Abstract
Ceramides are central intermediates of sphingolipid metabolism with critical functions in cell organization and survival. They are synthesized on the cytosolic surface of the endoplasmic reticulum (ER) and transported by ceramide transfer protein to the Golgi for conversion to sphingomyelin (SM) by SM synthase SMS1. In this study, we report the identification of an SMS1-related (SMSr) enzyme, which catalyses the synthesis of the SM analogue ceramide phosphoethanolamine (CPE) in the ER lumen. Strikingly, SMSr produces only trace amounts of CPE, i.e., 300-fold less than SMS1-derived SM. Nevertheless, blocking its catalytic activity causes a substantial rise in ER ceramide levels and a structural collapse of the early secretory pathway. We find that the latter phenotype is not caused by depletion of CPE but rather a consequence of ceramide accumulation in the ER. Our results establish SMSr as a key regulator of ceramide homeostasis that seems to operate as a sensor rather than a converter of ceramides in the ER.
Introduction
Sphingolipids are vital components of cellular membranes in organisms ranging from mammals to yeast. Besides providing a structural framework for plasma membrane (PM) organization, sphingolipids are dynamic regulators of a wide range of cellular processes. Sphingoid long-chain bases (LCBs), ceramides, and other intermediates of sphingolipid metabolism act as signaling molecules in the regulation of cell growth, death, migration, and membrane trafficking (Spiegel and Milstien, 2003; Hannun and Obeid, 2008). Notably, a sensitive balance between phosphorylated LCBs and ceramides, referred to as the LCBP/ceramide rheostat, appears critical for normal cell growth and lifespan (Mandala et al., 1998; Kobayashi and Nagiec, 2003). Moreover, sphingolipids form gradients along the secretory pathway that may affect protein sorting through hydrophobic matching of membrane spans (Bretscher and Munro, 1993; Holthuis et al., 2001; Patterson et al., 2008).
Given their impact on cell organization and survival, the local concentration of sphingolipids and their metabolic intermediates must be tightly controlled. Although the mechanisms that regulate membrane sterol concentrations are well established (Goldstein et al., 2006), little is known about the mechanisms controlling sphingolipid homeostasis. As ceramides constitute the backbone of all sphingolipids and directly participate in cellular life and death decisions (Morales et al., 2007), controlling their local concentration is critical. Ceramides can be generated by the breakdown of sphingomyelin (SM) through SMases at the PM (Andrieu-Abadie and Levade, 2002) or synthesized de novo by N-acylation of LCBs on the cytosolic surface of the ER (Mandon et al., 1992; Hirschberg et al., 1993). The latter reaction is controlled by components of TORC2 (target of rapamycin complex 2) presumably through phosphorylation of the ceramide synthase (Aronova et al., 2008).
In mammalian cells, the bulk of newly synthesized ceramides is converted to SM in the lumen of the Golgi (Tafesse et al., 2006). Efficient delivery of ER-derived ceramides to the site of SM production requires a cytosolic ceramide transfer protein (CERT; Hanada et al., 2003). Besides a START domain that binds ceramide, CERT contains an FFAT motif for interaction with the ER membrane and a pleckstrin homology domain targeting the Golgi. CERT is phosphorylated at a serine repeat motif, which down-regulates its ceramide transport function. Loss of SM and cholesterol from cells causes dephosphorylation of the motif, resulting in CERT activation (Kumagai et al., 2007). Thus, the local concentration of ceramides in cells is controlled by an intricate network of lipid-metabolizing enzymes, transfer proteins, kinases, and phosphatases.
SM production is mediated by a phosphatidylcholine (PC)/ceramide cholinephosphotransferase or SM synthase (SMS). Mammalian cells contain two SMS isoforms, SMS1 in the Golgi and SMS2 at the PM (Huitema et al., 2004). In addition, the mammalian genome encodes a third SMS-related (SMSr) protein of unknown function with homologues in nematodes and insects like Drosophila melanogaster (Fig. 1 A). Drosophila does not synthesize SM but produces the SM analogue ceramide phosphoethanolamine (CPE) as a major membrane constituent (Rao et al., 2007). CPE production also occurs in mammals and is catalyzed by a phosphatidylethanolamine (PE)/ceramide ethanolamine phosphotransferase or CPE synthase (CPES; Malgat et al., 1986, 1987). The identity of the responsible enzyme is not known. Production of SM and CPE involves a similar reaction chemistry (Malgat et al., 1986). Because SMS1, SMS2, and SMSr are structurally related and share two highly conserved sequence motifs with putative active site residues (Fig. 1 B; Huitema et al., 2004; Tafesse et al., 2006), SMSr is a prime candidate for the elusive CPES.
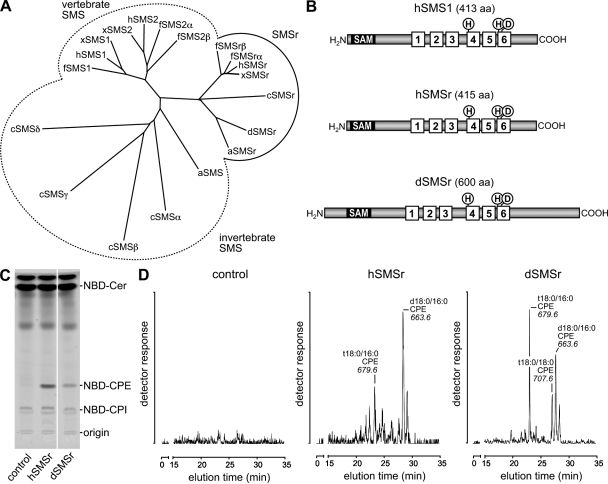
Figure 1.
