Abstract
Alternative lengthening of telomeres (ALT) is a recombination-mediated process that maintains telomeres in telomerase-negative cancer cells. In asynchronously dividing ALT-positive cell populations, a small fraction of the cells have ALT-associated promyelocytic leukemia nuclear bodies (APBs), which contain (TTAGGG)n DNA and telomere-binding proteins. We found that restoring p53 function in ALT cells caused p21 up-regulation, growth arrest/senescence, and a large increase in cells containing APBs. Knockdown of p21 significantly reduced p53-mediated induction of APBs. Moreover, we found that heterochromatin protein 1 (HP1) is present in APBs, and knockdown of HP1α and/or HP1γ prevented p53-mediated APB induction, which suggests that HP1-mediated chromatin compaction is required for APB formation. Therefore, although the presence of APBs in a cell line or tumor is an excellent qualitative marker for ALT, the association of APBs with growth arrest/senescence and with “closed” telomeric chromatin, which is likely to repress recombination, suggests there is no simple correlation between ALT activity level and the number of APBs or APB-positive cells.
Introduction
The telomeres of human cells contain a linear tandem array of TTAGGG repeats bound by telomere-associated proteins, and are essential for chromosome stability and genomic integrity (de Lange, 2002). The progressive erosion of telomeres in normal cells during DNA replication eventually leads to the permanent arrest of cell division, which is referred to as replicative senescence. Telomere shortening and senescence appears to be a potent tumor suppression mechanism (Hanahan and Weinberg, 2000; Reddel, 2000). Cancer cells bypass senescence and achieve unlimited replicative potential by activating a telomere length maintenance pathway, either telomerase (Greider and Blackburn, 1985) or alternative lengthening of telomeres (ALT; Bryan et al., 1995). Telomerase is active in ∼85% of cancers (Shay and Bacchetti, 1997), and an ALT mechanism is active in many telomerase-negative tumors (Bryan et al., 1997; Henson et al., 2005). Although molecular details of the ALT mechanism are just beginning to be understood (Muntoni and Reddel, 2005), previous studies have indicated that ALT in human cells involves telomere–telomere recombination (Murnane et al., 1994; Dunham et al., 2000). With a few exceptions (Cerone et al., 2005; Fasching et al., 2005; Marciniak et al., 2005; Brachner et al., 2006), the hallmarks of human ALT-positive cells include (1) a unique pattern of telomere length heterogeneity, with telomeres that range from very short to greater than 50-kb long (Bryan et al., 1995), and (2) the presence of ALT-associated promyelocytic leukemia (PML) nuclear bodies (APBs) containing (TTAGGG)n DNA and telomere-specific binding proteins (Yeager et al., 1999).
PML bodies are found in most somatic cells; they increase in size and number when cells undergo cellular senescence, and are thus regarded as a marker of senescence (Jiang and Ringertz, 1997; Pearson et al., 2000; Ferbeyre et al., 2000). APBs are a subset of PML bodies that are present only in ALT cells, and are not found in mortal cells or telomerase-positive cells (Yeager et al., 1999). In addition to constitutive components of PML bodies such as PML and Sp100, and telomeric DNA and telomere-associated proteins such as TRF1, TRF2, TIN2, and RAP1 (Yeager et al., 1999; Wu et al., 2003; Jiang et al., 2007), they also contain other proteins involved in DNA replication, recombination, and repair including RAD51, RAD52, and RPA (Yeager et al., 1999); RAD51D (Tarsounas et al., 2004); BLM (Yankiwski et al., 2000; Stavropoulos et al., 2002); WRN (Johnson et al., 2001); RAP1 and BRCA1 (Wu et al., 2003); MRE11, RAD50, and NBS1 (Wu et al., 2000; Zhu et al., 2000); ERCC1 and XPF (Zhu et al., 2003); hRAD1, hRAD9, hRAD17, and hHUS1 (Nabetani et al., 2004); Rif1 (Silverman et al., 2004); and hnRNP A2 (Moran-Jones et al., 2005). Formation of APBs requires NBS1, which recruits MRE11, RAD50, and BRCA1 into these structures (Wu et al., 2003; Jiang et al., 2005). We induced APB accumulation with methionine restriction, and used RNAi-based screening to extend the list of proteins required for APB formation to include PML, TRF1, TRF2, TIN2, RAP1, MRE11, and RAD50 (Jiang et al., 2007). It was recently found (Potts and Yu, 2007) that the structural maintenance of chromosomes SMC5/6 complex localizes to APBs in ALT cells and sumoylates TRF1 and TRF2, and this plays an essential role in APB formation. It has long been suggested that APBs may have an integral role in the ALT mechanism (Yeager et al., 1999; Grobelny et al., 2000; Wu et al., 2000; Molenaar et al., 2003; Wu et al., 2003), and, consistent with this suggestion, inhibition of ALT in some somatic cell hybrids formed by fusion of ALT and telomerase-positive cell lines resulted in a substantial decrease in APBs (Perrem et al., 2001). Furthermore, our recent study showed that inhibition of ALT is accompanied by suppression of APBs, providing evidence for a direct link between APBs and ALT activity (Jiang et al., 2005; Zhong et al., 2007).
Although we speculated that the increase in APB-positive cells after methionine starvation may have resulted from reduced levels of methylation at telomeric and subtelomeric regions, and a consequent increase in telomeric recombination events (Jiang et al., 2007), it also remained a possibility that the increase in APBs was instead related to cell cycle arrest. To clarify this, we examined the effect of activating wild-type (wt) p53 on APB formation in p53-negative ALT cells. We found that activation of p53 up-regulated p21, and caused growth arrest and senescence, accompanied by a very large increase in APB formation. This upsurge in APB numbers was substantially prevented by siRNA-mediated knockdown of p21, indicating that p21 is a major downstream p53 effector of APB induction. Moreover, both p21 and its binding partner proliferating cell nuclear antigen (PCNA) were found to be present inside APBs, but knockdown of PCNA did not affect p53/p21-mediated APB induction.
Because APBs contain telomeric chromatin, which is heterochromatic in nature, we have also investigated whether APBs are associated with the heterochromatin protein 1 (HP1) family. The HP1 family plays a critical role in establishing and maintaining transcriptionally inactive heterochromatin, including that of telomeres. Three mammalian HP1 proteins have been identified and are known as HP1α, HP1β, and HP1γ (Eissenberg and Elgin, 2000). They are encoded by distinct genes localized on three different chromosomal sites (Chevillard et al., 1993), and they are small proteins, with <200 amino acids and molecular masses of ∼25 kD. HP1 proteins are nonhistone chromatin components that interact with a variety of proteins that play a role in chromatin remodeling and transcriptional silencing (Ma et al., 2001). We found that all three members of the HP1 family—HP1α, β, and γ—are present in APBs. Knockdown of HP1α or HP1γ, but not HP1β, significantly decreased the p53/p21-mediated APB induction, which suggests that HP1α- and HP1γ-mediated chromatin compaction is required for APB formation.
These results indicate that APBs form in growth-arrested cells, and that in this context they contain “closed” telomeric chromatin, and are therefore not likely to be sites for telomere–telomere recombination. These data indicate that it is unlikely that there is a direct correlation between APB numbers and the ALT activity level.
Results
Activation of the p53 pathway in p53-negative ALT cells induces formation of APBs
Large APBs are usually found in ∼5% of cells within asynchronously growing ALT cell populations (Yeager et al., 1999). The proportion of APB-positive cells can be greatly increased by methionine starvation (Jiang et al., 2007) or by DNA-damaging agents (Fasching et al., 2007). In each of these cases, the treatments that induced APBs also caused growth arrest (Fasching et al., 2007; Jiang et al., 2007). We therefore addressed the question of whether the induction of APBs is directly related to growth arrest by restoring p53 function in p53-negative ALT cells. First, we examined the effect of activating wt p53 on APB formation in two p53–estrogen receptor (ER) fusion gene-transfected IIICF/c cell lines—c/p53ER/7 and c/p53ER/8 (abbreviated to C7 and C8)—in which p53 function can be activated by exposure to 4-hydroxytamoxifen (4OHT; Homer et al., 2005). IIICF/c is an ALT cell line (Rogan et al., 1995) derived from IIICF Li-Fraumeni syndrome fibroblasts containing one mutant (essentially null) and one wt TP53 allele (Warneford et al., 1992), which became immortalized spontaneously via a series of genetic changes that included loss of the wt TP53 allele (Rogan et al., 1995).
