Abstract
Mitotic spindle formation and chromosome segregation depend critically on kinetochore–microtubule (KT–MT) interactions. A new protein, termed Spindly in Drosophila and SPDL-1 in C. elegans, was recently shown to regulate KT localization of dynein, but depletion phenotypes revealed striking differences, suggesting evolutionarily diverse roles of mitotic dynein. By characterizing the function of Spindly in human cells, we identify specific functions for KT dynein. We show that localization of human Spindly (hSpindly) to KTs is controlled by the Rod/Zw10/Zwilch (RZZ) complex and Aurora B. hSpindly depletion results in reduced inter-KT tension, unstable KT fibers, an extensive prometaphase delay, and severe chromosome misalignment. Moreover, depletion of hSpindly induces a striking spindle rotation, which can be rescued by co-depletion of dynein. However, in contrast to Drosophila, hSpindly depletion does not abolish the removal of MAD2 and ZW10 from KTs. Collectively, our data reveal hSpindly-mediated dynein functions and highlight a critical role of KT dynein in spindle orientation.
Introduction
Error-free chromosome segregation during mitosis depends on the formation of a bipolar spindle and correct attachment of all kinetochores (KTs) to spindle microtubules (MTs) (O'Connell and Khodjakov, 2007; Tanaka, 2008). To prevent chromosome missegregation, the spindle assembly checkpoint (SAC) delays the onset of anaphase until all chromosomes are properly attached to MTs (Musacchio and Salmon, 2007).
Prominent among the MT-dependent motor proteins implicated in various mitotic functions is dynein/dynactin. This minus end–directed motor complex is required for proper spindle formation, plays a role in spindle pole focusing and separation, and controls spindle length (Vaisberg et al., 1993; Echeverri et al., 1996; Sharp et al., 2000; Gaetz and Kapoor, 2004; Goshima et al., 2005). Remarkably, dynein localizes to unattached KTs and disassociates upon MT attachment (King et al., 2000). It is involved in establishing the initial lateral contact between KTs and MTs, which results in rapid poleward movements of chromosomes during early prometaphase (Rieder and Alexander, 1990; Yang et al., 2007; Vorozhko et al., 2008). Subsequently, these lateral MT interactions mature into stable end-on attachments at the KT that are mediated by the KMN network, a group of evolutionarily conserved proteins comprising KNL-1 and the Mis12 and Ndc80 complexes (Cheeseman and Desai, 2008). Importantly, the dynein/dynactin complex also localizes to the cell cortex where it contributes to spindle orientation and positioning, most likely by providing pulling forces on astral MTs (Busson et al., 1998; O'Connell and Wang, 2000).
The recruitment of dynein/dynactin to KTs is dependent on the conserved Rod/Zw10/Zwilch (RZZ) complex (Karess, 2005), possibly through direct interaction between ZW10 and the dynactin subunit p50-dynamitin (Starr et al., 1998). Additional proteins, notably Nde1 and Ndel1, are also required for targeting dynein/dynactin to KTs (Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007). In recent years, manipulation of the RZZ complex has been extensively used to study the roles of KT-associated dynein. In fact, inhibition of the RZZ complex suppressed the rapid poleward movement of chromosomes and reduced KT tension (Savoian et al., 2000; Yang et al., 2007), consistent with results obtained upon direct disruption of dynein function (Howell et al., 2001; Vorozhko et al., 2008). However, the role of KT dynein in chromosome congression remains controversial. Although inhibition of dynein/dynactin during prometaphase did not impair metaphase plate formation in vertebrate cells (Howell et al., 2001; Vorozhko et al., 2008), depletion of ZW10 from human cells induced chromosome misalignment, suggesting that KT dynein is required for efficient congression (Li et al., 2007; Yang et al., 2007). Similarly, depletion of Rod from Drosophila S2 cells delayed chromosome alignment (Griffis et al., 2007), but Drosophila embryos and larval neuroblasts depleted of ZW10 showed no obvious defect in metaphase plate formation (Williams and Goldberg, 1994).
More recently, the dynein/dynactin complex has been implicated in the removal of SAC proteins from the outer KT, and this, in turn, has been proposed to play an important role in SAC silencing (Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004; Griffis et al., 2007; Mische et al., 2008; Varma et al., 2008; Whyte et al., 2008; Sivaram et al., 2009). However, studies on the role of dynein/dynactin in SAC silencing through interference with the RZZ complex are confounded by the fact that the RZZ is also essential for SAC activation (Basto et al., 2000; Chan et al., 2000), particularly for recruiting MAD1/MAD2 to KTs (Buffin et al., 2005; Kops et al., 2005).
A protein termed Spindly in Drosophila (Griffis et al., 2007) and SPDL-1 in C. elegans (Gassmann et al., 2008) has recently been identified as a new KT regulator of dynein. However, depletion of this protein yielded partially distinct phenotypes in the two invertebrate species, suggesting that some KT dynein functions may have diverged during evolution (Civril and Musacchio, 2008). Here, we report the characterization of the human homologue of Spindly/SPDL-1, which we identified originally as a putative component of the spindle apparatus (Sauer et al., 2005). Our results have important implications for the role of KT-associated dynein in spindle orientation and in the removal of outer KT components during SAC silencing.
Results
hSpindly localizes to KTs and spindle poles
Bioinformatic analysis of the human mitotic spindle proteome (Sauer et al., 2005) by the ENFIN consortium predicted CCDC99 (accession no. Q96EA4) to be a spindle protein (unpublished data). Independently, CCDC99 has been suggested as the potential human homologue of Drosophila Spindly (Griffis et al., 2007). Analysis of Hela S3 cells overexpressing myc-tagged CCDC99 by immunofluorescence microscopy revealed KT and spindle pole localization in mitosis (Fig. S1 A), identifying this protein as a bona fide spindle component. Thus, hereafter we refer to CCDC99 as human Spindly (hSpindly). Endogenous hSpindly localization was determined using an hSpindly-specific antibody that recognizes a single band of 70 kD, the predicted molecular weight of hSpindly (Fig. S1 B). No specific signal could be detected with the preimmune serum or after depletion of hSpindly by siRNA (Fig. S1, B–D), confirming antibody specificity. Interphase localization of hSpindly was mainly nuclear (Fig. 1 A; Fig. S1 A). This contrasts with Drosophila Spindly, which localized to MT plus-end tips (Griffis et al., 2007). In mitosis, hSpindly decorated KTs in early prometaphase before it relocalized to the spindle poles before metaphase (Fig. 1 B; Fig. S1, A and E). Colocalization with BubR1, adjacent to the CREST signal, indicated that hSpindly is an outer KT protein (Fig. 1 C). After all chromosomes achieved perfect alignment, no obvious staining on the spindle poles could be seen (Fig. S1 E), suggesting that hSpindly diffuses to the cytosol after moving to the poles. At later stages of mitosis (anaphase and telophase), no specific association of hSpindly with any spindle structures could be detected (Fig. 1 B). A parallel biochemical analysis revealed that hSpindly protein levels were reduced after release from nocodazole arrest (Fig. S1 F), but this degradation was blocked by addition of the proteasome inhibitor MG132 (Fig. S1 G), indicating that hSpindly is degraded upon mitotic exit. Furthermore, an upshift could be detected in nocodazole-treated cells, suggesting that hSpindly is modified during M phase (Fig. S1, F–H). This modification appeared to be phosphorylation dependent because the slower migrating form of hSpindly was not detected in lysates from nocodazole-arrested cells treated with λ-phosphatase (Fig. S1 H).
