Abstract
Ribosomes synthesizing secretory and membrane proteins are bound to translocons at the membrane of the endoplasmic reticulum (ER). Both the ribosome and translocon are complex macromolecular machines whose structural and functional interactions are poorly understood. A new study by Pool (Pool, M.R. 2009. J. Cell Biol. 185:889–902) has now shown that the structure of the translocon is dictated by the identity of the protein being synthesized by the ribosome, thereby demonstrating that the two macromolecular machines are structurally coupled for functional purposes. The study also identifies an unexpected component in the apparent molecular linkage that connects the two machines, a discovery that shows the current view of translocon structure is oversimplified.
The mammalian translocon is assembled from four core proteins (the heterotrimeric Sec61α, Sec61β, Sec61γ, and translocating nascent chain–associated membrane protein [TRAM]) and various accessory proteins, only some of which have well-defined functions (e.g., the signal peptidase and the oligosaccharyltransferase; Rapoport, 2007). This assembly forms the aqueous, gated pore that transports nascent proteins through the membrane (Crowley et al., 1994). Because two molecular machines, the ribosome and the translocon, function simultaneously on the same nascent protein during cotranslational protein trafficking, they presumably function together as a unit (the ribosome–translocon complex [RTC]) that includes all proteins regularly associated with the translocon. Yet these machines are commonly treated as separate entities in papers and talks; ribosomologists tend to assume that membrane-bound ribosomes are indistinguishable from free ribosomes except for their location, whereas translocophiles tend to consider the ribosome simply a source of substrate that docks on the translocon. However, the results published in this issue (see Pool on p. 889) demonstrate that the identity of the nascent chain being synthesized alters RTC structure, thereby revealing that the two machines are indeed coupled.
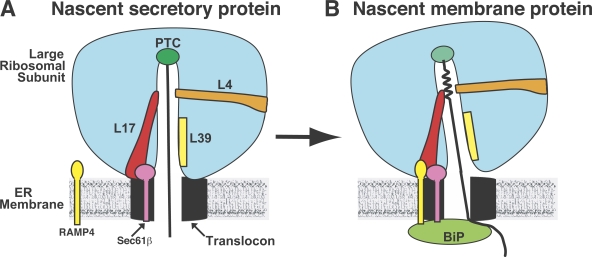
The nascent chain moves through the large ribosomal subunit via an ∼100-Å-long tunnel that is contiguous with the aqueous pore formed by the translocon (Fig. 1 A; Crowley et al., 1994; Beckmann et al., 1997; Nissen et al., 2000). Nascent chain control of protein trafficking from inside the ribosome was first identified when the location of a nascent chain transmembrane segment (TMS) in the tunnel was found to dictate whether the nascent chain was exposed to the cytosolic, lumenal, or neither side of the ER membrane (Liao et al., 1997). It was postulated that a weakly nonpolar patch in the tunnel nucleated the folding of the TMS into an α-helix, which in turn elicited conformational changes in the RTC that triggered complementary changes at each end of the pore to minimize ion passage/leakage through the translocon during integration. A later study revealed that ribosome-induced folding of a nascent chain TMS did occur, and this folding coincided with TMS photocrosslinking to Rpl17 at a constriction in the tunnel (Fig. 1 B; Woolhead et al., 2004), a site formed in part by a loop of Rpl17 that extends far into the large ribosomal subunit (Nissen et al., 2000; Berisio et al., 2003).
Figure 1.
Nascent chain control of translocon structure from inside the ribosome. (A and B) A nascent secretory protein is fully extended during synthesis (A), whereas the TMS in a nascent membrane protein (B) folds into an α-helix upon reaching a tunnel constriction formed by Rpl4 and Rpl17. Although Sec61β is always adjacent to Rpl17 in an RTC, RAMP4 is recruited to the RTC and is cross-linked to Rpl17 only when a TMS reaches the constriction. The cross-linking of Rpl17 to a TMS and to RAMP4 coincides with the BiP-mediated closure (either directly, as depicted, or indirectly) of the lumenal end of the aqueous pore and the subsequent opening of the ion-tight ribosome–translocon junction (depicted by a tilting of the ribosomal subunit). PTC, peptidyl transferase center.