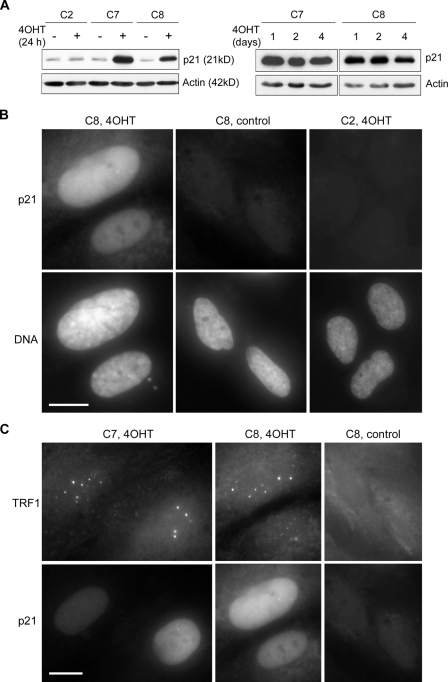
4OHT-treated C7 and C8 cells had up-regulated p21 levels at 24 h, and a high level of expression was maintained until day 4 of treatment (Fig. 1, A and B). Up-regulation of p21 was not seen in the ethanol vehicle-treated controls, nor in a 4OHT-treated IIICF/c control clone, c/ER/2 (abbreviated to C2), that had been transfected with an ER construct only (Fig. 1, A and B). Increased p21 expression in 4OHT-treated C7 and C8 cultures was accompanied by a significant increase in the proportion of cells containing APBs (detected here as large TRF1 foci), most of which were also p21 positive (Fig. 1 C and Table I). The basal levels of APB-positive cells in the vehicle controls were higher than those of most ALT cells under normal conditions of asynchronous growth. This was due in part to continuous selection of the cells in 1 µg/ml puromycin (unpublished data), and is presumably also partly due to a low level of leakiness of the p53ER inducible system.
Figure 1.
Induction of APBs in C7 and C8 cells upon 4OHT-mediated activation of p53. (A) p21 was up-regulated in C7 and C8 cells, which express a p53-ER fusion protein, after 1, 2, or 4 d of 4OHT-treatment, but not in C2 cells, which express a control ER protein. The Western blot was probed with the indicated antibodies. (B) p21 staining of C2 and C8 cells treated with 4OHT or ethanol vehicle for 4 d. Only 4OHT-treated C8 cells showed strong p21 staining. (C) Double immunostaining of TRF1 and p21 in C7 and C8 cells treated with 4OHT or ethanol vehicle for 4 d. Large TRF1 foci were induced in cells positive for p21. Bars, 20 µm.
Table I.
Proportion of APB-positive cells after treatment with 4OHT or vehicle
| Cell lines | Treatmenta | APB+/total (%) |
| c/ER/2 | EtOH | 26/239 (10.9) |
| c/ER/2 | 4OHT | 28/215 (13.0) |
| c/p53ER/7 | EtOH | 41/238 (17.2) |
| c/p53ER/7 | 4OHT | 110/234 (47.0) |
| c/p53ER/8 | EtOH | 47/258 (18.2) |
| c/p53ER/8 | 4OHT | 108/240 (45.0) |
Cells were treated with 1 µM 4OHT or 0.01% ethanol (EtOH) for 4 d before being fixed for immunostaining.
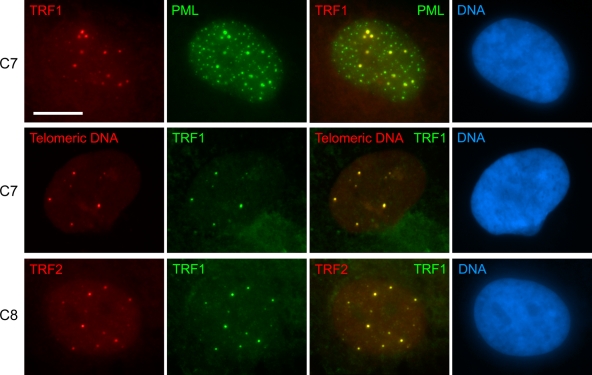
To demonstrate that the TRF1 foci were APBs, we showed that they also contained telomeric DNA, TRF2, and PML protein (Fig. 2). APBs are usually somewhat larger than the PML bodies in ALT cells that do not contain telomeric contents, and the quantity of telomeric DNA and telomeric binding proteins that they contain is often greater than the amount present at individual telomeres (Fig. 2). Because of a tight correlation between foci of telomeric DNA and of TRF1 or TRF2, APBs can be detected interchangeably by either telomeric FISH or immunostaining of TRF1 or TRF2 (Jiang et al., 2005, 2007). In this study, APBs were generally detected by visualizing TRF1 or TRF2 within a PML body, but were also identified as large, bright TRF1, TRF2, or telomeric DNA foci (Fig. 2). The 4OHT-treated C7 and C8 cultures contained 2.6-fold more APB-positive cells than the control cultures that were treated with ethanol vehicle alone (Table I). Changes in the proportion of APB-positive cells were minimal in 4OHT-treated C2 cells (Table I).
Figure 2.
APBs can be detected interchangeably by either telomeric FISH or immunostaining of TRF1 or TRF2. After 4 d of 4OHT treatment, APBs were induced in C7 and C8 cells, where colocalization was observed between prominent TRF1 foci and large PML bodies (top), between telomeric DNA and TRF1 foci (middle), and between TRF1 and TRF2 foci (bottom). Bar, 20 µm.
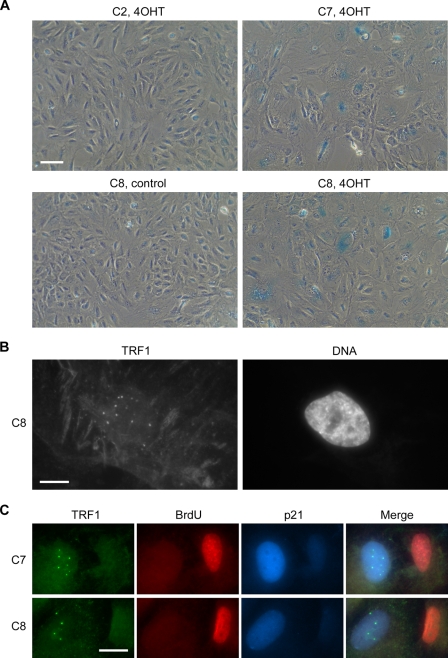
After 4 d of 4OHT treatment, many of the cells had undergone the characteristic morphological changes of senescence and stained positive for senescence-associated (SA) β-galactosidase (SA-β-gal) activity (Fig. 3 A). Most of the cells that were morphologically senescent contained APBs, as illustrated in Fig. 3 B, and triple staining for BrdU, p21, and TRF1 revealed that most of the APB-positive cells were p21 positive and BrdU negative (Fig. 3 C and Table II). Based on these data, we conclude that the APBs that were induced upon restoration of p53 activity in C7 and C8 cultures mostly occurred in growth-arrested or phenotypically senescent cells.
Figure 3.
Induction of APBs is associated with p53/p21-mediated senescence. (A) SA-β-gal staining of C2, C7, and C8 cells treated with 4OHT or ethanol vehicle for 4 d. SA-β-gal expression was found in 4OHT-treated C7 and C8 cells. (B) TRF1 and DAPI staining of C8 cells treated with 4OHT for 3 d. APBs were found in phenotypically senescent cells. (C) Triple immunostaining of TRF1, BrdU, and p21 in C7 and C8 cells after treatment with 4OHT for 4 d, and with BrdU for 24 h before the end of 4OHT treatment. APBs (visualized here as large TRF1 foci) were found mainly in cells staining positive for p21 and negative for BrdU. Bars: (A) 100 µm; (B and C) 20 µm.
Table II.
Frequencies of p21-positive and/or BrdU-negative cells within the APB-positive populations after a 4-d induction of p53 and a 24-h pulse of BrdU
| Cell lines (treatmenta) | Total APB+ countedb | p21+ (%) | BrdU− (%) | p21+ and BrdU− (%) |
| C7 (4OHT) | 150 | 132 (88.0) | 129 (86.0) | 122 (81.3) |
| C8 (4OHT) | 127 | 105 (82.7) | 106 (83.5) | 94 (74.0) |
| IIICF-T/B3(SV40T-siRNA) | 153 | 128 (83.7) | 131 (85.6) | 115 (75.2) |
Cells were treated with 1 µM 4OHT or 10 nM SV40T siRNA for 4 d, and BrdU was added 24 h before the end of treatment.