Figure 1.
hSpindly localizes at outer KTs and spindle poles, downstream of the RZZ and Ndc80 complexes. (A) HeLa S3 cells in interphase were stained with anti-hSpindly (red) and anti–α-tubulin antibodies (green), and DAPI (blue). (B) Cells at different mitotic stages were stained with anti-hSpindly antibody (red), CREST serum or α-tubulin antibody (green), and DAPI (blue). (C) Prometaphase cell stained with anti-hSpindly (red) and anti-BubR1 (green) antibodies and CREST serum (blue). The inset shows a magnification of the selected area (bar, 1 µm). (D) Cells treated for 72 h with GL2 (control) or two independent siRNAs targeting ZW10 (ZW10-1 or ZW10-2 siRNA) were stained with anti-hSpindly antibody (red), CREST serum (green), and DAPI (blue). (E) Western blotting of mitotic (nocodazole shake-off) cells treated for 72 h with GL2 (control), ZW10-1, or ZW10-2 siRNAs. Membranes were probed for the indicated antibodies and α-tubulin is shown as loading control. (F) Cells treated with GL2 (control) or Hec1 siRNAs for 48 h were stained with anti-Hec1 (green) and anti-hSpindly (red) antibodies, and DAPI (blue). (G) Western blotting of mitotic (nocodazole shake-off) cells treated with GL2 (control), or Hec1 siRNAs for 40 h. Membranes were probed for the indicated antibodies and α-tubulin is shown as loading control. Bars, 10 µm.
hSpindly acts downstream of the RZZ and Ndc80 complexes
The Drosophila and Caenorhabditis elegans homologues of hSpindly require the RZZ complex to localize to the KT (Griffis et al., 2007; Gassmann et al., 2008; Yamamoto et al., 2008). In the case of hSpindly we found that siRNA-mediated depletion of ZW10 (ZW10-1 [Kops et al., 2005] and ZW10-2) not only abolished KT localization (Fig. 1 D) but also caused a reduction in hSpindly protein levels (Fig. 1 E). Conversely, hSpindly depletion did not affect either localization or stability of ZW10 (Fig. S2 A and see Fig. 4 C). The observed effect of RZZ depletion on the stability of hSpindly appears to reflect a new level of regulation, as no such effect had been seen in either Drosophila (Griffis et al., 2007) or C. elegans (Gassmann et al., 2008). In agreement with the results observed in C. elegans (Gassmann et al., 2008), RZZ members (ZW10 and Rod), but not dynein, could be coimmunoprecipitated with hSpindly (Fig. S2 B) from mitotic HeLa S3 cells lysates. However, this interaction could only be observed under detergent-free conditions (see Materials and methods). In addition, glycerol gradient centrifugation revealed that a fraction of ZW10 co-migrated with hSpindly, and hSpindly depletion caused a slight shift in the sedimentation behavior of ZW10 (Fig. S2 C). Thus, our results are consistent with an interaction between hSpindly and the RZZ complex, albeit weak and likely dynamic.
Figure 4.
hSpindly targets both dynein and dynactin to KTs. (A) HeLa S3 cells were treated with GL2 or hSpindly siRNAs for 48 h. Nocodazole was added to the cells 2 h before they were stained with CREST serum (green), anti-p150Glued antibody (red), and DNA (blue). (B) Cells treated as in A were stained with CREST serum (green) and anti-dynein intermediate chain (DIC) antibody (red) and DNA (blue). (C) Cells were treated with GL2, hSpindly-1, or hSpindly-2 siRNAs for 48 h. Lysates from mitotic cells (nocodazole shake-off) were prepared and equal amounts of cell extracts were separated by SDS-PAGE and probed by Western blotting with the indicated antibodies. (D) Cells were transfected with hSpindly-2 siRNA together with either a myc-vector construct (V) or a myc-tagged hSpindly (siRNA-resistant) construct (WT) for 48 h. Cells were stained with anti-myc, anti-p150Glued antibodies (red), CREST serum (green), and DAPI (blue). (E) Cells were treated as in D. Lysates were prepared and equal amounts of cell extracts were separated by SDS-PAGE and probed by Western blotting with the indicated antibodies. (F) Table showing positive/negative localization of the indicated KT/centromere proteins after hSpindly depletion for 48 h (−, lost from KT; +, indistinguishable from control). For DIC and p150, experiments were repeated with treatment of nocodazole (2 h) to confirm their dependency on hSpindly. Bars, 10 µm.
Next, we asked whether KT localization was important for the stability of hSpindly. Upon depletion of Hec1/Ndc80 we found that both ZW10 and hSpindly were displaced from KTs (Fig. 1 F; Fig. S2 D; see also Lin et al., 2006), and yet, levels of hSpindly were unaffected (Fig. 1 G). This clearly demonstrates that hSpindly degradation does not result from its dissociation from KTs and instead suggests that the interaction with the RZZ stabilizes the protein.
Aurora B controls KT localization of hSpindly
Aurora B has been shown to promote the recruitment of the RZZ complex to the KT until the establishment of tension (Famulski and Chan, 2007). Thus, we asked whether the KT association of hSpindly is also regulated by Aurora B. First, we analyzed hSpindly localization after depletion of Aurora B or inhibition of the kinase by the small molecule inhibitor ZM447439 (Ditchfield et al., 2003). In both cases, hSpindly KT and spindle pole localization was lost (Fig. 2 A; Fig. S2 E), as expected, considering the dependency of the RZZ on Aurora B. Furthermore, the stability of hSpindly was clearly reduced (Fig. 2 B). We also observed a slight decrease in ZW10 protein levels, further pointing to an influence of Aurora B on both the RZZ and hSpindly. To examine the localization of hSpindly in response to lack of tension, MG132-treated metaphase cells were treated with taxol (1 µM). On most KTs of taxol-treated cells, hSpindly colocalized with BubR1 (Fig. 2, C and D), which had previously been reported to be enriched at tensionless KTs (Skoufias et al., 2001). Although this treatment resulted in few MAD2- (and hSpindly)-positive KTs, which were likely to be unattached (Hauf et al., 2003), many KTs were hSpindly-positive but MAD2-negative (Fig. 2 F; Fig. S3 C; see Fig. S3, A and B for MAD2 antibody characterization). Inhibition of Aurora B kinase by a short treatment with ZM447439, sufficiently short as to not reduce hSpindly levels (Fig. 2 G), inhibited the recruitment of both hSpindly and MAD2 in cells treated with taxol, but not nocodazole (Fig. 2, F and H; Fig. S3 C). Thus, Aurora B controls hSpindly recruitment to KTs in response to taxol, presumably through the RZZ complex.
Figure 2.
Aurora B regulates the localization of hSpindly in response to taxol. (A) HeLa S3 cells were treated with GL2 (control) or Aurora B siRNAs for 40 h, or with ZM447439 (ZM) for 16 h before being stained with anti-hSpindly (red) and anti–α-tubulin (green) antibodies and DAPI (blue). (B) Cells were treated with GL2 (control) or Aurora B siRNAs for 40 h. DMSO (control) or ZM447439 (ZM) was added to GL2-treated cells for the last 16 h. Mitotic cells (nocodazole shake-off) were collected and equal amounts of cell extracts were separated by SDS-PAGE and probed by Western blotting with the indicated antibodies; α-tubulin is shown as loading control. (C) Cells were treated with MG132 for 2 h. During the second hour, taxol was added. Cells were then stained with anti-hSpindly (red) and anti-BubR1 (green) antibodies and CREST serum (blue). (D) Graph showing the numbers of KTs from 10 cells treated as in C (399 KTs in total) that were positive for BubR1 (left circle) or hSpindly (right circle). The overlapping region represents the number of KTs that were positive for both BubR1 and hSpindly. Percentages of KTs positive for BubR1 or hSpindly are shown. (E) Graph showing the percentages of sister KT pairs with both KTs positive (+/+), only one sister KT positive (+/−), or both negative (−/−) for BubR1 and hSpindly, respectively (>90 sister KT pairs from 10 cells were counted). (F) Cells were treated with MG132 for 2 h. During the second hour, DMSO, nocodazole (Noc), or taxol with or without ZM447439 (ZM) was added. Cells were then stained with anti-hSpindly (red) and anti-MAD2 (green) antibodies and CREST serum (blue). (G) Cells were synchronized by sequential thymidine arrest (overnight) and release (9 h) before being treated as in F. Mitotic cells were collected by shake-off. Equal amounts of cell extracts were separated by SDS-PAGE and probed by Western blotting with the indicated antibodies; α-tubulin is shown as loading control. (H) Bar graph showing the quantification of hSpindly and MAD2 staining intensities at KTs (normalized against CREST) of cells in F (20 KTs were counted per cell and error bars indicate the standard deviation [SD] of measurements from 8 cells). Bars, 10 µm.
