Abstract
A remarkable instability at simple repeated sequences characterizes gastrointestinal cancer of the microsatellite mutator phenotype (MMP). Mutations in the DNA mismatch repair gene family underlie the MMP, a landmark for hereditary nonpolyposis colorectal cancer. These tumors define a distinctive pathway for carcinogenesis because they display a particular spectrum of mutated cancer genes containing target repeats for mismatch repair deficiency. One such gene is BAX, a proapoptotic member of the Bcl-2 family of proteins, which plays a key role in programmed cell death. More than half of colon and gastric cancers of the MMP contain BAX frameshifts in a (G)8 mononucleotide tract. However, the functional significance of these mutations in tumor progression has not been established. Here we show that inactivation of the wild-type BAX allele by de novo frameshift mutations confers a strong advantage during tumor clonal evolution. Tumor subclones with only mutant alleles frequently appeared after inoculation into nude mice of single-cell clones of colon tumor cell lines with normal alleles. In contrast, no clones of BAX-expressing cells were found after inoculation of homozygous cell clones without wild-type BAX. These results support the interpretation that BAX inactivation contributes to tumor progression by providing a survival advantage. In this context, survival analyses show that BAX mutations are indicators of poor prognosis for both colon and gastric cancer of the MMP.
Tumors of the microsatellite mutator phenotype (MMP) accumulate hundreds of thousands of clonal somatic mutations in simple repeated sequences (1). The MMP, also known as replication error or microsatellite instability, is characteristic of the majority of tumors from kindreds with hereditary nonpolyposis colorectal cancer and a minority of sporadic gastrointestinal cancers (1–4). Tumors of the MMP are paradigmatic for the cancer as a mutator phenotype hypothesis (5). This profound genomic instability is a result of mutations in the DNA mismatch repair gene family (6), germ-line mutations in hereditary nonpolyposis colorectal cancer, and somatic mutations in sporadic gastrointestinal cancer (7, 8).
Tumors of the MMP display striking differences in phenotype relative to tumors without microsatellite instability (1, 2, 7, 9). MMP colon cancers represent a distinct molecular pathway for carcinogenesis (10–12) because, as a result of their mutator phenotype, the mutated cancer genes are, in general, different from those mutated in tumors without mismatch repair deficiency (1, 8, 9–14). The proapoptotic gene BAX is one of these specific cancer genes of tumors of the mutator phenotype pathway. More than half of gastrointestinal MMP tumors contain BAX mutations, whereas these mutations are rarely found in tumors without enhanced microsatellite instability (15–19). BAX mutations have been also identified in hematological malignancies (20).
BAX belongs to the Bcl-2 family of proteins and is a key player in apoptosis (21, 22). The Bcl-2 gene was the first isolated oncogene that functions by altering the control for programmed cell death rather than by stimulating cell proliferation (23). BAX and Bcl-2 are antagonists and their balance regulates the apoptotic response by the cell to some apoptotic stimuli (24). BAX is localized in the cytoplasm and translocates to mitochondria in response to apoptotic stimuli, where it promotes cell death by inducing the formation of ion-permeable pores that disrupt the mitochondrial membrane barrier (25–27). BAX also contributes to the apoptotic response by binding to antiapoptotic proteins of the Bcl-2 family via its BH3 domain and inhibiting their functions (28, 29).
The common occurrence of frameshift mutations in the BAX gene in colon and gastric cancers of the MMP suggests that inactivation of BAX during tumorigenesis may contribute to tumor progression by enhancing escape from apoptosis. This hypothesis is supported by the effect of reduced BAX expression in some systems (30–32). However, the functional significance of these mutations in malignancy remains to be established.
In this study we have addressed this issue by taking advantage of the reversibility of the frameshift mutations in tumor cells with mismatch repair deficiency. In these tumor cells, insertions and deletions of one repeated unit occur with frequencies at least 2 orders of magnitude higher than in normal cells (33). This facilitates the isolation of single-cell clones with insertions and deletions of one nucleotide in the BAX (G)8 tract, which in turn can be used to compare their properties in tumorigenicity assays in nude mice. Frameshift mutations in BAX (and other genes) in tumors of the MMP present a unique feature. These mutations (insertions/deletions) are reversible, and thus, the existence of positive selection for, but not against, can be unambiguously demonstrated for an identical mutational event. By using this experimental system, we report here that frameshift mutations inactivating BAX are under a strong selective pressure in colon tumor cells of the MMP, establishing a functional role for the common BAX frameshift mutations in tumor progression. In support of this notion we also report the association between BAX mutations and poor prognosis for both colon and gastric cancers of the MMP.