SMSr proteins display CPES activity. (A) Phylogenetic tree of SMS family members from Homo sapiens (h), Xenopus tropicalis (x), Fugu rubripes (f), Drosophila (d), Apis mellifera (a), and C. elegans (c). The tree was constructed using ClustalX. (B) SMSr proteins share a common domain structure with vertebrate SMS1, which includes six transmembrane helices, an active site consisting of conserved His (H) and Asp (D) residues, and a N-terminal SAM domain. (C) TLC separation of reaction products formed when NBD-Cer was incubated with lysates of yeast strains expressing hSMSr or dSMSr or transfected with empty vector (control). White line indicates that intervening lanes have been spliced out. (D) Yeast strains expressing hSMSr or dSMSr produce CPE in vivo. Different molecular species of CPE were detected by electrospray LC/MS/MS (neutral loss of 141) after alkaline hydrolysis of glycerolipids. No CPE was detected in lipid extracts of control (untransfected) yeast.
In this study, we show that SMSr displays CPES activity and, contrary to SMS1 and 2, localizes to the ER. However, we find that SMSr produces only trace amounts of CPE and that bulk production of CPE in insect cells is mediated by a different enzyme. Unexpectedly, blocking SMSr activity causes a marked increase of ceramide levels in the ER. This is accompanied by a fragmentation of ER exit sites and a structural collapse of the Golgi. These morphological aberrations are not caused by a lack of CPE but rather a consequence of the ceramide accumulation in the ER. We propose that SMSr is a CPES with dual activity as ceramide sensor to control ceramide homeostasis in the ER and that the latter process is critical for the integrity of the early secretory pathway.
Results
SMSr proteins display CPES activity
To test whether SMSr proteins catalyze CPE production, human SMSr (hSMSr) and Drosophila SMSr (dSMSr) were expressed in budding yeast, an organism lacking endogenous CPES activity. SMSr-expressing cells were lysed and incubated with fluorescent C6-NBD–ceramide (NBD-Cer). TLC analysis of the reaction mixtures showed the presence of an NBD-labeled product with a retention factor value distinct from that of NBD–ceramide phosphoinositol and NBD-SM. The product was missing in reactions performed with control (empty vector) cells (Fig. 1 C). Liquid chromatography (LC) mass spectrometry (MS)/MS analysis identified the product as NBD-CPE (unpublished data), suggesting that SMSr proteins can synthesize CPE.
To determine whether SMSr proteins recognize natural ceramides as substrates for CPE production, a yeast strain was used in which the endogenous enzymes for ceramide production were replaced by mouse ceramide synthase CerS5 (Cerantola et al., 2007). As the ceramides produced in this strain structurally resemble those found in animal cells, they were expected to be suitable substrates for CPE biosynthesis. LC/MS/MS analyses revealed the presence of several molecular species of CPE in lipid extracts of both hSMSr- and dSMSr-expressing strains (Fig. 1 D). The extracts were devoid of SM. No CPE was detectable in control cells. Together, these results demonstrate that SMSr proteins function as CPESs.
SMSr represents a major CPES activity in mammalian cells
To investigate whether SMSr corresponds to the CPES activity previously described in mammalian cells (Malgat et al., 1986, 1987), lysates of HeLa cells in which hSMSr was overexpressed or depleted by RNAi were analyzed for CPES activity. Incubation of control cell lysates with NBD-Cer yielded three fluorescent products, corresponding to NBD-glucosylceramide (GlcCer), NBD-SM, and NBD-CPE (Fig. 2 A, left lane). Although formation of NBD-CPE was stimulated by addition of PE, addition of either PC or CDP-ethanolamine had no effect, suggesting that PE is the headgroup donor in the CPES reaction (Fig. 2 B; Fig. S1; and see Fig. 4 E). Heterologous expression of V5-tagged hSMSr led to a sixfold increase in CPES activity, leaving SMS activity unaffected (Fig. 2, A and C). Conversely, CPES activity dropped >50% after depletion of hSMSr by RNAi (Fig. 2 D). This shows that hSMSr represents a major CPES activity in HeLa cells.
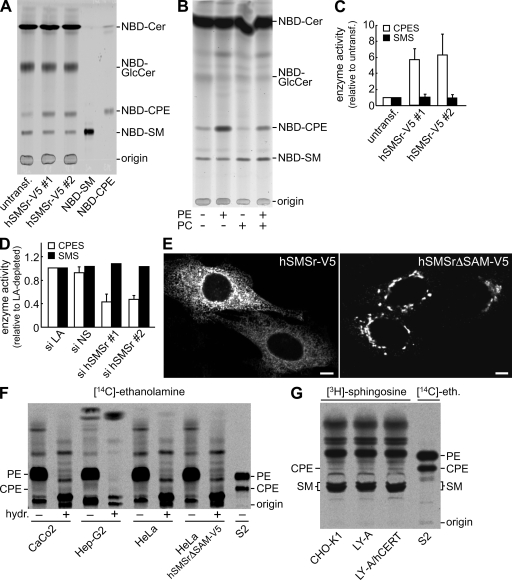
Figure 2.
Mammalian cells synthesize only trace amounts of CPE despite the ER residency of SMSr. (A) TLC analysis of reaction products formed when lysates of control (untransfected [untransf]) or hSMSr-V5–expressing HeLa cells were incubated with NBD-Cer. (B) TLC analysis of reaction products formed when NBD-Cer was incubated with lysates of HeLa cells expressing hSMSr-V5 in the presence or absence of externally added PE or PC. (C) Expression of hSMSr-V5 stimulates CPES activity in HeLa cells. CPES and SMS activity levels were determined by TLC analysis of reaction products formed when cell lysates were incubated with NBD-Cer and expressed relative to control cells. (D) CPES and SMS activity levels were determined in lysates of HeLa cells treated for 7 d with siRNA-targeting LA (si LA) or siRNA-targeting hSMSr (si hSMSr) or with nonsilencing siRNA (si NS) as in C and expressed relative to si LA–treated cells. (E) Confocal sections of HeLa cells were transfected with hSMSr-V5 or an SAM-deficient truncation mutant, hSMSrΔSAM-V5, and immunolabeled for V5. Note that removal of SAM causes hSMSr-V5 to redistribute from the ER to the Golgi. Bars, 5 µm. (F) Human Caco-2, Hep-G2, HeLa, hSMSrΔSAM-V5-expressing HeLa, and Drosophila S2 cells were labeled with [14C]ethanolamine for 48 h and subjected to lipid extraction, TLC analysis, and autoradiography. In some extracts, glycerolipids were deacylated by mild alkaline hydrolysis (hydr; +). (G) Chinese hamster CHO-K1, CHO-K1–derived LY-A (CERT mutant), and LY-A/human CERT cells (mutant expressing human CERT) were labeled with [3H]sphingosine for 24 h and subjected to lipid extraction, TLC analysis, and autoradiography. Error bars indicate SD; n = 3.