To further investigate the possible tension responsiveness of hSpindly, we analyzed hSpindly localization in cells treated with monastrol. Similar to taxol-treated cells, hSpindly was found on many MAD2-negative KTs (Fig. S3, D and E), suggesting that KT–MT attachment is not sufficient to remove hSpindly from KTs. However, a significant fraction of sister KT pairs clearly displayed asymmetric hSpindly localization (Fig. S3 F), and similar observations were made, albeit less frequently, in taxol-treated cells (Fig. 2 E). This suggests that hSpindly localization to KTs is not regulated by tension but, presumably, by some structural or dynamic aspect of KT–MT interaction. Interestingly, though, hSpindly can be retained at KTs under some conditions of MT attachment that still allow MAD2 removal.
hSpindly is essential for mitotic progression
To determine the function of hSpindly, we analyzed the depletion phenotype in HeLa S3 cells. Depletion of hSpindly with either one of two siRNAs (hSpindly-1 and hSpindly-2 siRNA) resulted in an increased mitotic index when compared with Photinus pyralis luciferase gene (GL2)–depleted cells (Fig. 3 A). Immunofluorescence analysis of hSpindly-depleted cells revealed elongated spindles and severe chromosome misalignments (Fig. 3, B and C; see Fig. 8 A). Strikingly, a significant proportion of hSpindly-depleted cells (∼25% and 32%; hSpindly-1 and -2, respectively, ∼10% GL2) showed a monopolar-like spindle with chromosomes arranged in a circle (Fig. 3 B, discussed in detail later).
Figure 3.
Depletion of hSpindly induces chromosome misalignment and mitotic delay. (A) Mitotic indices were determined for HeLa S3 cells after treatment for 48 h with GL2 (control) or two independent siRNAs targeting hSpindly (hSpindly-1 and hSpindly-2). Bar graphs show the results of three independent experiments (>100 cells each) and error bars indicate SD. The bottom panel shows Western blotting of cells treated for the indicated times with the above-mentioned siRNAs. Membranes were probed with anti-hSpindly antibody and α-tubulin is shown as loading control. (B) Cells treated with GL2 or hSpindly-2 siRNA for 48 h were stained with anti-hSpindly and anti–α-tubulin antibodies (green), and CREST serum (red) and DAPI (blue). (C) Cells were treated with GL2, hSpindly-1, or hSpindly-2 siRNAs for 48 h (and treated with or without MG132 for 2 h) and the percentage of mitotic cells with all chromosomes aligned was determined. Bar graphs represent the results of three independent experiments (>100 cells each, only prometaphase and metaphase cells were counted) and error bars indicate SD. (D) Representative stills from videos of H2B-GFP expressing HeLa S3 cells treated with GL2, hSpindly-2, and ZW10 siRNAs for 48 h before filming. Time is shown in h:min. t = 0 was defined as the time point one frame before chromosome condensation became evident. (E) Box-and-whisker plot showing the time cells spent in mitosis from nuclear envelope breakdown (NEBD) to anaphase after treatment with GL2 (control), hSpindly-1 and hSpindly-2 siRNA (3 experiments, >80 cells per experiment, P < 0.0001), or ZW10 siRNAs (2 experiments, >80 cells per experiment, P < 0.0001). Bars, 10 µm.
Figure 8.
Spindle misorientation and increased spindle length after hSpindly depletion. (A) HeLa S3 cells were treated with GL2 and hSpindly siRNAs for 48 h and stained with anti-hSpindly, anti–centrin-3 (green), and anti–α-tubulin (red) antibodies, and DNA (blue). Panels on the right show four representative sections along the z-axis (section numbers are indicated); z-stacks were taken every 0.2 µm. (B) Cells were seeded onto fibronectin-coated coverslips, treated with the indicated siRNAs for 48 h, and synchronized by a thymidine block (overnight) and release (9 h). MG132 was added to the cells 1 h before they were stained with anti-pericentrin (red) and anti–α-tubulin antibodies (green), and DAPI (blue). Bottom panels show the X-Z projection of the pericentrin signal. (C) Box-and-whisker plot showing the spindle angles (α) of cells treated as in B, calculated by measuring the pole-to-pole distance (x) and the vertical distance (z) between two poles after taking Z-stacks every 0.4 µm (illustrated in top panel). Only cells with well-separated spindle poles (x > 7 µm) were counted (>50 cells) (GL2 vs. hSpindly/ZW10/Nde1, P < 0.0002; GL2 vs. CenpE, P > 0.05). (D) Bar graph showing the pole-to-pole distances (x) of the cells in B. Error bars show the SD after measuring distances from >50 cells. Bars, 10 µm.
To examine the hSpindly depletion phenotype in real time, we performed live-cell imaging experiments using HeLa S3 cells stably expressing histone H2B-GFP. hSpindly-depleted cells spent an increased time in mitosis from nuclear envelope breakdown to anaphase onset (mean of 92 min and 148 min for hSpindly-1 and -2 siRNAs, respectively) when compared with GL2-depleted cells as control (mean of 36 min; Fig. 3, D and E; Videos 1–3). When the mitotic arrest caused by hSpindly depletion exceeded 4 h, we often observed cell death (unpublished data). hSpindly-depleted cells also showed many unaligned chromosomes, similar to the phenotype seen upon ZW10 depletion (Fig. 3 D; Videos 2–4). However, in agreement with reported functions for ZW10 (Kops et al., 2005; Yang et al., 2007), depletion of the latter protein produced no significant effect on overall mitotic timing, although it caused premature anaphase and chromosome missegregation (Fig. 3, D and E). Thus, although depletion of either hSpindly or ZW10 results in a chromosome congression defect, only depletion of hSpindly causes a substantial mitotic delay.
hSpindly recruits both dynein and dynactin to KTs, but is dispensable for removal of checkpoint proteins
Whereas Drosophila Spindly was reported to recruit dynein to KTs independently of dynactin (Griffis et al., 2007), C. elegans SPDL-1 targets both dynein and dynactin to unattached KTs (Gassmann et al., 2008). In view of these conflicting data, we asked what role hSpindly plays in the KT recruitment of dynein and dynactin in human cells. Although depletion of hSpindly did not affect the normal localization of either dynein or dynactin on the spindle (see Fig. 9 D below), it clearly abolished the KT association of both dynein intermediate chain (DIC) and the dynactin subunit p150Glued in nocodazole-treated cells (Fig. 4, A and B) without affecting their protein levels (Fig. 4 C). This localization could be restored by overexpressing an siRNA-resistant myc-hSpindly construct (Fig. 4, D and E). Thus, hSpindly is clearly required for KT loading of both dynein and dynactin, as reported for C. elegans (Gassmann et al., 2008), but unlike the situation in Drosophila (Griffis et al., 2007). Importantly, no other KT protein examined (except dynein/dynactin) was mislocalized after hSpindly depletion, strongly arguing against a general disruption of KT/centromere structure (Fig. 4 F; Fig. S4, A–C). Of particular interest, hSpindly depletion did not affect Nde1, another protein implicated in the KT recruitment of dynein (Stehman et al., 2007; Vergnolle and Taylor, 2007), nor did Nde1 depletion affect hSpindly (Fig. S4, D and E), indicating that the two proteins cooperate in localizing dynein to the KT.