Materials and Methods
Tumorigenicity Assays and Immunostaining.
HCT116 and LS180 cell lines were cultured in DMEM containing 10% FCS. Suspensions of trypsinized cells were diluted to an average of 0.3 cell per well and transferred into 96-well plates. Single colonies were isolated after 2 weeks. One million cells were inoculated s.c. in duplicate into nude mice. One to 2 months after inoculation, tumors were excised and fixed in Bouin's solution. Fixed tumors were embedded in paraffin. Immunostaining for BAX was done as described (34). In brief, dewaxed sections of tumors were exposed to a polyclonal antibody (1712-PAB, generated against a synthetic peptide) specific for BAX. Working dilutions for paraffin immunohistochemistry were 1:800 to 1:1,500 in 0.1 M Tris buffer (pH 7.6) containing 10% normal goat serum, 2% BSA, and 0.1% ovalbumin. The secondary antibody was biotinylated goat anti-rabbit IgG, supplemented with ovalbumin. Immunoreactivity was revealed with standard ABC techniques (Vector Laboratories) by using diaminobenzidine as the chromogen. By using BAX-stained slides as a template, microdissection was performed with a surgical scalpel by scraping the BAX-positive and BAX-negative cancer areas into Eppendorf tubes. To prevent contamination, a separate blade was used for scraping the stained and unstained areas of each specimen.
Analysis of BAX Frameshift Mutations.
DNA from cultures and paraffin-embedded tissues was isolated as described (35, 36). A 94-bp region encompassing the BAX (G)8 tract was amplified by PCR with primers 5′-ATCCAGGATCGAGCAGGGCG-3′ and 5′-ACTCGCTCAGCTTCTTGGTG-3′. PCR was carried out with Vent DNA polymerase (New England Biolabs) for one cycle of 94°C for 4 min followed by 30 cycles of 94, 55, and 72°C, for 30 s in each cycle in the presence of 0.2 μCi of [32P]dCTP (1 Ci = 37 GBq). PCR products were electrophoresed in a denaturing 6% polyacrylamide gel. The gel was dried on filter paper and subjected to autoradiography.
Tumors and Survival Analysis.
The origins of colorectal and gastric tumor samples have been described (15–18). In our previous studies, the MMP was detected in 50 of 385 (13%) tumors of the colon and 29 of 205 (14%) tumors of the stomach. Frameshift BAX mutations were exclusively present in 25 of 50 (50%) colon and 18 of 29 (62%) gastric tumors of the MMP (17, 18). An additional 123 colorectal and 75 gastric tumor samples were obtained from Sapporo Medical University. Informed consent was obtained from each subject. Of these, 14% (17 of 123) and 16% (12 of 75) were positive for BAX frameshift mutations, respectively. Frameshift mutations in transforming growth factor β type II receptor (TGFβRII) and Caspase 5 were determined by PCR in these additional samples as described (16, 17). Follow-up information was available for a subset of these tumors. The tumors from patients with follow-up data represent a mixed sample, with most colon cancers coming from patients from the southwestern United States and Sapporo, Japan, and most gastric cancers coming from Japanese patients from Sapporo and Tokyo (17). Survival curves were determined by the Kaplan–Meier method based on the follow-up data. The patient status was classified as dead whenever the tumor was directly involved in outcome. Other causes of death were considered as censored individuals with no further follow-up. Comparison of survival curves was accomplished by means of a log-rank test.
Results and Discussion
Selection for BAX Frameshift Mutations in Vivo.
Many cells from the colorectal carcinoma cell line HCT116 are heterozygous for BAX frameshift mutations at a (G)8 tract within the gene coding region (15). We took advantage of the persistent high instability in these tumor cells (33) to isolate single-cell clones that had undergone a second mutation in the wild-type allele. We isolated three homozygous single-cell clones, two with (G)7/(G)7 and one with (G)9/(G)7 mutant alleles. Several heterozygous (G)8/(G)7 single-cell clones were also isolated. For simplicity we designate clones with two mutant alleles as BAX(−/−) and heterozygous clones as BAX(+/−). Because these single-cell clones are the descendants of a single common progenitor cancer cell, BAX(+/−) and BAX(−/−) clones have almost identical genetic backgrounds. By comparing BAX(+/−) with BAX(−/−) single-cell clones, any growth or survival difference between these cells is probably a result of the difference in the BAX allelic status. We have shown by Western blot analyses that homozygous BAX(−/−) cells do not express the protein in contrast to heterozygous BAX(+/−) cells (15). Therefore, mechanisms of gene silencing do not appear to play a significant role in BAX gene inactivation.