Only cells that were APB positive were examined for BrdU and p21.
p21 is a key downstream effector of p53 for APB induction
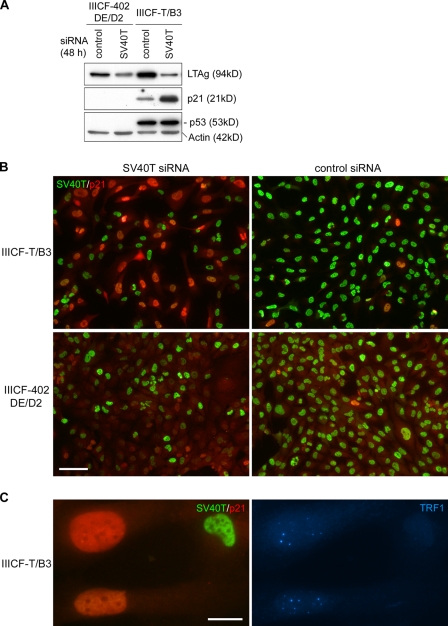
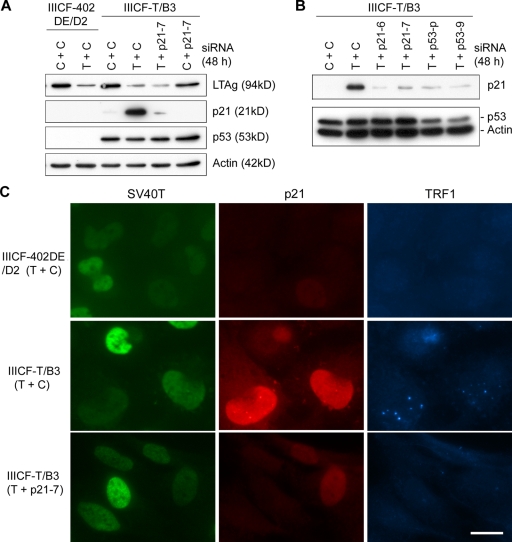
To confirm the involvement of the p53 pathway in APB induction, we also examined APB formation in an ALT cell line, IIICF-T/B3, that was established by transfecting IIICF cells with an SV40 early region expression plasmid (Maclean et al., 1994); these cells retain a wt TP53 allele. We restored p53 function by knockdown of SV40 large T antigen (LTAg) with an siRNA against the SV40 early region transcripts (SV40T siRNA). Because knockdown of LTAg in IIICF-T/B3 cells restored the function not only of p53 but also of other proteins, including the retinoblastoma protein (pRb) family members that also bind to LTAg, we used another ALT cell line, IIICF-402DE/D2, as a control to determine whether pathways other than p53 were involved. IIICF-402DE/D2 was, like IIICF-T/B3, derived from the same parental IIICF cells, but was transfected with a mutant SV40 early region plasmid, 402DE with a D→E mutation at LTAg amino acid 402 (Maclean et al., 1994). This mutant LTAg is disabled for p53 binding, and as a consequence, the wt TP53 allele was deleted spontaneously during immortalization of IIICF-402DE/D2 cells. Therefore, no p53 expression was detectable before or after LTAg knockdown (Fig. 4 A).
Figure 4.
Induction of APBs in SV40-immortalized ALT cells by siRNA-mediated knockdown of LTAg. (A) Western blots showed decreased LTAg in both IIICF-T/B3 (containing wt LTAg and one wt TP53 allele) and IIICF-402DE/D2 (containing mutant LTAg that does not bind p53, and no wt TP53 alleles) cells 48 h after SV40T siRNA transfection. Treatment with SV40T siRNA induced p21 in IIICF-T/B3 but not in IIICF-402DE/D2 cells. The blots were probed with the indicated antibodies. (B) Double immunofluorescence of SV40T and p21 in IIICF-T/B3 and IIICF-402DE/D2 cells treated with SV40T or control siRNAs for 4 d. Strong p21 staining was detected in IIICF-T/B3 cells depleted of SV40T. (C) IIICF-T/B3 cells were triple stained for TRF1, p21, and SV40T 4 d after SV40T siRNA transfection. APBs (visualized here as large TRF1 foci) were observed in cells with high levels of p21. Bars: (B) 100 µm; (C) 20 µm.
As expected, p21 expression increased in IIICF-T/B3 but not in IIICF-402DE/D2 cells after siRNA-mediated knockdown of LTAg (Fig. 4, A and B), and this induced a senescent phenotype in IIICF-T/B3 (Fig. S1) but not IIICF-402DE/D2 cells (unpublished data). Notably, induction of senescence was accompanied by a significant increase in APB formation in IIICF-T/B3 cells after a 4-d period of SV40T siRNA treatment (Fig. 4 C). Consistent with the results from 4OHT-treated C7 and C8 cells, most of the APB-positive IIICF-T/B3 cells were p21 positive and BrdU negative (Table II and Fig. S2). APBs were found in ∼55% of the IIICF-T/B3 cells that were depleted of LTAg, as compared with ∼10% of the IIICF-402DE/D2 cells where LTAg was undetectable (Fig. 5 C and Table III). This was not caused by an intrinsic difference in the ability of the two cell lines to form APBs because, despite the difference in p53 status, IIICF-402DE/D2 and IIICF-T/B3 cultures displayed a similar increase in the proportion of APB-positive cells in response to methionine starvation (Table S1). APBs were also induced in another SV40-immortalized ALT cell line, JFCF-6/T.1J/1D, upon knockdown of SV40 LTAg (Table III), but no APBs were found in the isogenic control telomerase-positive cell line, JFCF-6/T.1J/6B, which was SV40-immortalized from the same parental cell as the JFCF-6/T.1J/1D cells (Fig. S3).
Figure 5.
p21 is the major downstream effector of p53 for APB induction. (A and B) Induction of p21 in IIICF-T/B3 cells by SV40T siRNA was effectively abrogated by siRNAs against p21 (p21-6 and p21-7) or p53 (p53-p and p53-9). The Western blots were probed with the indicated antibodies. (C) Triple immunostaining of TRF1, p21, and SV40T in IIICF-T/B3 and IIICF-402DE/D2 cells treated with the indicated combinations of siRNAs for 4 d. p21 siRNA (p21-7) largely prevented SV40T siRNA-mediated induction of APBs. C, control siRNA; T, SV40T siRNA. Bar, 20 µm.
Table III.
Proportion of APB-positive cells after siRNA treatment
| Cell lines | siRNA treatmenta | APB+/SV40T−b (%) |
| IIICF-T/B3 | SV40T + C | 98/170 (57.6) |
| IIICF-402DE/D2 | SV40T + C | 20/176 (11.4) |
| IIICF-T/B3 | SV40T + p21-7 | 43/172 (25.0) |
| IIICF-402DE/D2 | SV40T + p21-7 | 18/179 (10.1) |
| JFCF-6/T.1J/1D | SV40T + C | 85/137 (62.0) |
| JFCF-6/T.1J/1D | SV40T + p21-6 | 36/132 (27.3) |
| IIICF-T/B3 | SV40T + C | 87/156 (55.8) |
| IIICF-T/B3 | SV40T + p21-6 | 34/158 (21.5) |
| IIICF-T/B3 | SV40T + p21-7 | 38/151 (25.2) |
| IIICF-T/B3 | SV40T + p53-9 | 29/153 (19.0) |
| IIICF-T/B3 | SV40T + p53-p | 38/160 (23.8) |
| IIICF-T/B3 | SV40T + PCNA-1 | 81/154 (52.6) |
| IIICF-T/B3 | SV40T + PCNA-6 | 82/149 (55.0) |
C, nonsilencing control siRNA.
Cells were treated with 10 nM siRNA per target for 4 d before being fixed for immunostaining.
Only cells that were negative by immunostaining for SV40T were examined for APBs.
Because a close correlation was found between APB formation and high levels of p21 (Figs. 1 C and 4 C; and Table II), we tested whether p21 plays an important role in p53-mediated induction of APBs in p53-negative ALT cells. p21 siRNAs (p21-6 or p21-7) were used in combination with SV40T siRNA to simultaneously knock down p21 and LTAg in IIICF-T/B3 cells. Western analysis showed that the induction of p21 by knockdown of LTAg was effectively blocked by either p21-6 or p21-7 siRNAs (Fig. 5, A and B). This resulted in a substantial reduction in the proportion of IIICF-T/B3 cells that became senescent (Fig. S1). Importantly, induction of APBs was reduced by >50% when IIICF-T/B3 cells were treated with siRNAs against both SV40T and p21 (Fig. 5 C and Table III). p53 siRNAs (p53-9 or p53-p) antagonized induction of APBs by SV40T siRNA to the same extent as p21 siRNAs (Fig. 5 B and Table III). These data indicate that p21 is a major downstream effector of p53 for APB formation.