Figure 9.
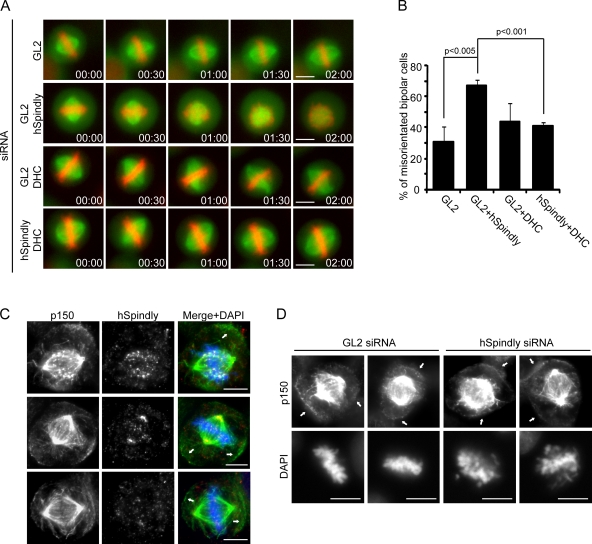
Spindle misorientation induced by hSpindly depletion depends on dynein. (A) Representative stills from videos of GFP-α-tubulin/cherry-H2B expressing HeLa Kyoto cells treated with the indicated combinations of siRNAs for 54 h and synchronized by double thymidine block and release. 9 h after the second release, MG132 was added for 1 h before filming. Time is shown in h:min. t = 0 was defined as the time point where chromosomes aligned in the metaphase plate. (B) Bar graphs showing the percentages of cells with spindle rotation defects. Error bars show the SD from three experiments (>15 cells per experiment). (C) Cells stained with anti-p150Glued (green) and anti-hSpindy antibodies (red), and DAPI (blue). Arrows indicate p150Glued cortical staining. (D) Cells were treated with GL2 or hSpindly siRNAs for 48 h and stained with anti-p150Glued antibody and DAPI. Arrows indicate p150Glued cortical staining. Bars, 10 µm.
Although the role of Spindly in recruiting dynein to KTs is apparently conserved among species, Drosophila Spindly and C. elegans SPDL-1 were described to impact differently on the SAC. Whereas SPDL-1 is required for MAD1/MAD2 recruitment to unattached KTs, Spindly is not required for this process, but instead is subsequently required for removal of MAD2 from aligned KTs (Griffis et al., 2007; Gassmann et al., 2008; Yamamoto et al., 2008). We therefore asked which of these roles is shared by hSpindly. In HeLa S3 cells, depletion of hSpindly did not affect the recruitment of MAD2 to prometaphase KTs (Fig. 5 A). Moreover, both MAD2 and ZW10 were detected only on unaligned KTs (Fig. 5, B and C), arguing that SAC proteins were normally displaced from KTs upon MT attachment. We conclude that hSpindly is neither required for the initial recruitment nor the subsequent removal of SAC components. So, the persistent SAC activation seen in hSpindly-depleted cells cannot be explained by impaired removal of SAC proteins.
Figure 5.
hSpindly is dispensable for removal of MAD2 and ZW10 from KTs. HeLa S3 cells were treated with GL2 or hSpindly siRNAs for 48 h and stained with anti-hSpindly (red) and anti-MAD2 antibodies (green) (A) or anti-hSpindly, anti-MAD2 antibodies (red) and CREST serum (green) (B), and DAPI (blue). (C) Cells were treated with GL2 or hSpindly siRNAs for 48 h and stained with anti-ZW10 (green) or anti-MAD2 (red) antibodies, and DAPI (blue). (D) Cells were transfected with either a myc-vector construct or a myc-tagged p50-dynamitin construct for 48 h and treated with MG132 for 1 h. Cells are then stained with anti-myc 9E10 serum, anti-hSpindly (red) and anti–α-tubulin antibodies (green), and DAPI (blue). Bars, 10 µm.
It has been previously shown that Spindly and SAC components are removed from KTs in a dynein-dependent manner (Howell et al., 2001; Wojcik et al., 2001; Griffis et al., 2007). To confirm that this also occurs in HeLa S3 cells, we overexpressed p50-dynamitin, a protein known to disrupt the dynein/dynactin complex (Echeverri et al., 1996). Indeed, we found that a much higher proportion of p50-overexpressing metaphase cells retained hSpindly and MAD2 on at least some KTs when compared with cells transfected with an empty vector (Figs. 5 D, 6 A, and 6 B), indicating that the removal of these proteins from KTs is inefficient upon disruption of the dynein/dynactin complex. In addition, the adoption of an ATP reduction assay that had previously been used to study dynein-dependent transport (Howell et al., 2001) revealed that hSpindly and SAC components relocalized from KTs to the spindle poles (Fig. 6 C), but were retained at KTs when cells were additionally treated with nocodazole (Fig. 6 D). Together, these results suggest that removal of SAC proteins from KTs occurs in an MT- and dynein-dependent manner but does not require hSpindly-mediated accumulation of KT dynein.
Figure 6.
Efficient removal of checkpoint proteins from KTs requires dynein/dynactin. (A) HeLa S3 cells were treated as in Fig. 5 D but stained with anti-MAD2 antibody instead of anti-hSpindly antibody. (B) The percentage of metaphase cells treated as in A with MAD2-positive KT(s) was determined. Bar graph showing the results of three independent experiments (>30 cells each), with error bars indicating SD. (C) To perform an ATP reduction assay, cells were rinsed once with isotonic salt solution and then incubated with either isotonic salt solution with glucose or with sodium azide (Az) and 2-deoxyglucose (DOG). Cells were stained with anti-MAD2 antibody and CREST serum (top), anti-ZW10 and anti-Hec1 antibodies (middle), and anti-hSpindly and anti-Hec1 antibodies (bottom). All cells were simultaneously stained also with anti–α-tubulin antibody and DAPI (blue). (D) Cells were treated as in C, except that 3.3 µM nocodazole was added to the isotonic salt solution. Bars, 10 µm.
hSpindly contributes to the establishment of tension and K-fiber stabilization
KT-associated dynein has been suggested to be required for the generation of inter-KT tension on metaphase chromosomes (Howell et al., 2001; Yang et al., 2007). To test whether depletion of hSpindly leads to a reduction of tension, we measured the inter-KT distance between Hec1-positive KT pairs in cells treated for 30 min with MG132. Inter-KT distances at unaligned chromosomes in hSpindly-depleted cells were similar to those seen in GL2-depleted cells treated with nocodazole (0.790 ± 0.124 µm, n = 58 KT pairs from 10 cells and 0.767 ± 0.111 µm, n = 100 KT pairs from 10 cells, respectively). On fully aligned chromosomes, however, the inter-KT distance in hSpindly-depleted cells was ∼26% shorter than in GL2-depleted metaphase cells (1.167 ± 0.188 µm, n = 70 KT pairs from 10 cells) as compared with 1.306 ± 0.178 µm (n = 94 KT pairs from 10 cells). These data are statistically significant (P < 10−5) and show that hSpindly depletion leads to reduced KT tension, consistent with the notion that KT dynein contributes to tension establishment (Yang et al., 2007).
As shown recently, antibody-mediated inhibition of Nde1/Ndel, depletion of ZW10 (Stehman et al., 2007; Yang et al., 2007), or overexpression of a dynein tail fragment (Varma et al., 2008) all affect K-fiber stability. Therefore, we assessed whether hSpindly is also required for K-fiber stability. Cells were exposed to 4°C for 20 min, a procedure known to test the stability of K-fibers (Rieder, 1981). Whereas Hec1/Ndc80-depletion, known to impair KT–MT attachments, resulted in cells devoid of cold-stable MTs, hSpindly-depletion caused only a partial reduction of K-fiber stability (Fig. 7, A and B). This indicates that K-fibers could form in the absence of hSpindly, although they were less stable.