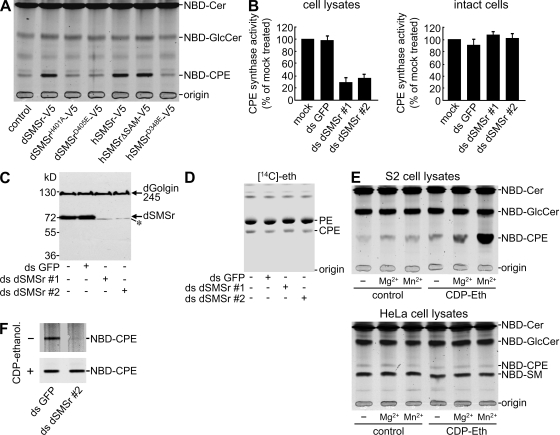
Figure 4.
Bulk production of CPE in insect cells is independent of SMSr. (A) TLC analysis of reaction products formed when NBD-Cer was incubated with lysates of Drosophila S2 cells expressing dSMSr-V5, active site mutants of dSMSr-V5 (H401A and D405E), hSMSr-V5, active site mutant of hSMSr-V5 (D348E), or SAM-deficient hSMSr-V5 (hSMSrΔSAM-V5). (B) S2 cells treated with ds GFP or dSMSr (ds dSMSr #1 or #2) for 7 d were metabolically labeled with NBD-Cer (intact cells) or lysed and incubated with NBD-Cer for 2 h (cell lysates). CPES activity levels were determined by TLC analysis and expressed relative to mock-treated cells. Error bars indicate SD; n = 3. (C) Immunoblots of S2 cells treated with dsRNA as in B were stained for dSMSr and dGolgin245. Note that the anti-dSMSr antibody cross reacts weakly with a dSMSr-unrelated protein (asterisk). (D) S2 cells treated with dsRNA as in B were metabolically labeled with [14C]ethanolamine ([14C]eth) for 2 h and subjected to TLC analysis and autoradiography. (E) TLC analysis of products formed in S2 (top) or HeLa cell lysates (bottom) when incubated with 50 µM NBD-Cer in the presence or absence of 500 µM CDP-ethanolamine (CDP-ethanol), 5 mM MgCl2, or 10 mM MnCl2. (F) TLC analysis of products formed in lysates of dsRNA-treated S2 cells incubated with NBD-Cer and MnCl2 in the presence or absence of CDP-ethanolamine as in E.
To determine the contribution of SMSr to CPE biosynthesis in vivo, endogenous CPE production levels were monitored by metabolic labeling of various cell lines. Labeling human colon carcinoma Caco-2, liver carcinoma Hep-G2, or HeLa cells for 48 h with [14C]ethanolamine yielded only trace amounts of radiolabeled CPE (Fig. 2 F). The same was true when HeLa or CHO-K1 cells were labeled with [3H]sphingosine (Fig. 2 G). In contrast, labeling Drosophila S2 cells with [14C]ethanolamine showed that the CPE production level in these cells is much higher (Fig. 2 F, right). Indeed, MS analyses revealed that although S2 cells contain a substantial amount of CPE (15.3 ± 1.1 mol% of total phospholipid; n = 3), the CPE levels in HeLa or CHO-K1 cells are exceedingly low (∼0.03 mol%, i.e., 300-fold lower than SM; see Fig. 5 A).
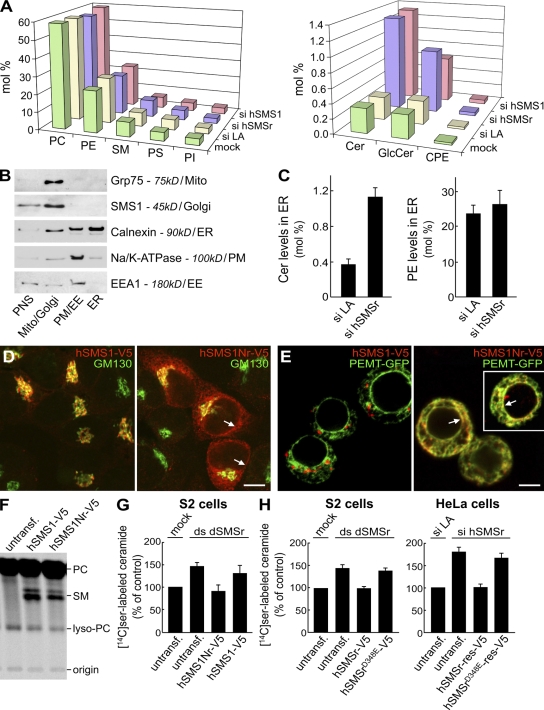
Figure 5.