No noticeable differences were observed between BAX(+/−) cell clones that express BAX protein and those BAX(−/−) clones that do not, in several in vitro parameters of cell growth or survival. We detected only a significant difference in the number of detached dead cells floating in the media when the cultures reached confluency. HCT116 BAX(+/−) single-cell clones had significantly higher numbers of detached apoptotic cells than HCT116 BAX(−/−) single-cell clones (not shown). This difference in the number of detached cells suggested a link between BAX function and the ability of cells to undergo apoptosis on detachment from the intestinal epithelial layer (“anoikis”) (37).
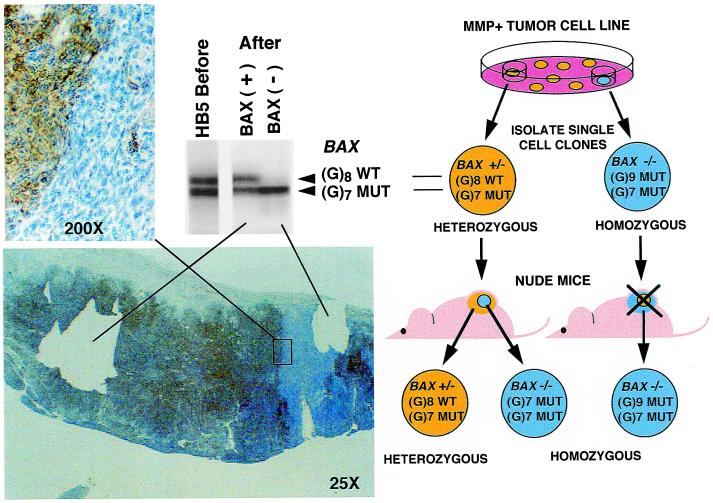
This prompted us to examine the behavior of these tumor cells in an in vivo environment. Tumor cells from heterozygous BAX(+/−) single-cell clones were inoculated into nude mice, and the resulting tumors were analyzed for the expression of BAX by immunostaining with anti-BAX antibodies. Insertion or deletion of one nucleotide in the (G)8 tract within the wild-type allele of a heterozygous clone generates a homozygous mutant clone that does not express the protein. This strategy (Fig. 1) permitted us to test whether or not inactivation of the wild-type BAX allele provides a survival advantage.
Figure 1.
In vivo selection of BAX frameshift mutation in HB5 BAX(+/−) single-cell clone of HCT116 cell line. (Right) The experimental strategy is shown (see text). (Left) Immunostaining with anti-BAX antibodies is shown with a higher magnification of the boundary between stained and unstained regions. The microslide shows the section of the tumor after microdissection of stained (+) and unstained (−) areas. At the top is shown the PCR amplification of the BAX region comprising the (G)8 tract from the single-cell clone culture before inoculation into nude mice and from microdissected tumor areas.
We detected areas in the tumors that were not stained with the anti-BAX antibody. These areas seemed to represent subclones expanded during the growth of the tumor (Fig. 1). PCR amplification of DNA from microdissected unstained areas confirmed that cells from these areas had lost the wild-type (G)8 allele and had become homozygous for the (G)7 allele. PCR amplification of DNA from stained areas showed retention of the PCR band corresponding to the wild-type allele (Fig. 1).
Directionality of BAX Mutations.
The BAX-negative clones could originate from the expansion of preexisting mutant cells present in a mixed-cell population. This possibility was ruled out because tumorigenicity assays with single-cell clones isolated from previous heterozygous single-cell clones yielded identical results (Fig. 2). As a control, we used homozygous BAX(−/−) single-cell clones to determine the directionality of the mutations. In this case, back mutations would result in the appearance of the wild-type allele detectable by immunostaining. However, in contrast to the results with heterozygous BAX(+/−) clones, no BAX-positive stained areas were observed in tumors originating after inoculation of the same number of cells from homozygous BAX(G)7/(G)7 single-cell clones (not shown).