Figure 7.
K-fiber stability after hSpindly depletion. HeLa S3 cells were treated with GL2, hSpindly, or Hec1 siRNAs for 48 h and left at 37°C (A) or placed at 4°C (B) for 20 min, before they were stained with anti–α-tubulin antibody (green), CREST serum (red), and DAPI (blue). Bars, 10 µm.
hSpindly inhibition causes spindle misorientation in a dynein-dependent manner
As described above, we frequently observed spindles with an apparently monopolar morphology in hSpindly-depleted cells (Fig. 3 B). However, closer examination of such spindles, through staining for the centrosomal protein centrin-3 and analysis of the z-dimension, revealed two well-separated spindle poles (Fig. 8 A). The x-z projection of the centrosomal signal obtained from hSpindly-, ZW10-, and Nde1-depleted cells seeded on fibronectin-coated coverslips indicated that spindles had frequently rotated when compared with either GL2- or CenpE-depleted cells, although similar to hSpindly-depleted cells, the latter showed prominent chromosome misalignments (Tanudji et al., 2004) (Fig. 8 B; the efficiency of CenpE siRNA is shown in Fig. S5 A). To quantify these observations, the angle between the axis of each metaphase spindle and the substrate plane (Fig. 8 C, top panel, α) was measured, showing a clear increase in hSpindly-, ZW10-, and Nde1-depleted cells (means of 28, 24, and 26 degrees, respectively) as compared with GL2- or CenpE-depleted cells (means of 12 and 17 degrees, respectively; Fig. 8 C). Similar results were obtained also when seeding cells on noncoated coverslips (Fig. S5 B). Considering that depletion of hSpindly, ZW10, or Nde1 all affect the subcellular localization of dynein, we conclude that the observed spindle rotation most likely reflects the impairment of dynein/dynactin function rather than chromosome misalignment, mitotic delay, or cell adhesion conditions.
Compared with GL2- or CenpE-depleted cells, we also noticed that the average spindle length of hSpindly-, ZW10-, or Nde1-depleted cells was increased, although to a variable extent (Fig. 8 D; Fig. S5 C). One plausible explanation is that KT dynein facilitates the formation of load-bearing KT–MT attachments that counteract the forces pulling on astral MTs, as in the case of dynein controlled by SPDL-1 in C. elegans (Gassmann et al., 2008). Alternatively, hSpindly may be involved in regulating MT polymerization/depolymerization, although our results do not favor the latter hypothesis because spindle pole accumulation of NuMA and Kif2a were not noticeably affected in hSpindly-depleted cells (unpublished data).
Next, we analyzed both astral MTs and the actin cytoskeleton, previously shown to be required for proper spindle orientation in HeLa S3 cells (Toyoshima and Nishida, 2007). We did not observe defects in astral MT formation or cortical actin pattern in hSpindly-depleted cells (Fig. S5, D and E). EB1 depletion and cytochalasin B treatment were used as positive controls. To examine whether hSpindly regulates spindle orientation through dynein, we analyzed the spindle rotation in HeLa Kyoto cells stably expressing GFP-α-tubulin/cherry-H2B by imaging metaphase (MG132-treated) cells for 2 h. More than 60% of hSpindly-depleted cells, but only ∼30% of GL2-treated cells, showed misoriented chromosomes and spindles. Strikingly, co-depletion of dynein heavy chain 1 (DHC) (Toyoshima et al., 2007) partially rescued the orientation defects induced by hSpindly depletion (∼40% of cells) (Fig. 9, A and B; Videos 5–7). As depletion of DHC affects spindle assembly (an example of a DHC-depleted cell that never forms a bipolar spindle is shown in Fig. S5 F), we selected only metaphase cells for analysis (Fig. 9 A; Video 8). Co-depletion of hSpindly and DHC effectively decreased the levels of both proteins (Fig. S5 G). Finally, we failed to detect cortical hSpindly (Fig. 9 C) and hSpindly depletion did not noticeably alter dynein/dynactin localization to the cell cortex (Fig. 9 D), suggesting that hSpindly does not regulate cortical dynein.
The above results suggested that it is the KT pool of dynein that contributes to control spindle orientation. According to this notion, depletion of bona fide KT core components should result in a similar rotation phenotype as the depletion of hSpindly. Indeed, clear spindle misorientation (comparable to hSpindly depletion) was observed upon Hec1 or CenpA depletion (Fig. S5, H–J). Collectively, our results suggest that spindle misorientation induced by hSpindly depletion is due primarily to mislocalization of KT dynein.
Discussion
In this study we dissect the diverse functions of human Spindly. We show that hSpindly localizes to the outer KTs during prometaphase and relocalizes to the spindle poles before metaphase, in an MT- and dynein/dynactin-dependent manner. Similar to Drosophila Spindly and C. elegans SPDL-1 (Griffis et al., 2007; Gassmann et al., 2008), the RZZ complex acts as an upstream regulator of hSpindly. However, in human cells, the RZZ complex not only controls localization of hSpindly but also its protein level, suggesting that hSpindly is stabilized through association with the RZZ complex. SPDL-1 could be coimmunoprecipitated with Zwilch from C. elegans extracts (Gassmann et al., 2008). Similarly, we were able to coimmunoprecipitate hSpindly with ZW10 and Rod, provided that detergents were omitted, and our glycerol gradient analysis is also consistent with an interaction between hSpindly and the RZZ complex. We propose that hSpindly forms transient and dynamic interactions with the RZZ complex, rather than being a stable subunit of the complex. The fact that Hec1/Ndc80 depletion mislocalized both ZW10 and hSpindly without affecting the corresponding protein levels indicates that KT localization is not a prerequisite for the stability of hSpindly. Similar to the RZZ complex (Famulski and Chan, 2007; and unpublished data), Aurora B activity controls hSpindly localization in response to taxol. Although this might be interpreted to suggest a tension-sensitive localization of hSpindly, we emphasize that hSpindly occasionally showed an asymmetric localization on sister KTs, especially in monastrol-treated cells, arguing that hSpindly localization responds to a particular state of KT–MT attachment (or MT dynamics) rather than tension across the inner centromeres. Interestingly, inhibition of Aurora B also caused a reduction in hSpindly levels, suggesting a regulatory relationship between the two proteins, likely through the RZZ complex. We found that hSpindly is phosphorylated on at least five Cdk1 sites both in vitro and in vivo (unpublished data), but have so far been unable to phosphorylate the protein by Aurora B. Similarly, we have not been able to phosphorylate ZW10 by Aurora B (unpublished data). Thus, if Aurora B regulates the interaction between hSpindly and the RZZ complex by direct phosphorylation of one of the subunits, this most likely involves other members of the RZZ complex.
The role of Spindly in the KT recruitment of dynein is apparently well conserved among species. In contrast, whereas Drosophila Spindly recruits dynein but not dynactin (Griffis et al., 2007), both hSpindly and C. elegans SPDL-1 recruit both complexes to KTs (Gassmann et al., 2008). KT dynein is believed to control the initial lateral interaction between KTs and MTs and facilitate the subsequent formation of end-on attachments mediated by the Ndc80 complex (Rieder and Alexander, 1990; Gassmann et al., 2008; Vorozhko et al., 2008). Consistent with this notion, chromosome misalignment was induced by either depletion of hSpindly or ZW10 (Li et al., 2007; Yang et al., 2007; this paper). Remarkably, however, only cells depleted of hSpindly showed an overall lengthening in the duration of M phase, likely because cells depleted of ZW10 compensate a prolonged prometaphase with a premature onset of anaphase (Yang et al., 2007), due to SAC defects (Buffin et al., 2005; Kops et al., 2005). Thus, the specific defects resulting from the impaired recruitment of dynein to KTs can more readily be visualized in hSpindly-depleted cells, as this protein is not required for SAC activity.