SMSr-depleted cells accumulate ceramides in the ER. (A) HeLa cells treated with si LA, si hSMSr, or siRNA-targeting hSMS1 (si hSMS1) for 3 d were subjected to lipid extraction. Lipid classes were quantified by MS/MS analyses and expressed in mole percent of total lipid analyzed. Data shown are representative of three independent experiments. (B) HeLa cells expressing hSMS1-V5 were subjected to subcellular fractionation, yielding a PNS and fractions enriched for mitochondria (mito)/Golgi, PM/early endosomes (EE), or ER. Equal amounts of protein from each fraction were analyzed by immunoblotting. Note that the ER fraction was virtually free of other organelles except for a minor contamination with PM (<10%). (C) MS/MS analysis of ceramide and PE levels in ER fractions prepared from HeLa cells treated with si LA or si hSMSr for 3 d. (D) HeLa cells transfected with hSMS1-V5 or an hSMS1 swap mutant carrying the N-terminal tail of hSMSr, hSMS1Nr-V5, were double labeled for V5 and cis-Golgi marker GM130. (E) S2 cells cotransfected with hSMS1-V5 or hSMS1Nr-V5 and ER marker PEMT-GFP were labeled for V5. The white box marks a cell captured from a different field. Note that hSMS1Nr-V5 localizes to the ER and nuclear envelope (D and E, arrows). (F) S2 cells transfected with hSMS1-V5 or hSMS1Nr-V5 were labeled with [14C]choline for 5 h and subjected to lipid extraction, TLC, and autoradiography. (G) dSMSr-depleted S2 cells were transfected with hSMS1-V5 or hSMS1Nr-V5 and labeled for 5 h with [14C]serine. Levels of radiolabeled ceramides were determined by TLC and autoradiography and expressed as a percent of mock-treated cells. (H) SMSr-depleted cells (S2/HeLa) were transfected with hSMSr-V5 or hSMSrD348E-V5 and labeled for 5 h with [14C]serine. Levels of radiolabeled ceramides were determined as in G. SMSr depletions were with ds dSMSr #2 (S2) or si hSMSr #2 (HeLa). Transfection of HeLa cells was with siRNA-resistant (res) hSMSr constructs. Untransf, untransfected. Error bars: (G) SD, n = 3; (C and H) range, n = 2.
SMSr produces only trace amounts of CPE
Although mammalian cells contain a readily detectable CPES activity (Fig. 2 A), their CPE content is very low. One explanation for this discrepancy could be that SMSr resides in a compartment that is not easily reached by newly synthesized ceramide. To test this, the subcellular distribution of hSMSr-V5 was analyzed in HeLa cells by immunofluorescence microscopy. hSMSr-V5 gave a reticular and nuclear envelope staining pattern and colocalized with the ER marker protein disulfide isomerase (Fig. 2 E and Fig. S2 A). The ER localization of hSMSr does not explain why mammalian cells contain only trace amounts of CPE, as the substrates for CPE synthesis, PE, and ceramide are made in the ER.
We tested whether these substrates have limited access to the enzyme's active site. SMSr and SMS1 share a common transmembrane domain organization with the active site facing the exoplasmic leaflet (Fig. 1 B; Huitema et al., 2004). SMSr-catalyzed CPE production would therefore occur in the ER lumen. As ceramide is synthesized on the cytosolic surface of the ER (Mandon et al., 1992; Hirschberg et al., 1993), it could be scavenged by CERT before being able to flip to the site of CPE production. To test this, we used CHO-K1–derived LY-A mutant cells, which are defective in CERT-mediated ceramide transport (Hanada et al., 2003). As shown in Fig. 2 G, [3H]sphingosine labeling of CHO-K1 and LY-A cells yielded similar levels of 3H-CPE, indicating that the low CPE content of mammalian cells is unlikely caused by scavenging of ceramides from the site of CPE production.
Another explanation for the low CPE content could be the presence of an inhibitory factor or environment in the ER. We found that removal of the N-terminal sterile α motif (SAM) domain of hSMSr caused its redistribution from the ER to the Golgi (Fig. 2 E and Fig. S2 B) but did not affect its enzymatic activity (see Fig. 4 A). However, metabolic labeling of HeLa cells expressing hSMSrΔSAM showed no significant increase in CPE production levels compared with cells expressing the full-length protein (Fig. 2 F). Thus, the ER does not seem to impose an inhibitory environment on SMSr enzymatic activity.
Last, we investigated whether CPE is readily converted to another product. Although SM production in mammalian cells mainly occurs through SMS1-mediated headgroup transfer from PC to ceramide (Tafesse et al., 2006), an alternative pathway involves methylation of CPE (Muehlenberg et al., 1972). To investigate the relative contribution of the latter pathway to SM production, HeLa cells were metabolically labeled with D4-ethanolamine and D9-choline simultaneously. MS/MS analysis showed that the D4/D9 ratio of SM after 24-h labeling was ∼1:100, indicating that headgroup transfer from PC is the major pathway of SM biosynthesis. Moreover, the D4-labeled SM pool most likely originated from headgroup transfer of D4-labeled PC, as this pool was reduced by depletion of human SMS1 (unpublished data). Finally, pulse-chase experiments with [14C]ethanolamine and [14C]choline showed that the turnover rate of CPE in HeLa cells was similar to that of SM (unpublished data). Thus, the low CPE content of mammalian cells is unlikely the result of CPE being a short-lived metabolic intermediate.
Together, these results suggest that SMSr proteins are intrinsically unable to produce bulk amounts of CPE. This is consistent with our finding that CPE levels in SMSr-expressing yeast strains are invariably low (≤0.5 mol% of total phosphosphingolipid). As bulk amounts of CPE are present in Drosophila, we next addressed the contribution of dSMSr to CPE production in Drosophila S2 cells.