Pool (2009) found that ribosomal protein Rpl17, located primarily at the ribosomal surface near the tunnel exit, was chemically cross-linked to Sec61β, thereby showing that Sec61β is adjacent to Rpl17 in all RTCs. But when the ribosome was synthesizing a membrane protein, Rpl17 also cross-linked to RAMP4, a small ribosome-associated membrane protein associated with the translocon (Schröder et al., 1999). Strikingly, Rpl17 cross-linking to RAMP4 was detected only after the TMS in the nascent chain reached the tunnel constriction (Fig. 1 B). The coincidence of these two cross-linking events, TMS to Rpl17 and Rpl17 to RAMP4, indicates that direct contact between the nascent chain TMS and Rpl17 inside the tunnel triggered a conformational change that was transmitted through the Rpl17 extension to the ribosomal surface and that stimulated RAMP4 association with the RTC close to Rpl17. Thus, a structural feature found only in nascent membrane proteins caused a structural realignment of the RTC. This change correlates with the transition of RTC operations from translocation to integration seen in earlier studies (Liao et al., 1997; Haigh and Johnson, 2002; Woolhead et al., 2004).
The data shown by Pool (2009) also demonstrate that the translocon, at least in mammals, is not as structurally defined as has been portrayed. Uncertainties in the composition, stoichiometry, exchangeability, and macromolecular arrangement of the core and associated translocon proteins, as well as translocon dynamics and homogeneity, have been recognized for some time (Johnson and van Waes, 1999), but only recently have translocon heterogeneity (Snapp et al., 2004; Shibatani et al., 2005), conformational changes (Hamman et al., 1997), and exchangeable membrane proteins (e.g., importin α-16 appears to be a sorting factor for inner nuclear membrane proteins; Saksena et al., 2004, 2006) been documented. The data now show that RAMP4 in the bilayer associates with the RTC near Rpl17 when a TMS binds Rpl17 inside the ribosome Pool, 2009). Thus, translocon structure is dynamic and involves more proteins than the core Sec61α, Sec61β, Sec61γ, and TRAM proteins. Equally important, as shown by Pool (2009), translocon structure can be altered by a nascent chain structural feature from inside the ribosome. Thus, the structures of the ribosome and translocon are intimately coupled, and the conversion of the RTC from one functional state to another involves structural changes (e.g., the introduction of a new protein) not accommodated in current models of translocon structure and function.
The study by Pool (2009) also emphasizes the importance of using multiple approaches to examine complex systems, especially those containing membranes. Protein crystallography and cryo-EM have made tremendous strides recently in describing the structures of molecular assemblies such as the ribosome and translocon (Beckmann et al., 1997; Nissen et al., 2000; Berisio et al., 2003; Van den Berg et al., 2004), and it is difficult to overstate how important those studies have been in terms of influencing subsequent RTC research. Yet no single technique is a panacea, and the data shown by Pool (2009) dramatically highlight the limitations of assuming that crystallographic and cryo-EM studies provide all that one needs to understand the structural aspects of how a machine works, much less the functional, mechanistic, and/or regulatory aspects. For example, RAMP4, the core protein TRAM, and other translocon-associated proteins have not been identified in cryo-EM images of the detergent-solubilized RTC as a result of their small size, flexibility, and/or weak association with the Sec61 core. Similarly, the existence of ribosome-induced TMS folding inside the ribosome tunnel and its regulation of nascent chain accessibility to alternate sides of the ER membrane would not have been detected with samples that had been detergent-treated and lacked water, the lipid bilayer, and some of the core and associated translocon proteins. Thus, although models derived from crystallographic and cryo-EM images of detergent-treated, incomplete, anhydrous, and nonfunctional RTC samples may prove to be accurate, well-designed experiments and controls using intact samples in aqueous solution must be performed to directly correlate structural and functional states and thereby fully appreciate subtle and sophisticated operational and regulatory mechanisms that do not withstand harsh treatment.
