and a defective mitotic spindle checkpoint in the pancreatic cancer cell line Hs766T.Hum. Mutat. 21:445. [DOI] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores.Mol. Biol. Cell. 12:1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells.Curr. Biol. 14:953–964 [DOI] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J.B., Liu S.T., Kao G.D., Muschel R., Chan G.K., Yen T.J. 2007. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments.J. Cell Biol. 177:413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis.J. Cell Biol. 179:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V.L., Scott M.I., Holt S.V., Hussein D., Taylor S.S. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression.J. Cell Sci. 117:1577–1589 [DOI] [PubMed] [Google Scholar]
- Kang J., Yang M., Li B., Qi W., Zhang C., Shokat K.M., Tomchick D.R., Machius M., Yu H. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1.Mol. Cell. 32:394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima S.A., Watanabe Y. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis.Nature. 427:510–517 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Hauf S., Ohsugi M., Yamamoto T., Watanabe Y. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting shugoshin localization.Curr. Biol. 15:353–359 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1.Dev. Cell. 13:663–676 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding.Dev. Cell. 16:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments.Nat. Cell Biol. 7:93–98 [DOI] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase.Dev. Cell. 16:118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity.EMBO J. 26:5033–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Sorger P.K. 2005. A dual role for Bub1 in the spindle checkpoint and chromosome congression.EMBO J. 24:1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression.Dev. Cell. 7:45–60 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time.Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- Niikura Y., Dixit A., Scott R., Perkins G., Kitagawa K. 2007. BUB1 mediation of caspase-independent mitotic death determines cell fate.J. Cell Biol. 178:283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K., Haraoka S., Yoshioka S., Hamasaki M., Fujiki T., Suzumiya J., Kawasaki C., Kanda M., Kikuchi M. 2000. Mutation analysis of mitotic checkpoint genes (hBUB1 and hBUBR1) and microsatellite instability in adult T-cell leukemia/lymphoma.Cancer Lett. 158:141–150 [DOI] [PubMed] [Google Scholar]
- Ouyang B., Knauf J.A., Ain K., Nacev B., Fagin J.A. 2002. Mechanisms of aneuploidy in thyroid cancer cell lines and tissues: evidence for mitotic checkpoint dysfunction without mutations in BUB1 and BUBR1.Clin. Endocrinol. (Oxf.). 56:341–350 [DOI] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint.Dev. Cell. 13:566–579 [DOI] [PubMed] [Google Scholar]
- Rischitor P.E., May K.M., Hardwick K.G. 2007. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes.PLoS ONE. 2:e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Farr K.A., Hoyt M.A. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase.Mol. Cell. Biol. 14:8282–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock K.L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A.L. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules.Cell. 78:761–771 [DOI] [PubMed] [Google Scholar]
- Ru H.Y., Chen R.L., Lu W.C., Chen J.H. 2002. hBUB1 defects in leukemia and lymphoma cells.Oncogene. 21:4673–4679 [DOI] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing.Curr. Biol. 14:942–952 [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H., Chen R.H. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity.J. Cell Biol. 153:1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichiri M., Yoshinaga K., Hisatomi H., Sugihara K., Hirata Y. 2002. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival.Cancer Res. 62:13–17 [PubMed] [Google Scholar]
- Shigeishi H., Oue N., Kuniyasu H., Wakikawa A., Yokozaki H., Ishikawa T., Yasui W. 2001. Expression of Bub1 gene correlates with tumor proliferating activity in human gastric carcinomas.Pathobiology. 69:24–29 [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. 2004a. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint.Mol. Cell. 16:387–397 [DOI] [PubMed] [Google Scholar]
- Tang Z., Sun Y., Harley S.E., Zou H., Yu H. 2004b. Human Bub1 protects centromeric sister-chromatid cohesion through shugoshin during mitosis.Proc. Natl. Acad. Sci. USA. 101:18012–18017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N.A., Mumby M.C., Yu H. 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation.Dev. Cell. 10:575–585 [DOI] [PubMed] [Google Scholar]
- Tange Y., Niwa O. 2008. Schizosaccharomyces pombe Bub3 is dispensable for mitotic arrest following perturbed spindle formation.Genetics. 179:785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage.Cell. 89:727–735 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase.J. Cell Biol. 142:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Harlow E. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control.Science. 262:2050–2054 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Valsdottir R., Javerzat J.P., Hardwick K.G. 2004. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p.Mol. Cell. Biol. 24:9786–9801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C.D., Brady D.M., Johnston R.C., Hanna J.S., Hardwick K.G., Spencer F.A. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins.Mol. Biol. Cell. 13:3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.L., Roberts T.M., Gjoerup O.V. 2007. Bub1: escapades in a cellular world.Cell Cycle. 6:1699–1704 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Decottignies A., Nurse P. 2003. Function of Cdc2p-dependent Bub1p phosphorylation and Bub1p kinase activity in the mitotic and meiotic spindle checkpoint.EMBO J. 22:1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]