Bulk production of CPE in insect cells is independent of SMSr
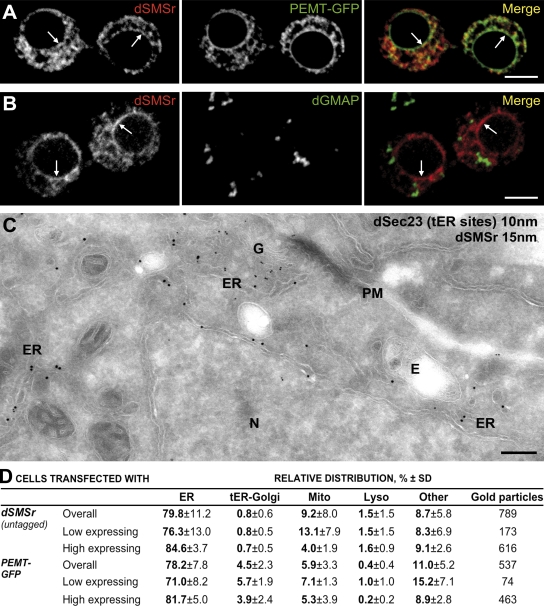
We first analyzed the subcellular distribution of dSMSr in S2 cells by using an antibody that specifically cross reacted with dSMSr on immunoblots of S2 cell extracts (see Fig. 4 C). As the endogenous protein was hardly detectable by immunofluorescence microscopy, S2 cells were transfected with a Cu+2-inducible dSMSr construct, and expression of the protein was enhanced fivefold (as indicated by immunoblotting). dSMSr colocalized extensively with a GFP-tagged version of the ER marker PE N-methyltransferase (PEMT; PEMT-GFP) but not with the Golgi microtubule-associated protein dGMAP (Fig. 3, A and B). The ER residency of dSMSr was confirmed by quantitative immunoelectron microscopy (Fig. 3, C and D). As observed for hSMSr, removal of its SAM domain caused dSMSr to redistribute from the ER to the Golgi (Fig. S2, C–E), which is consistent with a fundamental role of SAM in ER retention of SMSr proteins.
Figure 3.
dSMSr resides in the ER of insect cells. (A) Confocal sections of Drosophila S2 cells were double transfected with native dSMSr and ER marker PEMT-GFP and immunolabeled for dSMSr. (B) Confocal sections of S2 cells were transfected with untagged dSMSr and double labeled for dSMSr and cis-Golgi marker dGMAP. (A and B) Arrows indicate nuclear envelope staining. Bars, 5 µm. (C) Localization of dSMSr by immunoelectron microscopy is shown. Ultrathin cryosections of S2 cells transfected with dSMSr were double labeled for dSMSr (15-nm gold) and dSec23 (tER site marker; 10-nm gold). G, Golgi stack; N, Nucleus; E, endosome. Bar, 500 nm. (D) Quantification of immunogold labeling of S2 cells cotransfected with dSMSr and PEMT-GFP. Low-expressing cells are those with <40 gold particles per cell section; high-expressing cells are those with ≥40 gold particles per cell section.
We next investigated the contribution of dSMSr to CPE production in S2 cells. Incubation of S2 cell lysates with NBD-Cer led to formation of NBD-CPE (Fig. 4 A). NBD-CPE formation increased fivefold after overexpression of dSMSr. Mutation of active site residues His401 or Asp405 abolished dSMSr-mediated CPES activity (Fig. 4 A). Depletion of endogenous dSMSr resulted in a 60–70% drop in cell lysate–associated CPES activity (Fig. 4 B, left). Surprisingly, no significant reduction in CPE production levels was found when intact dSMSr-depleted S2 cells were metabolically labeled with NBD-Cer (Fig. 4 B, right) or [14C]ethanolamine (Fig. 4 D), although the RNAi treatment reduced dSMSr protein levels by ∼90% (Fig. 4 C). These results suggest that S2 cells are able to produce CPE through a second, dSMSr-independent enzyme.
One reason why this second enzyme is not readily detectable in cell lysates might be that it requires a soluble substrate that is continuously regenerated in living cells. CDP-ethanolamine is an attractive candidate for such a substrate given its role as headgroup donor in PE biosynthesis (Vance, 2008). Indeed, CPE production in S2 cell lysates increased dramatically after addition of CDP-ethanolamine provided that Mn2+ ions were present (Fig. 4 E, top). The CDP-ethanolamine–dependent CPES activity was unaffected by dSMSr depletion (Fig. 4 F) and appears unique for insect cells, as addition of CDP-ethanolamine did not enhance NBD-CPE production in HeLa cell lysates (Fig. 4 E, bottom).
In sum, insect cells contain two distinct CPESs: a PE/ceramide ethanolamine phosphotransferase corresponding to dSMSr and a dSMSr-unrelated CDP-ethanolamine/ceramide ethanolamine phosphotransferase that is likely responsible for bulk production of CPE. Importantly, these findings confirm that SMSr proteins in general lack the ability to produce bulk amounts of CPE.
Blocking SMSr catalytic activity causes ceramide accumulation in the ER
Although SMSr produces only trace amounts of CPE, SMSr-depleted HeLa cells showed a fourfold increase in ceramide levels (from 0.31 ± 0.01 to 1.29 ± 0.15 mol%; n = 3). This was accompanied by a threefold increase in GlcCer levels (from 0.32 ± 0.03 to 0.86 ± 0.09 mol%; n = 3; Fig. 5 A). Depletion of SMS1 caused a very similar rise in ceramide and GlcCer levels. In the latter case, however, this can be fully explained by a block in SM production (Tafesse et al., 2007). In contrast, the amount of ceramide and GlcCer accumulating in SMSr-depleted HeLa cells is 50-fold higher than can be ascribed to a block in CPE production (Fig. 5 A). The rise in ceramides affected all major species (Fig. S3 A and Table S1), could also be detected by [14C]serine labeling, and was not accompanied by an increase in sphingosine levels (Fig. S3 C). Moreover, it also occurred in SMSr-depleted hamster CHO-K1 and Drosophila S2 cells, although in the latter case to a lesser extent (Fig. S3 B).