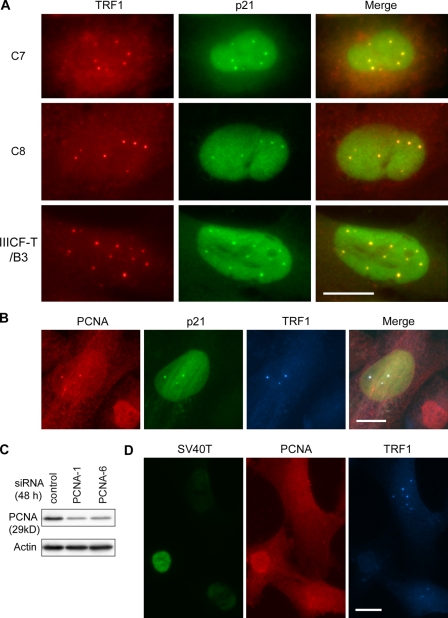
p21 and its binding partners PCNA and Cdk2 are localized in APBs
We found that p21 was present inside APBs in ∼30% of the APB-positive cells where the p53 pathway was restored (Fig. 6 A and Table S2), showing for the first time that a cell cycle regulatory protein was physically associated with APBs and PML bodies. To confirm the immunostaining results, we used two different p21 antibodies (see Materials and methods), and checked the specificity of each antibody by siRNA knockdown. This localization of p21 appears to be specific to ALT cells and APBs, as it was not seen in PML bodies in senescent normal IMR-90 fibroblasts (unpublished data). To determine what type of role p21 plays inside APBs, we first examined the localization of its binding partner PCNA in IIICF-T/B3 cells upon knockdown of LTAg because PCNA, along with p21, has been suggested to be involved in DNA repair (Li et al., 1994; Perucca et al., 2006). Triple immunostaining of p21, PCNA, and TRF1 revealed the coexistence of PCNA and p21 inside APBs, but only in a fraction of APB-positive cells (Fig. 6 B). The presence of PCNA in APBs did not require restoration of the p53 pathway because it was also found in APBs within the parental ALT cell line, IIICF/c, which is essentially p53 null (Fig. S4 A). In addition, p21 colocalized with another of its binding partners, Cdk2, inside APBs in a fraction of APB-positive cells (Fig. S4 B). However, in contrast to PCNA, Cdk2 was not present inside APBs in p53-negative IIICF/c cells (unpublished data), which indicates that it is unlikely to be involved in APB formation. Because of its involvement in DNA repair, we further investigated the role of PCNA by transfecting IIICF-T/B3 cells with a combination of SV40T siRNA and PCNA siRNAs (PCNA-1 or PCNA-6), the effectiveness of which was demonstrated by Western analysis (Fig. 6 C). We found that knockdown of PCNA did not block APB induction in cells treated with SV40T siRNA (Fig. 6 D), which contrasted with the results from the control where p21 siRNA was used instead of PCNA siRNA (Table III). These results indicate that PCNA is not required for APB formation.
Figure 6.
Presence of p21 and its binding partner, PCNA, in APBs. (A) Double immunostaining of TRF1 and p21 in C7 cells treated with 4OHT for 1 d, C8 cells treated with 4OHT for 4 d, and IIICF-T/B3 cells treated with a combination of SV40T and control siRNAs for 4 d. p21 was detected inside APBs. (B) Triple immunostaining of TRF1, p21, and PCNA in IIICF-T/B3 cells treated with a combination of SV40T and control siRNAs for 3 d. p21 and PCNA were colocalized in APBs. (C) The effectiveness of PCNA siRNAs (PCNA-1 and PCNA-6) was demonstrated by Western blots. (D) Triple staining of TRF1, SV40T, and PCNA in IIICF-T/B3 cells treated with combination of SV40T and PCNA-6 siRNAs for 4 d. APBs were still detected in cells depleted of both SV40T and PCNA. Bars, 20 µm.
HP1α and HP1γ, but not HP1β, are required for APB formation
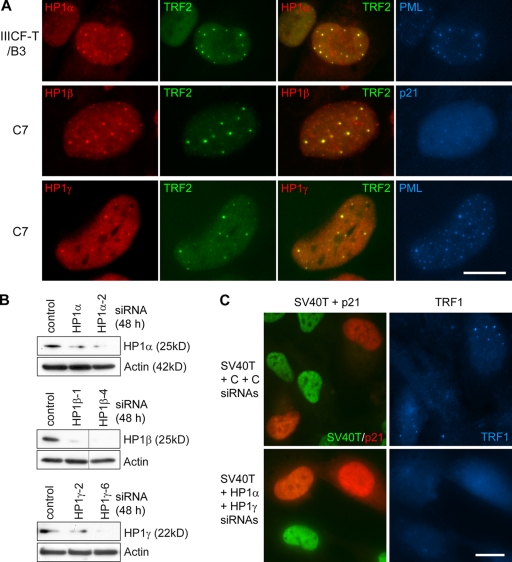
Upon cellular senescence, heterochromatin becomes highly compacted by a process that involves HP1 family members (Funayama and Ishikawa, 2007). APBs contain telomeric chromatin, which is heterochromatic in nature. We therefore addressed the question of whether chromatin compaction is required for APB formation upon p53/p21-mediated growth arrest/senescence. We examined the localization of HP1 family proteins in C7, C8, and IIICF-T/B3 cells upon restoration of the p53 pathway, and found that all three members of the HP1 family—HP1α, HP1β and HP1γ—are present in APBs (Fig. 7 A). This led to a further analysis to determine whether members of the HP1 family are required for APB formation. Double knockdown of LTAg and HP1 proteins was performed in IIICF-T/B3 cells with SV40T siRNA and the siRNAs against HP1α (HP1α or HP1α-2), HP1β (HP1β-1 or HP1β-4), or HP1γ (HP1γ-2 or HP1γ-6), the effectiveness of which was demonstrated by Western analysis (Fig. 7 B). We found that knockdown of HP1α or HP1γ inhibited the p53/p21-mediated induction of APBs by ∼40%, whereas knockdown of HP1β only slightly reduced the increase in APBs (Table IV). Moreover, simultaneous knockdown of HP1α and HP1γ had an additive effect in inhibiting APB formation by ∼60% (Fig. 7 C and Table IV). To know whether HP1 proteins are involved in APB formation in the absence of p53, we performed a similar set of experiments on IIICF/c cells that were p53 negative. HP1 proteins were found to be present in APBs in exponentially dividing or methionine-restricted IIICF/c cells (Fig. S5 and unpublished data). Also, simultaneous knockdown of HP1α and HP1γ largely prevented induction of APBs in methionine-restricted cells (Fig. S5 B and Table S3). In summary, the data indicate that HP1α and HP1γ are required for APB formation.
Figure 7.
HP1 localization in APBs and effects on APB formation of HP1 depletion. (A) Triple immunostaining showed colocalization between HP1α and APBs in IIICF-T/B3 cells treated with SV40T siRNA for 4 d (top), between HP1β and APBs (middle), and between HP1γ and APBs (bottom) in C7 cells treated with 4OHT for 4 d. (B) The effectiveness of individual siRNAs for HP1α (HP1α and HP1α-2), for HP1β (HP1β-1 and HP1β-4), and for HP1γ (HP1γ-2 and HP1γ-6) was demonstrated by immunoblotting. The black lines indicate that redundant lanes within the same gel have been spliced out. (C) IIICF-T/B3 cells were triple stained for TRF1, p21, and SV40T 4 d after transfection of the indicated combinations of siRNAs. Simultaneous treatment with HP1α (HP1α-2) and HP1γ (HP1γ-6) siRNAs prevented SV40T siRNA-mediated induction of APBs. C, control siRNA; T, SV40T siRNA. Bars, 20 µm.
Table IV.
Proportion of APB-positive IIICF-T/B3 cells after siRNA treatment
| siRNA treatmenta | Total (SV40T−)b | APB+ (%) |
| SV40T + C | 117 | 63 (53.8) |
| SV40T + HP1α | 111 | 40 (36.0) |
| SV40T + HP1α-2 | 120 | 38 (31.7) |
| SV40T + HP1β-1 | 123 | 56 (45.5) |
| SV40T + HP1β-4 | 124 | 61 (49.2) |
| SV40T + HP1γ-2 | 109 | 38 (34.9) |
| SV40T + HP1γ-6 | 128 | 41 (32.0) |
| SV40T + C + C | 94 | 53 (56.4) |
| SV40T + C + p21-6 | 98 | 25 (25.5) |
| SV40T + C + HP1α | 97 | 33 (34.0) |
| SV40T + C + HP1β-1 | 100 | 47 (47.0) |
| SV40T + C + HP1γ-6 | 106 | 30 (28.3) |
| SV40T + HP1α + HP1β-1 | 105 | 32 (30.5) |
| SV40T + HP1α + HP1γ-6 | 110 | 24 (21.8) |
| SV40T + HP1β-1 + HP1γ-6 | 99 | 31 (31.3) |
| SV40T + C + C | 111 | 58 (52.3) |
| SV40T + C + HP1α-2 | 106 | 32 (30.2) |
| SV40T + HP1α-2 + HP1α-2 | 119 | 33 (27.7) |
| SV40T + HP1α-2 + HP1γ-6 | 107 | 21 (19.6) |
C, nonsilencing control siRNA.