There is much left to learn about the RTC. Rpl17 acts as a direct communication conduit between a TMS in the tunnel and the translocon, but it remains to be seen whether RAMP4 then mediates transmembrane communication and triggers BiP binding. Similarly, RAMP4 interactions with the RTC have not been characterized nor have the protein composition, arrangement, and dynamics of the mammalian ER translocon in intact membranes. Thus, additional intriguing surprises are ahead.
References
- Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. 1997. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex.Science. 278:2123–2126 [DOI] [PubMed] [Google Scholar]
- Berisio R., Schlünzen F., Harms J., Bashan A., Auerbach T., Baram D., Yonath A. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation.Nat. Struct. Biol. 10:366–370 [DOI] [PubMed] [Google Scholar]
- Crowley K.S., Liao S., Worrell V.E., Reinhart G.D., Johnson A.E. 1994. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore.Cell. 78:461–471 [DOI] [PubMed] [Google Scholar]
- Haigh N.G., Johnson A.E. 2002. A new role for BiP: gating the aqueous ER translocon pore during membrane protein integration.J. Cell Biol. 156:261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B.D., Chen J.-C., Johnson E.E., Johnson A.E. 1997. The aqueous pore through the translocon has a diameter of 40-60 Å during cotranslational protein translocation at the ER membrane.Cell. 89:535–544 [DOI] [PubMed] [Google Scholar]
- Johnson A.E., van Waes M.A. 1999. The translocon: a dynamic gateway at the ER membrane.Annu. Rev. Cell Dev. Biol. 15:799–842 [DOI] [PubMed] [Google Scholar]
- Liao S., Lin J., Do H., Johnson A.E. 1997. Both lumenal and cytosolic gating of the aqueous ER translocon pore is regulated from inside the ribosome during membrane protein integration.Cell. 90:31–41 [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis.Science. 289:920–930 [DOI] [PubMed] [Google Scholar]
- Pool M.R. 2009. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase.J. Cell Biol. 185:889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes.Nature. 450:663–669 [DOI] [PubMed] [Google Scholar]
- Saksena S., Shao Y., Braunagel S., Summers M.D., Johnson A.E. 2004. Cotranslational integration and initial sorting at the endoplasmic reticulum translocon of proteins destined for the inner nuclear membrane.Proc. Natl. Acad. Sci. USA. 101:12537–12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Summers M.D., Burks J.K., Johnson A.E., Braunagel S.C. 2006. Importin α-16: a translocon associated protein that may facilitate sorting of integral membrane proteins to the nuclear envelope.Nat. Struct. Mol. Biol. 13:500–508 [DOI] [PubMed] [Google Scholar]
- Schröder K., Martoglio B., Hofmann M., Hölscher C., Hartmann E., Prehn S., Rapoport T.A., Dobberstein B. 1999. Control of glycosylation of MHC class II-associated invariant chain by translocon-associated RAMP4.EMBO J. 18:4804–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatani T., David L.L., McCormack A.L., Frueh K., Skach W.R. 2005. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits.Biochemistry. 44:5982–5992 [DOI] [PubMed] [Google Scholar]
- Snapp E.L., Reinhart G.A., Bogert B.A., Lippincott-Schwartz J., Hegde R.S. 2004. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells.J. Cell Biol. 164:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W.M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S.C., Rapoport T.A. 2004. X-ray structure of a protein-conducting channel.Nature. 427:36–44 [DOI] [PubMed] [Google Scholar]
- Woolhead C.A., McCormick P.J., Johnson A.E. 2004. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins.Cell. 116:725–736 [DOI] [PubMed] [Google Scholar]