As SMSr resides in the ER, the ceramide accumulation in SMSr-depleted cells likely originates from this organelle. This was confirmed by subcellular fractionation, which showed that the ER of SMSr-depleted HeLa cells contains nearly 300% more ceramide than the ER of lamin A (LA)–depleted control cells (Fig. 5, B and C). To verify this also for S2 cells, we created an ER-resident form of hSMS1 and analyzed its ability to reduce ceramide levels in dSMSr-depleted S2 cells. Consistent with a role of SMSr-derived SAM in ER retention, an hSMS1 swap mutant carrying the SAM-containing cytosolic tail of hSMSr, hSMS1Nr, localized primarily to the ER (Fig. 5, D and E). hSMS1Nr retained its SMS activity, as its expression in S2 cells supported production of SM (Fig. 5 F). Expression of ER resident hSMS1Nr but not Golgi-associated hSMS1 prevented the accumulation of [14C]serine-labeled ceramides in dSMSr-depleted S2 cells (Fig. 5 G). Together, these results indicate that SMSr acts as a negative regulator of ceramide levels in the ER.
To investigate whether SMSr requires its catalytic activity to prevent ceramide accumulation, dSMSr-depleted S2 cells were transfected with wild type or an enzyme-dead version of hSMSr. Expression of wild-type hSMSr effectively reduced the ceramide pool in dSMSr-depleted S2 cells (Fig. 5 H). However, mutation of active site residue Asp348 to Glu was sufficient to abrogate this effect. Transfection of hSMSr-depleted HeLa cells with siRNA-resistant hSMSr or hSMSrD348E yielded the same results (Fig. 5 H). Thus, even though SMSr produces only trace amounts of CPE, this appears essential to control the ceramide pool in the ER.
Loss of SMSr perturbs the organization of the early secretory pathway
Recent work revealed that CERT−/− mouse embryos accumulate ceramide in the ER and display a chronic state of ER stress characterized by vesiculation and engorgement of the ER and up-regulation of downstream components of the unfolded protein response pathway (Wang et al., 2009). Even though SMSr-depleted S2 and HeLa cells show a substantial rise in ER ceramide levels, they did not display any of the morphological (ER dilation and vesiculation) or biochemical (XBP-1 splicing, induction of CHOP/GADD153, and up-regulation of ER chaperones) features of ER stress (unpublished data). Instead, we found that the organization of ER exit sites and the Golgi complex was greatly perturbed.
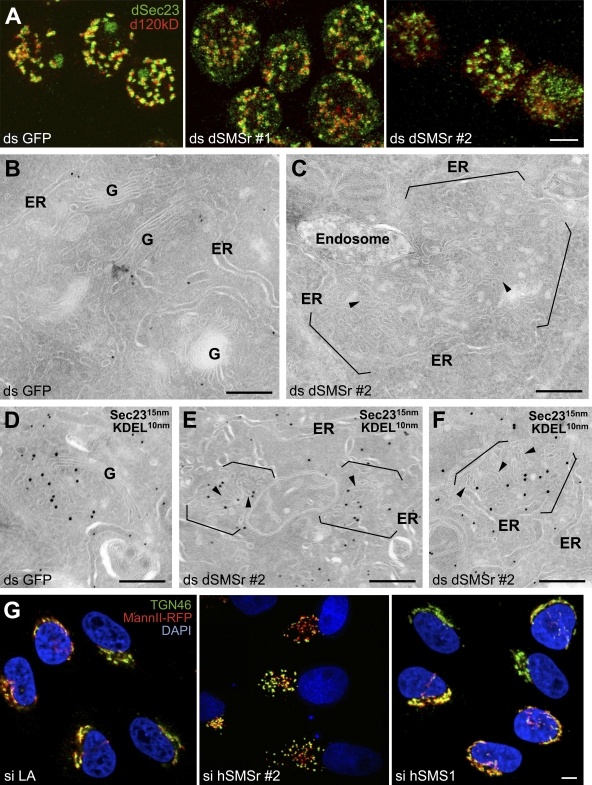
Golgi stacks in S2 cells do not form a large Golgi ribbon capping the nucleus as in mammalian cells. Instead, they remain discrete in the cytoplasm and are found in close proximity to transitional ER (tER) sites, forming 17–25 tER–Golgi units (Kondylis and Rabouille, 2003; Kondylis et al., 2007). In dSMSr-depleted S2 cells, the tER–Golgi units were much smaller and more numerous than in mock-depleted cells. Though the spatial relationship between the Golgi (marked by d120kd) and tER sites (marked by the COPII subunit dSec23) was maintained, both structures had a fragmented, hazy appearance (Fig. 6 A; and Fig. S4, A–I). This effect was quantitative, as more than half of the depleted cells displayed a fragmented Golgi (Fig. 7 C). However, the global architecture of the ER was unaffected by the depletion.
Figure 6.
SMSr depletion disrupts tER–Golgi units in insect and mammalian cells. (A) S2 cells were treated for 7 d with ds GFP, ds dSMSr #1, or ds dSMSr #2 and double labeled for dSec23 (tER sites) and d120kd (Golgi stacks). Confocal projections of the merge are presented. The single channels are presented in Fig. S5 A. Bar, 5 µm. (B–F) S2 cells treated for 5 d with ds GFP (B and D) or dSMSr #2 (C, E, and F) were fixed and processed for immunoelectron microscopy. Ultrathin cryosections were visualized before (B and C) or after double labeling for dSec23 (15-nm gold) and KDEL (10-nm gold; D–F). Note that Golgi stacks in most dSMSr-depleted cells are converted to a cluster of short tubular profiles (arrowheads) in cytoplasmic regions normally occupied by tER–Golgi units (between brackets). These short tubules are positively labeled for Sec23, suggesting that they comprise membrane of both compartments. G, Golgi stack. (G) Mannosidase II-RFP–expressing HeLa cells were treated with si LA, si hSMSr #2, or hSMS1 for 3 d and labeled for TGN46 and DNA (DAPI). Confocal projections of the merge are presented. The single channels are presented in Fig. S5 (J–L). Bars: (A and G) 5 µm; (B–F) 200 nm.