Cells were treated with 10 nM siRNA per target for 4 d before being fixed for immunostaining.
Only cells that were negative by immunostaining for SV40T were examined for APBs.
Discussion
Nearly all ALT cell lines have a dysfunctional p53 pathway, and consequently it has been speculated that mutation of p53 might be a contributing factor for ALT activation (Henson et al., 2002; Razak et al., 2004). In the present study, we have shown that restoration of p53 function in ALT cells causes up-regulation of p21, growth arrest, and senescence, as well as a large increase in APB formation. These observations suggest that methionine restriction–induced APB induction (Jiang et al., 2007) results from growth arrest rather than some other mechanism such as decreased DNA methylation. They also indicate that the use of cell cycle blocking agents may not be an appropriate approach for determining whether there is a relationship between the cell cycle phase and expression of APBs. In this study, restoration of p53 function resulted in APB induction in cells that were arrested predominantly in the G1 phase of the cell cycle (unpublished data), but because APBs are also found in cells arrested in late S/G2/M phases of the cell cycle (Grobelny et al., 2000; Wu et al., 2000), it seems likely that the correlation is between APB formation and growth arrest rather than with a particular cell cycle phase. Although large APBs are found in ∼5% of cells within asynchronously dividing ALT cell populations (Yeager et al., 1999), it seems likely that most of these have spontaneously undergone growth arrest and are either quiescent or senescent, as most APB-positive cells in these populations do not incorporate BrdU within a time period exceeding the mean cell doubling time, and many of these also display the enlarged, flattened morphology characteristic of senescence (unpublished data).
It has previously been shown that overexpression of p53 can induce a senescence-like growth arrest in tumor cells (Sugrue et al., 1997; Ling et al., 2000), and that activation of endogenous p53 in the U-2 OS ALT cell line can induce senescence (Stagno D'Alcontres et al., 2007). Among the multiple genes activated by p53, p21 is a crucial transcriptional target of p53 and a mediator of p53-dependent senescence (Brown et al., 1997). p21 is a pleiotropic inhibitor of different cyclin/Cdk complexes (Dotto, 2000), the induction of which can cause cell cycle arrest and senescence in normal cells (Noda et al., 1994) and phenotypic senescence in tumor cells (Chang et al., 1999; Fang et al., 1999; Kagawa et al., 1999; Wang et al., 1999). It has also been shown that infection of spontaneously immortalized Li-Fraumeni syndrome cells, either telomerase positive or negative, with a p21 retroviral vector resulted in senescence (Vogt et al., 1998). Consistent with these results, our data showed that high levels of p21 correlated with p53-mediated senescence in ALT cells. Moreover, knockdown of p21 inhibited induction of the senescent phenotype and suppressed p53-mediated induction of APBs, which is consistent with p21 being a major regulator of p53-mediated senescence, and indicates that p21 is a major downstream effector of p53 for APB induction.
Our finding that p21 is present inside APBs was not entirely unexpected, as APBs contain substantial amounts of telomeric DNA, some of which is linear and may be recognized as DNA damage (Fasching et al., 2007). Although it is a cell cycle regulatory protein, p21, along with PCNA, has been suggested to play a role in DNA repair (Li et al., 1994; Perucca et al., 2006). The coexistence of PCNA and p21 inside APBs in a small fraction of ALT cells suggests that p21 and PCNA may be involved in DNA repair inside APBs. Nevertheless, PCNA was not required for formation of APBs because knockdown of PCNA did not affect p53/p21-mediated APB induction. Moreover, the presence of p21 and another of its binding partners, Cdk2, inside APBs in p53-reconstituted but not p53-negative ALT cells suggests that localization of both proteins into PML bodies is unlikely to be a prerequisite for APB formation.
We have also shown here for the first time that all three members of the HP1 protein family, HP1α, β, and γ, were present in APBs. This is in agreement with previous findings that human and mouse telomeres are enriched for HP1 (Koering et al., 2002; Garcia-Cao et al., 2004; Gonzalo et al., 2005, 2006). Upon cellular senescence, heterochromatin becomes highly compacted by a process that involves HP1 family members (Funayama and Ishikawa, 2007), which have previously been shown to associate with PML bodies (Seeler et al., 1998), including in normal senescent fibroblasts (Zhang et al., 2005). HP1 proteins have also been found in the giant PML body that associates with juxtacentromeric satellite DNA during G2 phase in cells from individuals with immunodeficiency, centromeric instability, and facial dysmorphy (ICF) syndrome, and on the basis of this cell cycle timing, the authors suggested that the HP1 proteins are most likely involved in reestablishment of the heterochromatic state of late-replicating juxtacentromeric satellite DNA (Luciani et al., 2006). Our demonstration that knockdown of HP1α and/or HP1γ significantly inhibited p53/p21-mediated APB induction and also inhibited formation of APBs in methionine-restricted cell populations shows for the first time that these proteins are not only present in APBs but are also required for their formation, and suggests that HP1α- and HP1γ-mediated chromatin compaction is involved in this process. It should be pointed out that knockdown of HP1β has only minor effects on APB formation as compared with HP1α and HP1γ. This could be due to HP1 isoform-specific effects on telomeres, which have been demonstrated by a previous study on overexpression of HP1 isoforms in telomerase-positive cells (Sharma et al., 2003).
It has previously been shown that the MRE11/RAD50/NBS1 (MRN) complex (Wu et al., 2003; Jiang et al., 2005) and shelterin proteins (Jiang et al., 2007) are required for APB formation, although under different experimental conditions, depletion of TRF2 did not always inhibit APB formation (Stagno D'Alcontres et al., 2007). We therefore proposed a model in which telomeric DNA binds to the MRN complex via the shelterin component RAP1, and then translocates to PML bodies to form APBs (Jiang et al., 2007). The data presented here showing that HP1 proteins are also required for APB formation raise the question of what role they play in this process, and whether shelterin and MRN proteins may interact with HP1 proteins at telomeres. HP1 proteins are usually recruited to chromatin through their affinity for trimethylated H3K9 residues (Lachner et al., 2001; Garcia-Cao et al., 2004), but it is also possible that this occurs through the interactions between TRF1 and the HP1-interacting developmental regulator SALL1 (Netzer et al., 2001) or between TIN2 and HP1 (Kaminker et al., 2005). Another interesting possibility is suggested by the observation that MRN is required for recruitment of HP1 proteins to the Drosophila telomere (Ciapponi et al., 2004); at mammalian telomeres, although HP1 and MRN proteins are known to be present (for review see Blasco, 2007), a role for MRN in recruitment of HP1 has not yet been demonstrated. Based on these data and the known role of HP1 proteins in compaction of chromatin, we propose that the role of HP1 proteins in formation of APBs may be to compact the telomeric DNA, possibly as a prerequisite for its translocation to PML bodies, and this may be mediated at least in part by some indirect interactions of HP1 proteins with telomeric DNA via shelterin or MRN proteins. Furthermore, as proposed for juxtacentromeric satellite DNA in G2 (Luciani et al., 2006), it seems possible that HP1 may tether telomeric DNA into PML bodies by interacting with sumoylated proteins via ATRX and DAXX.
APBs have long been suggest to play an integral role in the ALT mechanism (Yeager et al., 1999; Grobelny et al., 2000; Wu et al., 2000; Molenaar et al., 2003; Wu et al., 2003), based on the observations that they contain telomeric DNA and proteins involved in recombination and DNA repair (Yeager et al., 1999; Wu et al., 2000; Yankiwski et al., 2000; Zhu et al., 2000, 2003; Johnson et al., 2001; Stavropoulos et al., 2002; Wu et al., 2003; Nabetani et al., 2004; Tarsounas et al., 2004), and that they are sites of DNA synthesis (Wu et al., 2000, 2003; Nabetani et al., 2004). However, the results from our study show that APBs are induced in growth-arrested, phenotypically senescent cells, and that HP1, which compacts heterochromatic DNA, is required for this process, suggesting that the telomeric DNA inside the APBs associated with growth arrest is in a state that is unlikely to permit telomere–telomere recombination. These data indicate that it is not likely that there is a simple correlation between the number of APB-positive cells in an ALT population and the level of ALT activity.