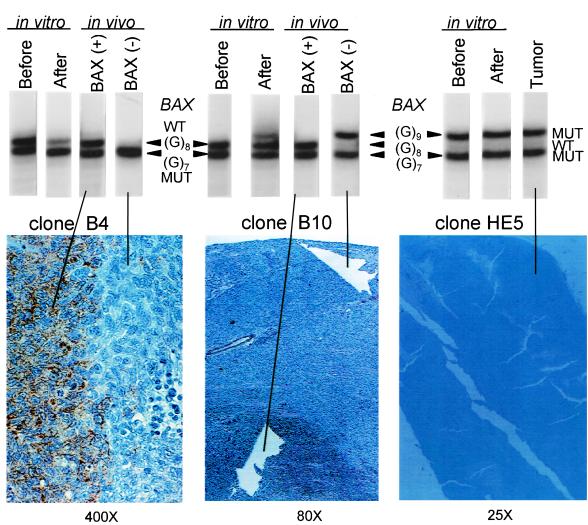
Figure 2.
In vivo and in vitro selection of BAX frameshift mutations of BAX(+/−) single-cell clones isolated from single-cell clones of the HCT116 cell line. (Upper) BAX allelic pattern determined by PCR amplification of the region containing the (G)8 tract from single-cell clones before and after tissue culture and from microdissected tissues from stained and unstained areas of the tumor sections. (Lower) Tumor sections from two BAX(+/−) heterozygous clones and one BAX(−/−) homozygous clone immunostained with anti-BAX antibodies. PCR amplification also was carried out with cells before inoculation and after prolonged culture in vitro. The results of the PCR amplification reveal the enrichment of mutant over wild-type alleles after long-term culture in heterozygous clones B4 and B10. The directionality of the mutations, always from wild type to mutant, was confirmed by the absence of detectable wild-type allele after long-term culture of the homozygous (G)9/(G)7 HE5 clone. This shows that loss of wild-type allele also confers a subtle selective advantage to the cells in vitro.
The directionality of mutations, from wild type to mutant, but not from mutant to wild type, could be caused by an intrinsic difference in the mutability of these alleles. DNA slippage mutations by strand misalignment are strongly dependent on the length of the repeat (33, 38). Thus, it was possible that (G)8 → (G)7 mutations were more frequent than (G)7 → (G)8 mutations. However, this was not the case because the same result was obtained on inoculation of a homozygous clone containing mutant alleles with 7 and 9 nucleotides. The (G)9 → (G)8 mutation, which was never observed (Fig. 2C), could be no less likely to occur than the frequently observed (G)8 → (G)7 or (G)8 → (G)9 mutations. These results do not imply that back mutations in the homozygous BAX(−/−) clones do not occur, but only that cells with the reverted wild-type allele are not detectable as clonal overgrowths within the surrounding BAX-negative cells.
Altogether, loss of the wild-type allele was observed in animals inoculated with 8 of 10 heterozygous clones, but reversion to the wild-type allele was not observed in any of 7 homozygous clones (Table 1, P = 0.001, Fisher exact test). Thus, despite their reversibility, all frameshift mutations detected within the polyguanine tract of the BAX gene resulted in ablation of BAX expression.
Table 1.
BAX frameshift mutations in single-cell clones of HCT116 cell line in vitro and in vivo
| Single-cell clones | BAX alleles (8: wild type/7,9: mutations) | Mutations in vitro* | Mutations in vivo† | BAX alleles before → after‡ |
|---|---|---|---|---|
| HB5§ | 8/7 | + | + | 8/7 → 7/7 |
| B2 | 8/7 | NA | − | NA |
| B3 | 8/7 | NA | − | NA |
| B4 | 8/7 | + | + | 8/7 →7/7 |
| B7 | 8/7 | NA | + | NA |
| B8 | 8/7 | + | + | NA |
| B9 | 8/7 | + | + | 8/7 → 7/7 |
| B10 | 8/7 | + | + | 8/7 → 9/7 |
| HA5§ | 8/7 | + | + | 8/7 → 7/7 |
| A5 | 8/7 | + | + | NA |
| HE5§ | 9/7 | − | − | 9/7 → 9/7 |
| E1 | 9/7 | NA | − | NA |
| E2 | 9/7 | NA | − | NA |
| E3 | 9/7 | NA | − | NA |
| E4 | 9/7 | NA | − | NA |
| HC4§ | 7/7 | − | − | 7/7 → 7/7 |
| HD7§ | 7/7 | − | − | 7/7 → 7/7 |
Single-cell clones were kept in culture by serial passages of 1∶100 once a week. After 4 months, DNA was prepared from the cultivated single-cell clones and from the initial frozen stock cultures. Positive selection for inactivating BAX mutations in vitro was detected by the increase of intensity of the PCR band corresponding to the BAX(G)7 mutant allele vs. the band of the BAX(G)8 normal allele or by the appearance of the new PCR band corresponding to the BAX(G)9 mutant allele (Fig. 3).