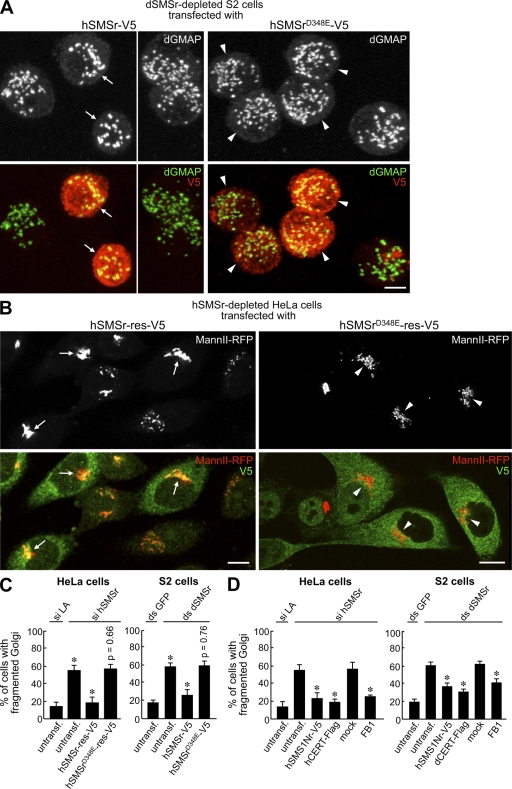
Figure 7.
Structural integrity of tER–Golgi units requires an enzyme-active form of SMSr. (A) S2 cells were treated with ds SMSr #2 and transfected with hSMSr-V5 or hSMSrD348E-V5. After 7 d, cells were double labeled for Golgi marker dGMAP and V5 to mark transfected cells. Confocal projections are presented. Note that cells expressing hSMSr (arrows) mostly show the typical wild-type Golgi organization of 17–25 large spots. In contrast, nontransfected cells or those expressing enzyme-dead hSMSr (arrowheads) typically have more numerous and smaller Golgi spots. (B) Mann II-RFP–expressing HeLa cells were treated with si hSMSr #2 and transfected with siRNA-resistant (res) hSMSr-V5 or hSMSrD348E-V5. After 3 d, cells were labeled for V5 to mark transfected cells. Confocal projections are presented. Note that cells expressing hSMSr-V5 contain an intact Golgi (arrows). In contrast, cells expressing enzyme-dead hSMSr still have fragmented Golgi (arrowheads). (C) Quantitation of the rescue of the Golgi fragmentation phenotype in SMSr-depleted cells (HeLa/S2) after transfection of wild-type or enzyme-dead SMSr. SMSr-depleted cells are compared with control cells (si LA or ds GFP treated), and SMSr-depleted cells transfected with wild-type or enzyme-dead SMSr are compared with untransfected (untransf) SMSr-depleted cells. (D) Quantification of the rescue of the Golgi fragmentation phenotype in SMSr-depleted cells (Hela/S2) after transfection with hSMS1Nr-V5 or CERT-Flag or after treatment with 50 µg/ml fumonisin B1 (FB1) for 2 d (HeLa) or 5 h (S2) before fixation. SMSr-depleted cells expressing hSMS1Nr-V5 or CERT-Flag are compared with untransfected cells, and mock-treated cells are compared with FB1-treated cells. Errors bars indicate SD, n = 3. *, P < 0.001 by two-tailed unpaired Student's t test. Bars, 5 µm.
At the ultrastructural level, Golgi morphology in dSMSr-depleted S2 cells was profoundly affected. Instead of showing the typical organization of stacked cisternae (Fig. 6, B and D), the Golgi was converted into many short tubular structures in the majority of the cells (Fig. 6, C, E, and F). These tubular clusters were mostly devoid of KDEL-containing proteins but were positive for dSec23 (Fig. 6, E and F), suggesting that they represent a mixture of Golgi and tER membranes.
hSMSr-depleted HeLa cells showed a very similar phenotype. The Golgi ribbon was completely fragmented in more than half of the cells as indicated by immunostaining against markers of medial (MannII-RFP) and trans-Golgi compartments (TGN46; Fig. 6 G; Fig. 7 C; Fig. S4, J–L). These results further strengthen the functional similarity between hSMSr and dSMSr. Moreover, the requirement of SMSr for Golgi integrity appears specific because the Golgi ribbon was unperturbed in hSMS1-depleted HeLa cells (Fig. 6 G).
We next tested whether fragmentation of the early secretory pathway could result in or be the result of inhibition of anterograde transport. To this end, we monitored the transport efficiency of the transmembrane protein Delta from the ER to the PM in S2 cells (Kondylis and Rabouille, 2003). Brefeldin A treatment blocked PM delivery of Delta and caused its accumulation in the ER. In contrast, PM delivery of Delta was not significantly impaired in dSMSr-depleted S2 cells, suggesting that the fragmented tER–Golgi units in these cells are fully competent for anterograde transport (Fig. S5 A). Similar results were obtained in hSMSr-depleted HeLa cells using the well-established VSV-045G transport assay (Fig. S5, C and D). Moreover, SMSr depletion had no major impact on protein glycosylation (Fig. S5 E). Together, these findings indicate that SMSr proteins play a critical role in the acquisition of Golgi stack morphology without affecting the functional integrity of the organelle. This adds further evidence to the notion that transport of cargo to, through, and out of the Golgi complex does not require a fully developed organelle and can proceed under conditions in which the cisternal stacks are severely disorganized (Kondylis and Rabouille, 2003; Zolov and Lupashin, 2005).
Golgi fragmentation in SMSr-depleted cells is caused by ceramide accumulation in the ER
To test whether catalytic activity of SMSr is required to prevent Golgi fragmentation, we monitored the rescue of this phenotype by transfecting dSMSr-depleted S2 cells with wild-type or enzyme-dead hSMSr. Expression of wild-type hSMSr almost completely restored Golgi integrity. However, mutation of active site residue Asp348 to Glu was sufficient to abolish the rescuing effect (Fig. 7, A and C). These results completely parallel those obtained with hSMSr-depleted HeLa cells after transfection with siRNA-resistant hSMSr or hSMSrD348E (Fig. 7, B and C).