Materials and methods
Cell culture
The spontaneously immortalized Li-Fraumeni syndrome fibroblast line IIICF/c (Rogan et al., 1995) and SV40-immortalized human fibroblast lines JFCF-6/T.1J/1D and JFCF-6/T.1J/6B were cultured in DME (Invitrogen). The p53-ER fusion gene or ER-only transfected IIICF/c cell lines c/p53ER/7 (C7), c/p53ER/8 (C8), and c/ER/2 (C2) were cultured in phenol red–free DME (Invitrogen) containing 1 µg/ml puromycin (Homer et al., 2005). IIICF cells immortalized with SV40 LTAg that was wt (IIICF-T/B3) or mutant (IIICF-402DE/D2) (Maclean et al., 1994) were cultured in RPMI 1640 medium (Invitrogen). All culture media contained 10% fetal bovine serum (FBS) and 50 µg/ml gentamicin, and cultures were incubated in a 5% CO2 humidified atmosphere at 37°C.
For p53 induction experiments, C2, C7, and C8 cells were seeded in phenol red–free DME and grown to 30–40% confluency. The cell cultures were treated with 1 µm 4OHT or 0.01% ethanol as a vehicle control. After various time periods, cells were either fixed for immunostaining or harvested for isolation of protein.
For methionine restriction, cells were seeded in normal medium and grown to 30–40% confluency. Cells were washed once with methionine-free medium before changing to this medium. After 4 d, cells were fixed for immunostaining. Methionine-deficient medium was reconstituted from methionine- and cystine-deficient DME (Invitrogen) by adding l-cystine (48 mg/liter; Sigma-Aldrich).
Antibodies
The following antibodies were used in this study: mouse anti-p21, anti-TRF2, anti-Cdk2, and anti-BrdU (BD); goat anti-p21 (R&D Systems); rabbit anti-p53 (FL-393), goat anti-PCNA (C-20), goat anti-PML (N-19), and mouse anti-PML (Santa Cruz Biotechnology, Inc.); rabbit anti-PML and anti-Sp100 (Millipore); mouse anti-TRF2 and anti-HP1α (Millipore); rabbit anti-HP1α, anti-HP1β, and anti-HP1γ (Cell Signaling Technology); and rabbit anti-HP1β and anti-HP1γ (Proteintech Group). Mouse anti-SV40T (PAb108) was purified from the supernatant of hybridoma TIB-230 (American Type Culture Collection), and polyclonal anti-TRF1 rabbit serum was raised against a TRF1 peptide, residues 13–35.
RNAi
The following siRNAs were designed and synthesized by QIAGEN: for p21, 5′-CAGTTTGTGTGTCTTAATTAT-3′ (p21-6) and 5′-CTGGCATTAGAATTATTTAAA-3′ (p21-7); for p53, 5′-AAGGAAATTTGCGTGTGGAGT-3′ (p53-9); for PCNA, 5′-ATGGATTTAGATGTTGAACAA-3′ (PCNA-6); for Sp100, 5′-CAGGAAATTATGATAAACTCA-3′ (Sp100-1); for HP1β, 5′-AAGGGAAGGAGTTCTACTTGT-3′ (HP1β-1) and 5′-AAGGACTAAGCCTGTTCATAA-3′ (HP1β-4); and for HP1γ, 5′-AAAGTACTAGATCGACGTGTA-3′ (HP1γ-2) and 5′-CTGGTTACTTTGAACAAATAA-3′ (HP1γ-6). The following siRNAs were synthesized by QIAGEN: for SV40T, 5′-AAAATTGTGTACCTTTAGCTT-3′ (Harborth et al., 2001); for p53 (p53-p), 5′-CGGCATGAACCGGAGGCCCAT-3′ (Martinez et al., 2002); for PCNA (PCNA-1), 5′-GAGGAGGAAGCTGTTACCATA-3′ (Senga et al., 2006); and for HP1α (HP1α and HP1α-2), 5′-AACCTGAGAAAAACTTGGATT-3′ (Obuse et al., 2004) and 5′-GAGGAGCACAATACTTGGGAA-3′ (Sripathy et al., 2006). The nonsilencing control siRNA was obtained from QIAGEN.
To determine the extent of knockdown, cells were transfected with 10 nM siRNA per target gene using HiPerFect transfection reagent according to the manufacturer's instructions (QIAGEN). For Western analysis, cells were seeded into 6-well plates 1–2 d before siRNA transfection. After transfection for 48 h, cells were harvested for protein isolation.
APB screening in SV40-immortalized cell lines by RNAi
Cells were seeded into 4-well chamber slides (Thermo Fisher Scientific) 2 d before transfection of siRNAs. For double or triple knockdown experiments, 10 nM siRNA per target gene, along with 10 nM SV40T siRNA, was transfected into cells using HiPerFect. 4 d later, cells were fixed and immunostained for SV40T, p21, and TRF1 (large foci of which are recognized as APBs). Finally, APB positivity was scored for the cells in which SV40T was depleted.
Immunostaining, BrdU labeling, telomere FISH, and fluorescence microscopy
Cells grown in 4-well chamber slides were fixed for 15 min in 2% paraformaldehyde at room temperature, then permeated with methanol/acetone (1:1) at −20°C for 15 min. Cells were incubated overnight with primary antibodies at 4°C, then incubated with fluorescently conjugated secondary antibodies at room temperature for 40 min. In some cases, DAPI (Sigma-Aldrich) was included in the secondary incubation to visualize DNA. Finally, the preparations were mounted in anti-fading medium containing DABCO (Sigma-Aldrich) or medium consisting of glycerol/PBS (70%:30%). The secondary antibodies used were as follows: FITC- or Texas red–conjugated goat anti–mouse; FITC- or Texas red–conjugated goat anti–rabbit; 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-, FITC-, or Texas red–conjugated donkey anti–mouse; AMCA-, FITC- or Texas red–conjugated donkey anti–rabbit; and AMCA- or Texas red–conjugated donkey anti–goat (Jackson ImmunoResearch Laboratories).
For BrdU labeling of growth-arrested cells, cells were grown in 4-well chamber slides to 30–40% confluency and then treated with 1 µM 4OHT or 10 nM SV40T siRNA for 4 d. 10 µM BrdU (Roche) was added to the culture medium for 24 h before fixation. The cells were fixed as described in the previous paragraph, then incubated with 2 µg/ml DNase I (Sigma-Aldrich) for 30 min at 37°C before incubation with primary antibodies against BrdU, p21, and TRF1.
Double staining of telomeric DNA and APB-associated proteins was performed as described previously (Henson et al., 2005). In brief, slides were first immunostained with primary and secondary antibodies, then cross-linked with 4% formaldehyde and dehydrated. Telomere FISH was done by using a Cy3-conjugated telomere-specific peptide nucleic acid probe (Applied Biosystems).
The samples were examined at room temperature on a fluorescence microscope (DMLB; Leica). A Plan-Fluotar 40×/0.7 NA objective lens and a Plan-Fluotar 10×/0.3 NA objective lens (Leica) were used in this study. Images were recorded using a cooled charge-coupled device camera (SPOT2; Diagnostic Instruments, Inc.) with SPOT image acquisition software (Diagnostic Instruments, Inc.), and analyzed with Photoshop 6.0 (Adobe). The contrast/brightness of images was adjusted uniformly across the field.
SA-β-gal activity assay
Cells were grown in four-well chamber slides to 30–40% confluency and then treated with 1 µM 4OHT or 10 nM SV40T siRNA for 3 or 4 d. The SA-β-gal staining was performed with a SA-β-gal staining kit (Cell Signaling Technology) according to the manufacturer's instructions. The samples were examined on an inverted microscope (IMT-2; Olympus) with an A10PL 10×/0.25 NA objective lens (Olympus). Images were recorded using a digital camera (DP12; Olympus) and analyzed with Photoshop 6.0.
Immunoblotting
For immunoblotting analyses, cell lysates were prepared, electrophoretically separated on SDS-PAGE gels, and electrotransferred to a nylon membrane as described previously (Toouli et al., 2002). Immunoblotting procedures were performed as recommended by the antibody suppliers. HRP-conjugated goat anti–mouse, goat anti–rabbit, swine anti–rabbit, or rabbit anti–goat IgG (Dako) were used as secondary antibodies.