One million cells of each single-cell clone were injected s.c. into nude mice. When tumors were about 1 cm in diameter (after 2–3 months), mice were killed, and the tumors were fixed in Bouin's solution and embedded in paraffin. Selection for BAX frameshift mutations in vivo was detected by the presence of unstained areas in the tumor sections after staining with anti-BAX antibodies (Figs. 2 and 3).
Selection for frameshift mutations by insertion (8/7 → 9/7) or deletion (8/7 → 7/7) of one base pair in the (G)8 tract of the wild-type BAX allele (Figs. 2 and 3).
These single-cell clones were isolated from mass cultures of the HCT116 cell line. From single-cell clone HB5 we isolated 10 single cell clones named B1–B10. From HA5 we isolated single-cell clone A5. From single-cell clone HE5 we isolated 4 single-cell clones named E1–E4. The boldface clones are homozygous for the mutation, which serve as controls for the directionality of the in vivo selection for BAX inactivation.
Selection in vivo for BAX mutational inactivation was also demonstrated for the LS180 colon tumor cell line, homozygous for the (G)7 mutant BAX allele (15). We transfected LS180 cells with a retrovirus encoding a wild-type BAX gene. Single-cell clones were isolated and inoculated into nude mice. Unstained tumor areas were observed, which had lost protein expression (data not shown). These results support the existence of a positive selection for BAX gene mutation during in vivo tumorigenesis.
BAX Mutations and Cancer Survival.
To examine the relevance of the nude mice assays to human cancer pathogenesis, we determined the relationship between BAX mutations and the survival of patients with colon and gastric cancers of the microsatellite mutator phenotype. These tumors are less invasive, and patients exhibit improved survival relative to those with tumors without enhanced microsatellite instability (1, 2, 17, 39–41). We analyzed 508 tumors of the colon and 280 of the stomach for the MMP. The positive tumors were analyzed for BAX frameshift mutations.
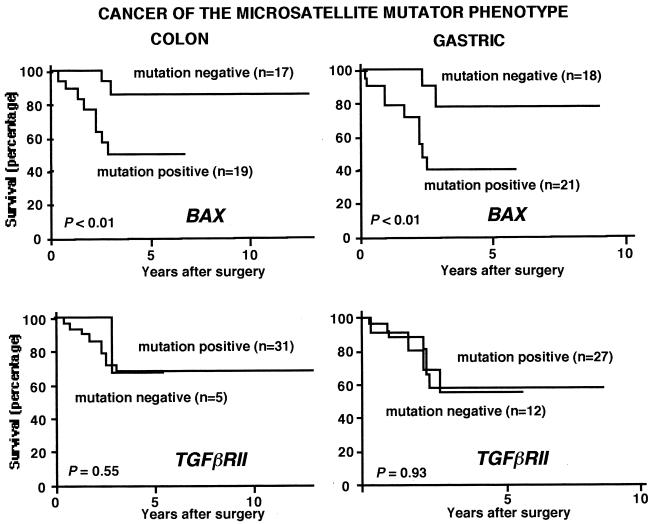
The frequency of the MMP was 13% for colon (67 of 508) and 15% for gastric (41 of 280) cancer. BAX mutations accounted for 51% (34 of 67) and 61% (25 of 41) of tumors of the colon and stomach, respectively. Follow-up survival data were available for 36 patients with colorectal cancer with tumors of the MMP and 38 patients with gastric cancer with tumors of the MMP. Patients with BAX mutations had a significantly shorter survival than those without these mutations, for both colorectal (P < 0.01) and gastric (P < 0.01) cancer (Fig. 3). Simultaneous analysis in these tumors for frameshift mutations in TGFβRII and Caspase 5, other genes that target the MMP, revealed no significant differences in survival (Fig. 3 and data not shown).
Figure 3.