KT dynein was suggested to be required for the generation of tension across sister KTs (Howell et al., 2001; Yang et al., 2007). The decreased inter-KT distance observed upon hSpindly depletion clearly supports this notion. Furthermore, the increase in spindle length upon hSpindly knock-down is in agreement with the idea that KT dynein facilitates the formation of load-bearing attachment, as is the case for SPDL-1 (Gassmann et al., 2008). For C. elegans it was further proposed that the RZZ complex negatively regulates the MT-binding activity of the Ndc80 complex at tensionless KTs (Gassmann et al., 2008). The data obtained here for HeLa S3 cells suggest that the functions of hSpindly and ZW10 differ significantly between species. First, in human cells the RZZ complex controls not only the localization but also the stability of hSpindly, whereas in invertebrates it apparently controls only localization. Second, the depletion of Hec1/Ndc80 abolished KT localization of both ZW10 and hSpindly in human cells, but not in C. elegans (Gassmann et al., 2008; Yamamoto et al., 2008). Third, KNL-1 in C. elegans is required for KT targeting of both Ndc80 and RZZ complexes (Desai et al., 2003; Gassmann et al., 2008), whereas Blinkin, its human homologue, is not required for the KT localization of Hec1/Ndc80 (Kiyomitsu et al., 2007). Together with data from the literature, our present results suggest that in human cells hSpindly and the RZZ complex act downstream of the Ndc80 complex to recruit dynein/dynactin.
Interference with Spindly function in different species also revealed fundamental differences with regard to the role of KT dynein during SAC inactivation. Several studies have implicated dynein/dynactin in the silencing of the SAC through the removal of checkpoint proteins, such as MAD2 and the RZZ complex, from KTs and their transport along MTs to spindle poles (Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004; Griffis et al., 2007; Mische et al., 2008; Varma et al., 2008; Whyte et al., 2008; Sivaram et al., 2009). In contrast, hSpindly depletion did not block the removal of MAD2 and ZW10 from aligned chromosomes, and yet this process required dynein/dynactin. This suggests that the dynein/dynactin complex is able to remove SAC components from KTs without the need for an hSpindly-mediated accumulation of dynein at these structures. It is worth mentioning in this context that we seldom observed a complete inhibition of SAC protein removal upon overexpression of p50-dynamitin in metaphase cells. This could either reflect an incomplete disruption of the dynein/dynactin complex, or alternatively, the existence of a dynein-independent removal mechanism that is able to compensate for the loss of dynein. Regardless, hSpindly-depleted cells clearly showed a prolonged prometaphase delay, indicating persistent SAC activity despite ongoing removal of MAD2.
Importantly, we also found that depletion of hSpindly or ZW10 induced frequent spindle rotation, similar to what had previously been observed upon depletion of the dynein regulator Nde1 (Feng and Walsh, 2004) and depletion or overexpression of LIS1 (Faulkner et al., 2000; Yingling et al., 2008). These proteins are well known to determine the stability of astral MTs, the cortical localization of dynein and proper spindle assembly (Faulkner et al., 2000; Feng et al., 2000; Feng and Walsh, 2004; Yingling et al., 2008). In addition, ZW10, Nde1, and dynein are also required for the integrity of the Golgi apparatus (Hirose et al., 2004; Liang et al., 2004), but this is not the case for hSpindly (unpublished data). These studies thus suggest that ZW10, Nde1/Ndel1, and LIS1 regulate dynein activity both at the KT and elsewhere, whereas hSpindly exclusively controls the function of dynein at KTs.
So, how could hSpindly depletion lead to spindle misorientation? hSpindly was undetectable at the cortex or on astral MTs. Furthermore, neither the actin cytoskeleton nor the cortical localization of dynein was detectably impaired in hSpindly-depleted cells. Yet, remarkably, the spindle rotation phenotype could be rescued by co-depletion of dynein. The observation that a comparable spindle orientation defect could also be induced by depletion of Hec1 or CenpA highlights a role of KTs in spindle orientation and supports the notion that spindle misorientation induced by hSpindly depletion is due to interference with KT-associated but not cortical dynein. It seems plausible, therefore, that KT forces cooperate with cortical forces essential for spindle orientation.
In summary, our characterization of hSpindly helps to understand which dynein-mediated processes require dynein accumulation at the KT (notably chromosome congression) and which processes do not (notably SAC silencing). Both chromosome alignment and KT tension are now confirmed to be regulated by KT-associated dynein/dynactin complexes, which in turn are regulated by Spindly and the RZZ complex in all metazoans. In contrast, the mechanism by which hSpindly and KT dynein control spindle orientation awaits elucidation.
Materials and methods
Cloning procedures
For cloning hSpindly (Q96EA4), a cDNA clone (IRAUp969B0653D) was obtained from the Deutsches Ressourcenzentrum für Genomforschung (RZPD), and was cloned in-frame into a pcDNA3.1 vector (Invitrogen) encoding an N-terminal 3xmyc tag. The siRNA-resistant version of hSpindly containing six silent mutations was produced by mutating the siRNA-corresponding region with the following pair of primers (5′-TTACAGAATCAATTGGACAAGTGCCGCAACGAGATGATGACCATG-3′ and its antisense) using QuickChange Site-Directed Mutagenesis kit (Stratagene). Underlined letters correspond to the six silent point mutations made. For cloning of p50-dynamitin, a cDNA clone (IMAGE: 3504109) was obtained from Geneservice and cloned into pcDNA3.1 vector encoding an N-terminal 3xmyc tag.
Protein purification and antibody production
Antibodies against hSpindly were generated by immunization of rabbits (Charles River Laboratories) with five injections of 250 µg of N-terminal His6-tagged hSpindly (aa 1–444) produced in Escherichia coli and purified by Ni-NTA agarose beads (QIAGEN). Anti-hSpindly antibodies were purified by Affi-Prep protein A Matrix (Bio-Rad Laboratories). A MAD2 monoclonal antibody (IgG1κ) was generated in-house against GST-tagged full-length MAD2.
Cell culture and synchronization
HeLa S3 cells were cultured at 37°C, in a 5% CO2 atmosphere in DME (Invitrogen), supplemented with 10% heat-inactivated fetal calf serum (FCS) and penicillin/streptomycin (100 IU/ml and 100 µg/ml, respectively). Nocodazole (100 ng/ml), taxol (1 µM), monastrol (150 µM), cytochalasin B (4 µM), G418 (0.5 mg/ml), puromycin (0.5 µg/ml), 2-deoxy-d-glucose (1 mM), sodium azide (5 mM), and thymidine (2 mM) were obtained from Sigma-Aldrich. ZM447439 (10 µM) was obtained from Tocris. MG132 (10 µM) was obtained from EMD. Fibronectin-coated coverslips were purchased from BD Biosciences. ATP reduction assays were performed as described previously (Howell et al., 2001).
Transient plasmids and siRNA transfection
Plasmid transfections were performed using TransIT-LT1 reagent (Mirus Bio Corporation) according to the manufacturer's instructions. siRNA duplexes were transfected using Oligofectamine (Invitrogen) as described previously (Elbashir et al., 2001). The following siRNA duplexes were purchased from QIAGEN: hSpindly-1, 5′-GGAGAAAUUUAAGAAUUUA-3′; hSpindly-2, 5′-GGAUAAAUGUCGUAAUGAA-3′ (due to a more efficient depletion [see Fig. 3 A] hSpindly-2 was used throughout our study unless otherwise stated); ZW10-2, 5′-CCACGAAGUGAUGAAUUUA-3′; and EB1, 5′-UUGCCUUGAAGAAAGUGAA-3′. The siRNAs ZW10-1 (Kops et al., 2005; used throughout our study unless otherwise stated), Hec1, MAD2, CenpE (Martin-Lluesma et al., 2002), Aurora B, CenpA (Klein et al., 2006), Nde1 (Vergnolle and Taylor, 2007), and DHC (DHC1; Toyoshima et al., 2007) were published previously.