Online supplemental material
Fig. S1 shows induction of a senescent phenotype in SV40-immortalized ALT cells upon treatment with SV40T siRNA. Fig. S2 shows the association of APB induction with p53/p21-mediated growth arrest/senescence. Fig. S3 shows that there is no APB induction in SV40-immortalized telomerase-positive cells. Fig. S4 shows the presence of p21, PCNA, and Cdk2 within APBs. Fig. S5 shows the requirement of HP1 for APB formation in p53-negative IIICF/c cells. Table S1 shows the proportion of APB-positive cells after methionine starvation. Table S2 shows the proportion of APB+ cells containing p21+ APBs after induction of p53. Table S3 shows the proportion of APB-positive IIICF/c cells after siRNA-treatment and methionine restriction. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810084/DC1.
Acknowledgments
We thank Christine Smyth and Axel Neumann for technical advice and help.
This work was supported by a Program Grant from the Cancer Council New South Wales and a Senior Principal Research Fellowship from the National Health and Medical Research Council of Australia (to R.R. Reddel), and a Cancer Institute New South Wales Cancer Research Leaders Program Grant (to A.W. Braithwaite).
Footnotes
Abbreviations used in this paper: 4OHT, 4-hydroxytamoxifen; ALT, alternative lengthening of telomeres; APB, ALT-associated PML body; ER, estrogen receptor; HP1, heterochromatin protein 1; LTAg, SV40 large T antigen; PCNA, proliferating cell nuclear antigen; PML, promyelocytic leukemia; SA, senescence associated; wt, wild type.
References
- Blasco M.A. 2007. The epigenetic regulation of mammalian telomeres.Nat. Rev. Genet. 8:299–309 [DOI] [PubMed] [Google Scholar]
- Brachner A., Sasgary S., Pirker C., Rodgarkia C., Mikula M., Mikulits W., Bergmeister H., Setinek U., Wieser M., Chin S.F., et al. 2006. Telomerase- and alternative telomere lengthening-independent telomere stabilization in a metastasis-derived human non-small cell lung cancer cell line: effect of ectopic hTERT.Cancer Res. 66:3584–3592 [DOI] [PubMed] [Google Scholar]
- Brown J.P., Wei W., Sedivy J.M. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts.Science. 277:831–834 [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R. 1995. Telomere elongation in immortal human cells without detectable telomerase activity.EMBO J. 14:4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines.Nat. Med. 3:1271–1274 [DOI] [PubMed] [Google Scholar]
- Cerone M.A., Autexier C., Londono-Vallejo J.A., Bacchetti S. 2005. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT.Oncogene. 24:7893–78901 [DOI] [PubMed] [Google Scholar]
- Chang B.D., Xuan Y., Broude E.V., Zhu H., Schott B., Fang J., Roninson I.B. 1999. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs.Oncogene. 18:4808–4818 [DOI] [PubMed] [Google Scholar]
- Chevillard C., Reik W., McDermott M., Fontes M., Mattei M.G., Singh P.B. 1993. Chromosomal localization of human homologs of the Drosophila heterochromatin protein 1 (HP1) gene.Mamm. Genome. 4:124–126 [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., Gorski M.M., Engels W.R., Gatti M. 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage.Curr. Biol. 14:1360–1366 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2002. Protection of mammalian telomeres.Oncogene. 21:532–540 [DOI] [PubMed] [Google Scholar]
- Dotto G.P. 2000. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta. 1471:M43–M56 [DOI] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R. 2000. Telomere maintenance by recombination in human cells.Nat. Genet. 26:447–450 [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., Elgin S.C. 2000. The HP1 protein family: getting a grip on chromatin.Curr. Opin. Genet. Dev. 10:204–210 [DOI] [PubMed] [Google Scholar]
- Fang L., Igarashi M., Leung J., Sugrue M.M., Lee S.W., Aaronson S.A. 1999. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53.Oncogene. 18:2789–2797 [DOI] [PubMed] [Google Scholar]
- Fasching C.L., Bower K., Reddel R.R. 2005. Telomerase-independent telomere length maintenance in the absence of ALT-associated PML bodies.Cancer Res. 65:2722–2729 [DOI] [PubMed] [Google Scholar]
- Fasching C.L., Neumann A.A., Muntoni A., Yeager T.R., Reddel R.R. 2007. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA.Cancer Res. 67:7072–7077 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G., De Stanchina E., Querido E., Baptiste N., Prives C., Lowe S.W. 2000. PML is induced by oncogenic ras and promotes premature senescence.Genes Dev. 14:2015–2027 [PMC free article] [PubMed] [Google Scholar]
- Funayama R., Ishikawa F. 2007. Cellular senescence and chromatin structure.Chromosoma. 116:431–440 [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M., O'Sullivan R., Peters A.H., Jenuwein T., Blasco M.A. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases.Nat. Genet. 36:94–99 [DOI] [PubMed] [Google Scholar]
- Gonzalo S., Garcia-Cao M., Fraga M.F., Schotta G., Peters A.H., Cotter S.E., Eguia R., Dean D.C., Esteller M., Jenuwein T., Blasco M.A. 2005. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin.Nat. Cell Biol. 7:420–428 [DOI] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A. 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells.Nat. Cell Biol. 8:416–424 [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts.Cell. 43:405–413 [DOI] [PubMed] [Google Scholar]
- Grobelny J.V., Godwin A.K., Broccoli D. 2000. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G2/M phase of the cell cycle.J. Cell Sci. 113:4577–4585 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2000. The hallmarks of cancer.Cell. 100:57–70 [DOI] [PubMed] [Google Scholar]
- Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs.J. Cell Sci. 114:4557–4565 [DOI] [PubMed] [Google Scholar]
- Henson J.D., Neumann A.A., Yeager T.R., Reddel R.R. 2002. Alternative lengthening of telomeres in mammalian cells.Oncogene. 21:598–610 [DOI] [PubMed] [Google Scholar]
- Henson J.D., Hannay J.A., McCarthy S.W., Royds J.A., Yeager T.R., Robinson R.A., Wharton S.B., Jellinek D.A., Arbuckle S.M., Yoo J., et al. 2005. A robust assay for alternative lengthening of telomeres (ALT) in tumors demonstrates the significance of ALT in sarcomas and astrocytomas.Clin. Cancer Res. 11:217–225 [PubMed] [Google Scholar]
- Homer C., Knight D.A., Hananeia L., Sheard P., Risk J., Lasham A., Royds J.A., Braithwaite A.W. 2005. Y-box factor YB1 controls p53 apoptotic function.Oncogene. 24:8314–8325 [DOI] [PubMed] [Google Scholar]
- Jiang W.Q., Ringertz N. 1997. Altered distribution of the promyelocytic leukemia-associated protein is associated with cellular senescence.Cell Growth Differ. 8:513–522 [PubMed] [Google Scholar]
- Jiang W.Q., Zhong Z.H., Henson J.D., Neumann A.A., Chang A.C., Reddel R.R. 2005. Suppression of alternative lengthening of telomeres by Sp100-mediated sequestration of MRE11/RAD50/NBS1 complex.Mol. Cell. Biol. 25:2708–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.Q., Zhong Z.H., Henson J.D., Reddel R.R. 2007. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference.Oncogene. 26:4635–4647 [DOI] [PubMed] [Google Scholar]
- Johnson F.B., Marciniak R.A., McVey M., Stewart S.A., Hahn W.C., Guarente L. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase.EMBO J. 20:905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa S., Fujiwara T., Kadowaki Y., Fukazawa T., Sok-Joo R., Roth J.A., Tanaka N. 1999. Overexpression of the p21sdi1 gene induces senescence-like state in human cancer cells: implication for senescence-directed molecular therapy for cancer.Cell Death Differ. 6:765–772 [DOI] [PubMed] [Google Scholar]
- Kaminker P., Plachot C., Kim S.H., Chung P., Crippen D., Petersen O.W., Bissell M.J., Campisi J., Lelievre S.A. 2005. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2.J. Cell Sci. 118:1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koering C.E., Pollice A., Zibella M.P., Bauwens S., Puisieux A., Brunori M., Brun C., Martins L., Sabatier L., Pulitzer J.F., Gilson E. 2002. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity.EMBO Rep. 3:1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins.Nature. 410:116–120 [DOI] [PubMed] [Google Scholar]
- Li R., Waga S., Hannon G.J., Beach D., Stillman B. 1994. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair.Nature. 371:534–537 [DOI] [PubMed] [Google Scholar]
- Ling Y.H., Zou Y., Perez-Soler R. 2000. Induction of senescence-like phenotype and loss of paclitaxel sensitivity after wild-type p53 gene transfection of p53-null human non-small cell lung cancer H358 cells.Anticancer Res. 20:693–702 [PubMed] [Google Scholar]