BAX mutations and cancer patient survival. TGFβRII mutations are also shown for comparative purposes. No correlation was found between BAX mutations and other clinical or histopathological parameters, including the age, gender, differentiation degree, anatomical localization, or invasiveness of the tumors. The presence of BAX mutations thus appears to be an independent indicator of poor prognosis.
Some of the tumors had biallelic BAX mutations, but others had monoallelic mutations (15–18). We did not distinguish between them because, in some cases, it was difficult to determine whether the mutations were biallelic, monoallelic, or (as shown in Figs. 1 and 2) monoallelic in some areas of the tumors and biallelic in others. Therefore, for the association between mutation and survival, the distinction between monoallelic and biallelic mutations in primary tumor specimens may not be as critical as whether they are present or not. Heterozygous mutations in the BAX gene elicit an alteration in cell growth phenotype in knockout mice (31). Moreover, the discrimination between biallelic mutations and monoallelic mutations in BAX (or any other gene) may not be appropriate in the context of MMP tumors. Because of their exacerbated mutator phenotype, these tumor cells elude not only the premise that the occurrence of biallelic mutations is a very rare event in tumorigenesis (1, 10–11), but also the premise that biallelism is a requirement for mutation functionality (Knudson “two hit” hypothesis) (42). Although MMP tumors accumulate numerous (>105) clonal biallelic mutations in nonfunctional sequences such as microsatellite repeats (1, 10–13), they also can carry multiple (>10) monoallelic mutations in functional sequences such as BAX (14–19). We address this apparent paradox by postulating an accumulative haploinsufficiency mechanism (18, 43).
The tumorigenicity and survival results together support the hypothesis that mutations in the proapoptotic gene BAX contribute to tumor progression in the mutator phenotype pathway for gastrointestinal cancer. In addition to frameshift mutations at the (G)8 tract, we found in colon and gastric cancers a hot spot for missense mutations at codon 169 of BAX, which impairs its apoptotic activity (44). These results add further support to the functional significance of the common BAX gene mutations and the escape from apoptosis for cancer progression in gastrointestinal cancer of the MMP. In the MMP pathway for gastrointestinal cancer, inactivation of TGFβRII occurs relatively early (45, 46). In contrast, the tumor suppressor function of BAX appears to occur later in tumor progression, as shown by the lack of clonality of these mutations in some tumors (15–19, 47). However, the mutations that occur in BAX at a late stage of tumorigenesis are independent prognostic indicators of poor survival. They also define a useful difference in genotype and phenotype among tumors of the MMP.
Acknowledgments
This work was supported by the National Institutes of Health National Cancer Institute Grants R01CA63585 and R01CA38579 (to M.P.).
Abbreviations
- MMP
microsatellite mutator phenotype
- TGFβRII
transforming growth factor β type II receptor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190210897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190210897
References
- 1.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Thibodeau S, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Javinen H, Powell S M, Jen J, Hamilton S R, Petersen G M, Kinzler K W, Vogelstein B, de la Chapelle A. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 4.Boland C R, Thibodeau S N, Hamilton S R, Sidransky D, Eshleman J R, Burt R W, Meltzer S J, Rodriguez-Bigas M A, Fodde R, Ranzani G N, Srivastava S A. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 5.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 6.Kolodner R D, Marsischky G T. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 7.Marra G, Boland C R. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 8.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim H, Jen J, Vogelstein B, Hamilton S R. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 10.Perucho M, Peinado M A, Ionov Y, Casares S, Malkhosyan S, Stanbridge E. Cold Spring Harbor Symp Quant Biol. 1994;59:339–348. doi: 10.1101/sqb.1994.059.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 12.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, Rycke Y D, Li Y J, Muzeau F, Girodet J, Salmon R-J, Thomas G. Proc Natl Acad Sci USA. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, Ushio K, Nomizu S, Konishi F, Utsunomiya J, Miyaki M. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 14.Markowitz S D, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Wilson J K V. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 15.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Sawai H, Perucho M. Cancer Res. 1997;57:4420–4426. [PubMed] [Google Scholar]