Cell extracts and Western blotting
Preparation of lysates and Western blotting analysis were performed as described previously (Sillje et al., 2006). For coimmunoprecipitation between hSpindly and the RZZ complex, cells were first resuspended in Hepes buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM DTT, 30 µg/ml RNase A, 30 µg/ml DNase, protease inhibitors, and phosphatase inhibitors) before being opened by nitrogen cavitation (1000 PSI, 20 min; Parr Instrument). For phosphatase assay, 2 µl of λ-phosphatase (Roche) was added to 100 µg of cell extracts and these were incubated at 37°C for 30 min.
Glycerol gradient centrifugation
Cell lysates (10 mg) were applied on a 10–25% glycerol cushion (10 ml) and spun down at 28,000 rpm (SW 40 Ti rotor; Beckman Coulter) for 14 h. 27 fractions of 400 µl each were collected for Western blotting analysis. Albumin, aldolase, ferritin, and thyroglobulin were used as controls to determine the sedimentation coefficient.
Immunofluorescence microscopy
HeLa S3 cells were grown on coverslips and simultaneously fixed and permeabilized for 10 min at RT in PTEMF buffer, as described previously (Sillje et al., 2006). For astral MTs, detection cells were fixed with cold methanol for 10 min. For cortical p150Glued detection, cells were first pre-extracted with 0.5% Triton X-100 in PEM (20 mM PIPES, pH 6.8, 10 mM EGTA, and 1 mM MgCl2), followed by cold methanol fixation for 10 min. For phalloidin staining, cells were fixed with 3.7% formaldehyde for 10 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min, followed by incubation with Alexa Fluor 546–phalloidin (Invitrogen) for 1 h. Primary and secondary antibodies used in this study are listed below. DNA was stained with DAPI (2 µg/ml). Immunofluorescence microscopy was preformed as described previously (Sillje et al., 2006). Deconvolved images and X-Y or Z-X projections were obtained by using Softworx (Applied Precision, LLC).
Antibodies
The following antibodies were used: rabbit anti-hSpindly (1:100 for Western blotting; 1:1,000 for immunofluorescence), rabbit anti-histone H3 (pS10-H3, 1:1,000; Millipore), mouse anti–p50-dynamitin (1:1,000; BD Biosciences), mouse anti-MAD2 (1:200; tissue culture supernatant), rabbit anti-MAD2 (1:1,000; Bethyl Laboratories), goat anti-CenpE (1:200; Santa Cruz Biotechnology, Inc.), mouse anti-CDC27 (1:1,000; BD Biosciences), mouse anti-Rod (1:100; Abnova), mouse anti-CenpA (1:1,000; MBL International); mouse anti–α-tubulin (1:5,000; Sigma-Aldrich), sheep anti–α-tubulin (1:400; Santa Cruz Biotechnology, Inc.), rabbit anti-ZW10 (1:400; Abcam), mouse anti-DIC (clone 70.1; 1:1,000 for Western blotting; 1:200 for immunofluorescence; Sigma-Aldrich), mouse anti-p150Glued (1:1,000; BD Biosciences), mouse anti-myc (1:5; 9E10 tissue culture supernatant), rabbit anti-myc (1:500; Santa Cruz Biotechnology, Inc.), mouse anti-cyclin B1 (1:1,000; Millipore), mouse anti-Hec1 (1:1,000; GeneTex GTX70268), rabbit anti-Nde1 (1:1,000; Proteintech Group), mouse anti-Aurora B (1:500; Abcam) and rabbit anti-DHC 1 (1:100; Santa Cruz Biotechnology, Inc.), mouse anti-BubR1 (1:5; Elowe et al., 2007), human CREST autoimmune serum (1:2,000; Immunovision), mouse anti-GM130 (1:500; BD Biosciences), rabbit anti-pericentrin (1:2,000; Abcam), and goat anti–centrin-3 (1:200; Thein et al., 2007). For immunofluorescence analysis, primary antibodies were detected with Cy2-, Cy3-, and Cy5-conjugated donkey anti–mouse, anti–rabbit, anti–goat, or anti–human IgGs (1:1,000; Dianova).
Time-lapse microscopy
Time-lapse analysis was preformed as described previously (Gaitanos et al., 2009). In brief, culture plates (Ibidi) were placed onto a sample stage within an incubator chamber (EMBLEM, Heidelberg, Germany) maintained at a temperature of 37°C, humidity 50%, in an atmosphere of 5% CO2. Imaging was performed using a microscope (Axio Observer Z1; Carl Zeiss, Inc.) equipped with Plan Neofluar 20x and 40x objectives. MetaMorph 7.1 software (MDS Analytical Technologies) was used to collect and process data. For analysis of mitotic timing, a HeLa S3 cell line stably expressing histone H2B-GFP was used (Sillje et al., 2006). Imaging was performed using a 20x objective and images were captured using 10-ms exposure times for GFP every 3 min, for 16 h. For analysis of spindle rotation, a HeLa Kyoto cell line with a FRT-site and stably expressing GFP-α-tubulin/cherry-H2B cell line was used (provided by D. Gerlich, ETH Zurich, Switzerland). Imaging was performed using a 40x objective and images were captured using 10- and 80-ms exposure time for cherry and EGFP, respectively, every 3 min.
Image processing, quantification, and statistical analysis
Images taken at identical exposure times within each experiment, acquired as 8-bit RGB images, were processed in Adobe Photoshop. Quantification of KT intensities was performed with ImageJ (http://rsb.info.nih.gov/ij/) as described previously (Elowe et al., 2007). Inter-KT distances were determined by measuring the distance between the center of sister Hec1 dots in the same optical section. In the box-and-whisker plot, the boxes represent 25–75% of the cells. Within the box the line indicates the median, and a black dot indicates the mean. The whiskers represent the 5th and 95th percentiles. The crosses represent the maximum and the minimum values measured. Statistical significance was verified by Student's t test.
Online supplemental material
Figure S1 shows the characterization of hSpindly. Figure S2 shows the analysis of hSpindly and ZW10 by immunoprecipitation, glycerol gradient centrifugation, and immunofluorescence. Figure S3 shows the characterization of the generated MAD2 antibody and the analysis of the KT localization of hSpindly in response to monastrol. Figure S4 shows the immunofluorescence analysis of several KT proteins upon hSpindly depletion. Figure S5 shows the analysis of spindle misorientation after depletion of hSpindly and other KT components. Videos 1–4 show the mitotic timing of HeLa S3 cells stably expressing H2B-GFP treated with GL2, hSpindly-1, hSpindly-2, or ZW10 siRNAs, respectively. Videos 5–8 show the spindle rotation of HeLa Kyoto cells stably expressing GFP-α-tubulin/cherry-H2B treated with GL2, GL2+hSpindly-2, hSpindly-2+DHC, or GL2+DHC siRNAs, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812167/DC1.
Acknowledgments
We thank Anja Wehner for technical assistance. We gratefully acknowledge Thomas Gaitanos for critical reading of the manuscript and insightful discussion and all the members of the Nigg laboratory for helpful suggestions. Y.W. Chan performed the experiments, A. Uldschmid, and L.L. Fava generated and characterized the MAD2 antibody, respectively; M.H.A. Schmitz and D.W. Gerlich provided the HeLa Kyoto GFP-α-tubulin/cherry-H2B cell line, A. Santamaria and E.A. Nigg supervised the work; and Y.W. Chan, E.A. Nigg, and A. Santamaria wrote the manuscript.
This work was supported by the Max Planck Society and the ENFIN, Network of Excellence funded by the European Commission within its FP6 Program (contract number LSHG-CT-2005-518254). A. Santamaria is supported by the Spanish Education and Science Ministry.