- Luciani J.J., Depetris D., Usson Y., Metzler-Guillemain C., Mignon-Ravix C., Mitchell M.J., Megarbane A., Sarda P., Sirma H., Moncla A., et al. 2006. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase.J. Cell Sci. 119:2518–2531 [DOI] [PubMed] [Google Scholar]
- Ma J., Hwang K.K., Worman H.J., Courvalin J.C., Eissenberg J.C. 2001. Expression and functional analysis of three isoforms of human heterochromatin-associated protein HP1 in Drosophila.Chromosoma. 109:536–544 [DOI] [PubMed] [Google Scholar]
- Maclean K., Rogan E.M., Whitaker N.J., Chang A.C.M., Rowe P.B., Dalla-Pozza L., Symonds G., Reddel R.R. 1994. In vitro transformation of Li-Fraumeni syndrome fibroblasts by SV40 large T antigen mutants.Oncogene. 9:719–725 [PubMed] [Google Scholar]
- Marciniak R.A., Cavazos D., Montellano R., Chen Q., Guarente L., Johnson F.B. 2005. A novel telomere structure in human alternative lengthening of telomeres cell line.Cancer Res. 65:2730–2737 [DOI] [PubMed] [Google Scholar]
- Martinez L.A., Naguibneva I., Lehrmann H., Vervisch A., Tchenio T., Lozano G., Harel-Bellan A. 2002. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways.Proc. Natl. Acad. Sci. USA. 99:14849–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar C., Wiesmeijer K., Verwoerd N.P., Khazen S., Eils R., Tanke H.J., Dirks R.W. 2003. Visualizing telomere dynamics in living mammalian cells using PNA probes.EMBO J. 22:6631–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Jones K., Wayman L., Kennedy D.D., Reddel R.R., Sara S., Snee M.J., Smith R. 2005. hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere.Nucleic Acids Res. 33:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni A., Reddel R.R. 2005. The first molecular details of ALT in human tumor cells.Hum. Mol. Genet. 14:R191–R196 [DOI] [PubMed] [Google Scholar]
- Murnane J.P., Sabatier L., Marder B.A., Morgan W.F. 1994. Telomere dynamics in an immortal human cell line.EMBO J. 13:4953–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Yokoyama O., Ishikawa F. 2004. Localization of hRad9, hHus1, hRad1 and hRad17, and caffeine-sensitive DNA replication at ALT (alternative lengthening of telomeres)-associated promyelocytic leukemia body.J. Biol. Chem. 279:25849–25857 [DOI] [PubMed] [Google Scholar]
- Netzer C., Rieger L., Brero A., Zhang C.D., Hinzke M., Kohlhase J., Bohlander S.K. 2001. SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin.Hum. Mol. Genet. 10:3017–3024 [DOI] [PubMed] [Google Scholar]
- Noda A., Ning Y., Venable S.F., Pereira-Smith O.M., Smith J.R. 1994. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen.Exp. Cell Res. 211:90–98 [DOI] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1.Nat. Cell Biol. 6:1135–1141 [DOI] [PubMed] [Google Scholar]
- Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P.P., Pelicci P.G. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras.Nature. 406:207–210 [DOI] [PubMed] [Google Scholar]
- Perrem K., Colgin L.M., Neumann A.A., Yeager T.R., Reddel R.R. 2001. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells.Mol. Cell. Biol. 21:3862–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucca P., Cazzalini O., Mortusewicz O., Necchi D., Savio M., Nardo T., Stivala L.A., Leonhardt H., Cardoso M.C., Prosperi E. 2006. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen.J. Cell Sci. 119:1517–1527 [DOI] [PubMed] [Google Scholar]
- Potts P.R., Yu H. 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins.Nat. Struct. Mol. Biol. 14:581–590 [DOI] [PubMed] [Google Scholar]
- Razak Z.R., Varkonyi R.J., Kulp-McEliece M., Caslini C., Testa J.R., Murphy M.E., Broccoli D. 2004. p53 differentially inhibits cell growth depending on the mechanism of telomere maintenance.Mol. Cell. Biol. 24:5967–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel R.R. 2000. The role of senescence and immortalization in carcinogenesis.Carcinogenesis. 21:477–484 [DOI] [PubMed] [Google Scholar]
- Rogan E.M., Bryan T.M., Hukku B., Maclean K., Chang A.C.M., Moy E.L., Englezou A., Warneford S.G., Dalla-Pozza L., Reddel R.R. 1995. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts.Mol. Cell. Biol. 15:4745–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J.S., Marchio A., Sitterlin D., Transy C., Dejean A. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment.Proc. Natl. Acad. Sci. USA. 95:7316–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T., Sivaprasad U., Zhu W., Park J.H., Arias E.E., Walter J.C., Dutta A. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination.J. Biol. Chem. 281:6246–6252 [DOI] [PubMed] [Google Scholar]
- Sharma G.G., Hwang K.K., Pandita R.K., Gupta A., Dhar S., Parenteau J., Agarwal M., Worman H.J., Wellinger R.J., Pandita T.K. 2003. Human heterochromatin protein 1 isoforms HP1(Hsα) and HP1(Hsß) interfere with hTERT-Telomere interactions and correlate with changes in cell growth and response to ionizing radiation.Mol. Cell. Biol. 23:8363–8376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W., Bacchetti S. 1997. A survey of telomerase activity in human cancer.Eur. J. Cancer. 33:787–791 [DOI] [PubMed] [Google Scholar]
- Silverman J., Takai H., Buonomo S.B., Eisenhaber F., de Lange T. 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint.Genes Dev. 18:2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy S.P., Stevens J., Schultz D.C. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression.Mol. Cell. Biol. 26:8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno D'Alcontres M., Mendez-Bermudez A., Foxon J.L., Royle N.J., Salomoni P. 2007. Lack of TRF2 in ALT cells causes PML-dependent p53 activation and loss of telomeric DNA.J. Cell Biol. 179:855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos D.J., Bradshaw P.S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M.S. 2002. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis.Hum. Mol. Genet. 11:3135–3144 [DOI] [PubMed] [Google Scholar]
- Sugrue M.M., Shin D.Y., Lee S.W., Aaronson S.A. 1997. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53.Proc. Natl. Acad. Sci. USA. 94:9648–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M., Munoz P., Claas A., Smiraldo P.G., Pittman D.L., Blasco M.A., West S.C. 2004. Telomere maintenance requires the RAD51D recombination/repair protein.Cell. 117:337–347 [DOI] [PubMed] [Google Scholar]
- Toouli C.D., Huschtscha L.I., Neumann A.A., Noble J.R., Colgin L.M., Hukku B., Reddel R.R. 2002. Comparison of human mammary epithelial cells immortalized by simian virus 40 T-antigen or by the telomerase catalytic subunit.Oncogene. 21:128–139 [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Yeargin J., Christiansen-Weber T., Haas M. 1998. Independent induction of senescence by p16INK4a and p21CIP1 in spontaneously immortalized human fibroblasts.Cell Growth Differ. 9:139–146 [PubMed] [Google Scholar]
- Wang Y., Blandino G., Givol D. 1999. Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin.Oncogene. 18:2643–2649 [DOI] [PubMed] [Google Scholar]
- Warneford S.G., Witton L.J., Townsend M.L., Rowe P.B., Reddel R.R., Dalla-Pozza L., Symonds G. 1992. Germ-line splicing mutation of the p53 gene in a cancer-prone family.Cell Growth Differ. 3:839–846 [PubMed] [Google Scholar]
- Wu G., Lee W.H., Chen P.L. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phrases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres.J. Biol. Chem. 275:30618–30622 [DOI] [PubMed] [Google Scholar]
- Wu G., Jiang X., Lee W.H., Chen P.L. 2003. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen breakage syndrome 1.Cancer Res. 63:2589–2595 [PubMed] [Google Scholar]
- Yankiwski V., Marciniak R.A., Guarente L., Neff N.F. 2000. Nuclear structure in normal and Bloom syndrome cells.Proc. Natl. Acad. Sci. USA. 97:5214–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager T.R., Neumann A.A., Englezou A., Huschtscha L.I., Noble J.R., Reddel R.R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body.Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- Zhang R., Poustovoitov M.V., Ye X., Santos H.A., Chen W., Daganzo S.M., Erzberger J.P., Serebriiskii I.G., Canutescu A.A., Dunbrack R.L., et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA.Dev. Cell. 8:19–30 [DOI] [PubMed] [Google Scholar]
- Zhong Z.H., Jiang W.Q., Cesare A.J., Neumann A.A., Wadhwa R., Reddel R.R. 2007. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres.J. Biol. Chem. 282:29314–29322 [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Kuster B., Mann M., Petrini J.H., de Lange T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres.Nat. Genet. 25:347–352 [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J.H., de Lange T. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes.Mol. Cell. 12:1489–1498 [DOI] [PubMed] [Google Scholar]