- 17.Yamamoto H, Perez-Piteira J, Yoshida T, Terada M, Itoh F, Imai K. Gastroenterology. 1999;116:1348–1357. doi: 10.1016/s0016-5085(99)70499-3. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S, Yamamoto H, Navarro M, Maestro M, Reventos J, Perucho M. Cancer Res. 1999;59:2995–3002. [PubMed] [Google Scholar]
- 19.Yagi O K, Akiyama Y, Nomizu T, Iwama T, Endo M, Yuasa Y. Gastroenterology. 1998;114:268–274. doi: 10.1016/s0016-5085(98)70477-9. [DOI] [PubMed] [Google Scholar]
- 20.Meijerink J P P, Mensink E J B M, Wang K, Sedlak T W, Sloetjes A W, de Witte T, Waksman G, Korsmeyer S J. Blood. 1998;91:2991–2997. [PubMed] [Google Scholar]
- 21.Reed J C. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 22.Chao D T, Korsmeyer S J. Annu Rev Immunol. 1998;16:385–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 23.Tsujimoto Y, Finger L R, Yunis J, Nowell P C, Croce C M. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 24.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 25.Wolter K G, Hsu Y T, Smith C L, Nechushtan A, Xi X G, Youle R J. J Cell Biol. 1998;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goping I S, Gross A, Lavoie J N, Nguyen M, Jemmerson R, Roth K, Korsmeyer S J, Shore G C. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzo I, Brenner C, Zamzami N, Jurgensmeier J M, Susin S A, Vieira H L, Prevost M C, Xie Z, Matsuyama S, Reed J C, Kroemer G. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 28.Zha H, Aime-Sempe C, Sato T, Reed J C. J Biol Chem. 1996;271:7440–7444. doi: 10.1074/jbc.271.13.7440. [DOI] [PubMed] [Google Scholar]
- 29.Hanada M, Aime-Sempe C, Sato T, Reed J C. J Biol Chem. 1995;270:11962–11969. doi: 10.1074/jbc.270.20.11962. [DOI] [PubMed] [Google Scholar]
- 30.Knudson C M, Tung K S, Tourtellotte W G, Brown G A, Korsmeyer S J. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 31.Yin C, Knudson C M, Korsmeyer S J, Van Dyke T. Nature (London) 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 32.Bargou R C, Wagener C, Bommer K, Mapara M Y, Daniel P T, Arnold W, Dietel M, Guski H, Feller A, Royer H D, Dorken B. J Clin Invest. 1996;97:2651–2659. doi: 10.1172/JCI118715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 34.Krajewski S, Mai J K, Krajewska M, Sikorska M, Mossakowski M J, Reed J C. J Neurosci. 1995;15:6364–6376. doi: 10.1523/JNEUROSCI.15-10-06364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 36.Shimizu H, Burns J C. In: PCR Strategies. Innis M A, Gelfand D H, Sninski J J, editors. London: Oxford Univ. Press; 1995. [Google Scholar]
- 37.Frisch S M, Ruoslahti E. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 38.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Halling K C, French A J, McDonnell S K, Burgart L J, Schaid D J, Peterson B J, Moon-Tasson L, Mahoney M R, Sargent D J, O'Connell M J, et al. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 40.Gryfe R, Kim H, Hsieh E T, Aronson M D, Holowaty E J, Bull S B, Redston M, Gallinger S. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 41.dos Santos N R, Seruca R, Constancia M, Seixas M, Sorbinho-Simoes M. Gastroenterology. 1996;110:38–44. doi: 10.1053/gast.1996.v110.pm8536886. [DOI] [PubMed] [Google Scholar]
- 42.Perucho M. Cancer Res. 1999;59:249–256. [PubMed] [Google Scholar]
- 43.Yamamoto H, Gil J, Schwartz S, Perucho M. Cell Death Differ. 2000;7:238–239. doi: 10.1038/sj.cdd.4400651. [DOI] [PubMed] [Google Scholar]
- 44.Gil J, Yamamoto H, Zapata J M, Reed J C, Perucho M. Cancer Res. 1999;59:2034–2037. [PubMed] [Google Scholar]
- 45.Akiyama Y, Iwanaga R, Saitoh K, Shiba K, Ushio K, Ikeda E, Iwama T, Nomizu T, Yuasa Y. Gastroenterology. 1997;112:33–39. doi: 10.1016/s0016-5085(97)70216-6. [DOI] [PubMed] [Google Scholar]
- 46.Grady W M, Rajput A, Myeroff L, Liu D F, Kwon K, Willis J, Markowitz S. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 47.Abdel-Rahman W M, Georgiades I B, Curtis L J, Arends M J, Wyllie A H. Oncogene. 1999;18:2139–2142. doi: 10.1038/sj.onc.1202589. [DOI] [PubMed] [Google Scholar]