Footnotes
Abbreviations used in this paper: DHC, dynein heavy chain; DIC, dynein intermediate chain; GL2, Photinus pyralis luciferase gene; hSpindly, human Spindly; KT, kinetochore; MT, microtubule; RZZ, Rod/Zw10/Zwilch; SAC, spindle assembly checkpoint.
References
- Basto R., Gomes R., Karess R.E. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila.Nat. Cell Biol. 2:939–943 [DOI] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis.Curr. Biol. 14:56–61 [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex.Curr. Biol. 15:856–861 [DOI] [PubMed] [Google Scholar]
- Busson S., Dujardin D., Moreau A., Dompierre J., De Mey J.R. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells.Curr. Biol. 8:541–544 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., Yen T.J. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores.Nat. Cell Biol. 2:944–947 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface.Nat. Rev. Mol. Cell Biol. 9:33–46 [DOI] [PubMed] [Google Scholar]
- Civril F., Musacchio A. 2008. Spindly attachments.Genes Dev. 22:2302–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Muller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans.Genes Dev. 17:2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores.J. Cell Biol. 161:267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal B.M., Vaughan K.T., Vallee R.B. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis.J. Cell Biol. 132:617–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells.Nature. 411:494–498 [DOI] [PubMed] [Google Scholar]
- Elowe S., Hummer S., Uldschmid A., Li X., Nigg E.A. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions.Genes Dev. 21:2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Chan G.K. 2007. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores.Curr. Biol. 17:2143–2149 [DOI] [PubMed] [Google Scholar]
- Faulkner N.E., Dujardin D.L., Tai C.Y., Vaughan K.T., O'Connell C.B., Wang Y., Vallee R.B. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function.Nat. Cell Biol. 2:784–791 [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C.A. 2004. Mitotic spindle regulation by Nde1 controls cerebral cortical size.Neuron. 44:279–293 [DOI] [PubMed] [Google Scholar]
- Feng Y., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A. 2000. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome.Neuron. 28:665–679 [DOI] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T.M. 2004. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles.J. Cell Biol. 166:465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos T.N., Santamaria A., Jeyaprakash A.A., Wang B., Conti E., Nigg E.A. 2009. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3.EMBO J. doi:10.1038/emboj.2009.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O'Rourke S.M., Bowerman B., McLeod I., Yates J.R., III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex.Genes Dev. 22:2385–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R.D. 2005. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins.J. Cell Biol. 171:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., Vale R.D. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore.J. Cell Biol. 177:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint.J. Cell Biol. 161:281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., Tagaya M. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi.EMBO J. 23:1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation.J. Cell Biol. 155:1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint.Trends Cell Biol. 15:386–392 [DOI] [PubMed] [Google Scholar]
- King J.M., Hays T.S., Nicklas R.B. 2000. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension.J. Cell Biol. 151:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1.Dev. Cell. 13:663–676 [DOI] [PubMed] [Google Scholar]
- Klein U.R., Nigg E.A., Gruneberg U. 2006. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP.Mol. Biol. Cell. 17:2547–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R., III, Tagaya M., Cleveland D.W. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore.J. Cell Biol. 169:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yu W., Liang Y., Zhu X. 2007. Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment.Cell Res. 17:701–712 [DOI] [PubMed] [Google Scholar]
- Liang Y., Yu W., Li Y., Yang Z., Yan X., Huang Q., Zhu X. 2004. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein.J. Cell Biol. 164:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Yu W., Li Y., Yu L., Zhang Q., Wang F., Yang Z., Du J., Huang Q., Yao X., Zhu X. 2007. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase.Mol. Biol. Cell. 18:2656–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.T., Chen Y., Wu G., Lee W.H. 2006. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control.Oncogene. 25:6901–6914 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., Nigg E.A. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2.Science. 297:2267–2270 [DOI] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., Hays T.S. 2008. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation.Mol. Biol. Cell. 19:4918–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time.Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- O'Connell C.B., Wang Y.L. 2000. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion.Mol. Biol. Cell. 11:1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell C.B., Khodjakov A.L. 2007. Cooperative mechanisms of mitotic spindle formation.J. Cell Sci. 120:1717–1722 [DOI] [PubMed] [Google Scholar]
- Rieder C.L. 1981. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells.Chromosoma. 84:145–158 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Alexander S.P. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells.J. Cell Biol. 110:81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G., Korner R., Hanisch A., Ries A., Nigg E.A., Sillje H.H. 2005. Proteome analysis of the human mitotic spindle.Mol. Cell. Proteomics. 4:35–43 [DOI] [PubMed] [Google Scholar]
- Savoian M.S., Goldberg M.L., Rieder C.L. 2000. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants.Nat. Cell Biol. 2:948–952 [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Brown H.M., Kwon M., Rogers G.C., Holland G., Scholey J.M. 2000. Functional coordination of three mitotic motors in Drosophila embryos.Mol. Biol. Cell. 11:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje H.H., Nagel S., Korner R., Nigg E.A. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes.Curr. Biol. 16:731–742 [DOI] [PubMed] [Google Scholar]
- Sivaram M.V., Wadzinski T.L., Redick S.D., Manna T., Doxsey S.J. 2009. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint.EMBO J. 28:902–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., Andreassen P.R., Lacroix F.B., Wilson L., Margolis R.L. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints.Proc. Natl. Acad. Sci. USA. 98:4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Hays T.S., Goldberg M.L. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore.J. Cell Biol. 142:763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman S.A., Chen Y., McKenney R.J., Vallee R.B. 2007. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores.J. Cell Biol. 178:583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U. 2008. Bi-orienting chromosomes: acrobatics on the mitotic spindle.Chromosoma. 117:521–533 [DOI] [PubMed] [Google Scholar]
- Tanudji M., Shoemaker J., L'Italien L., Russell L., Chin G., Schebye X.M. 2004. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay.Mol. Biol. Cell. 15:3771–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein K.H., Kleylein-Sohn J., Nigg E.A., Gruneberg U. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity.J. Cell Biol. 178:345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Nishida E. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner.EMBO J. 26:1487–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Matsumura S., Morimoto H., Mitsushima M., Nishida E. 2007. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells.Dev. Cell. 13:796–811 [DOI] [PubMed] [Google Scholar]
- Vaisberg E.A., Koonce M.P., McIntosh J.R. 1993. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation.J. Cell Biol. 123:849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Monzo P., Stehman S.A., Vallee R.B. 2008. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment.J. Cell Biol. 182:1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle M.A., Taylor S.S. 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes.Curr. Biol. 17:1173–1179 [DOI] [PubMed] [Google Scholar]
- Vorozhko V.V., Emanuele M.J., Kallio M.J., Stukenberg P.T., Gorbsky G.J. 2008. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin.Chromosoma. 117:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J., Bader J.R., Tauhata S.B., Raycroft M., Hornick J., Pfister K.K., Lane W.S., Chan G.K., Hinchcliffe E.H., Vaughan P.S., Vaughan K.T. 2008. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis.J. Cell Biol. 183:819–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Goldberg M.L. 1994. Determinants of Drosophila zw10 protein localization and function.J. Cell Sci. 107(Pt 4):785–798 [DOI] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaerou F., Karess R., Hays T. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein.Nat. Cell Biol. 3:1001–1007 [DOI] [PubMed] [Google Scholar]
- Yamamoto T.G., Watanabe S., Essex A., Kitagawa R. 2008. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans.J. Cell Biol. 183:187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U.S., Wadsworth P., Rieder C.L. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint.Curr. Biol. 17:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J., Youn Y.H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. 2008. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division.Cell. 132:474–486 [DOI] [PMC free article] [PubMed] [Google Scholar]