One possible interpretation of these results is that the structural integrity of the early secretory pathway depends on the trace amounts of CPE produced by SMSr. Alternatively, as an enzyme-dead mutant of SMSr is unable to prevent ceramide accumulation in the ER (Fig. 5 H), fragmentation of the early secretory pathway in SMSr-depleted cells could be caused by the rise in ceramide levels in the ER. To distinguish between these possibilities, ER resident SMS hSMS1Nr was expressed in SMSr-depleted S2 or HeLa cells and analyzed for its ability to rescue Golgi fragmentation. Expression of hSMS1Nr partially restored Golgi integrity in both cell types (Fig. 7 D), indicating that acquisition of Golgi stack morphology is not strictly dependent on production of CPE. As expression of hSMS1Nr was effective in preventing ceramide accumulation in SMSr-depleted cells (Fig. 5 G), we asked whether blocking ceramide production or stimulating ceramide export from the ER would also suppress Golgi fragmentation. Treatment with ceramide synthase inhibitor fumonisin B1 or overexpression of CERT partially but reproducibly restored Golgi integrity in both SMSr-depleted HeLa and S2 cells (Fig. 7 D). This indicates that the structural collapse of the early secretory pathway in SMSr-depleted cells is the result of ceramide accumulation in the ER. Thus, these results further support a primary role of SMSr in controlling ceramide homeostasis in the ER.
Discussion
SMSr proteins define an unconventional class of sphingolipid synthases
In this study, we show that SMSr, the most conserved member of the multigenic SMS family, catalyzes production of the SM analogue CPE. SMSr likely accounts for the CPES activity described two decades ago in rat brain and liver microsomes (Malgat et al., 1986, 1987). Common features between these enzymes include the use of PE as headgroup donor, their ER residency, and a membrane topology in which the active site is facing the ER lumen. Although mammalian cells contain a readily detectable CPES activity, their CPE content is very low (i.e., 300-fold lower than SM). This enigma is neither caused by a limited supply of substrates to the enzyme's active site nor by CPE being a short-lived metabolic intermediate. Instead, we find that SMSr intrinsically lacks the ability to synthesize significant amounts of CPE. This is radically different from closely related SMS1, which catalyses bulk production of SM (Tafesse et al., 2007).
What makes SMSr so different from SMS1? SMS1 utilizes a lipid phosphate phosphatase–type reaction mechanism, which involves a single lipid-binding site and proceeds via transfer of phosphocholine from PC to a conserved histidine in the enzyme's active site (Huitema et al., 2004; Tafesse et al., 2006). When DAG is replaced by ceramide, the phosphocholine headgroup is transferred onto ceramide to form SM. The latter is then released from the active site to allow another round of catalysis. Mutational analysis of putative active site residues indicates that SMSr-mediated CPE production involves a very similar reaction mechanism. However, although SM readily dissociates from SMS1, we propose that CPE remains bound to the active site of SMSr, thus blocking a further round of catalysis. This could be the result of several factors. First, SMSr might have a higher affinity for CPE than for PE. Second, dissociation of SM from SMS1 might be facilitated by its preferential interaction with cholesterol. Contrary to SM, CPE lacks this interaction (Terova et al., 2005). Thus, in the presence of cholesterol, SM production might be thermodynamically more favorable than CPE production.
The notion that SMSr is unable to produce significant amounts of CPE is further substantiated by our finding that bulk production of CPE in insects is mediated by a different enzyme. This second CPES uses CDP-ethanolamine instead of PE as headgroup donor, which is analogous to the ethanolamine phosphotransferases of the Kennedy pathway (Vance, 2008). This implies that, contrary to SM synthesis in mammals, bulk production of CPE occurs in the cytosolic leaflet of the membrane. As insects require CERT for efficient CPE production (Rao et al., 2007), the second enzyme likely resides in the Golgi.
SMSr qualifies for a ceramide sensor in the ER
Although SMSr is a poor CPES, we find that the enzyme has a major impact on ceramide levels in the ER. Indeed, SMSr-depleted mammalian cells display a rise in ceramide levels that is one order of magnitude too high to be explained by a reduced ceramide consumption for CPE biosynthesis. SMSr-depleted insect cells and SMSr-null mutants in Caenorhabditis elegans also show a marked increase in ceramide levels (this study; unpublished data). Strikingly, a single point mutation in the active site of SMSr proved sufficient to induce ceramide accumulation. This shows that SMSr must be catalytically active to keep ER ceramide levels low, even if the enzyme lacks the ability to consume significant amounts of ceramide. Together, these results establish SMSr as a novel regulator of ceramide homeostasis in the ER.
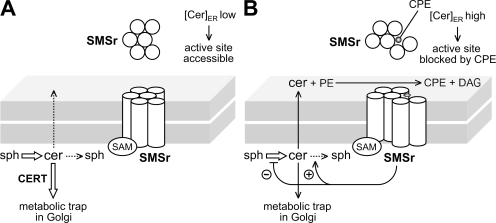
How does SMSr control ceramide levels in the ER? We anticipate that under normal conditions, only small amounts of ceramide will reach the enzyme's active site. This is because ceramides synthesized on the cytosolic surface of the ER are continuously removed by CERT (Fig. 8 A). Consequently, only a minor fraction of SMSr enzymes will be trapped in a CPE-bound form. Yet when the rate of ceramide production exceeds ceramide removal by CERT, the pool of CPE-bound enzymes will expand. It is possible that CPE-bound SMSr adopts a conformation that relays a signal to inhibit ceramide biosynthesis and/or stimulate ceramide degradation (Fig. 8 B). Alternatively, SMSr-derived CPE might directly influence the activity of ceramide-metabolizing enzymes in the ER. Either feedback mechanism would ensure maintenance of ceramide homeostasis in the ER. Distinguishing between these possibilities will require reconstitution experiments with purified enzymes.
Figure 8.
Model of how SMSr might control ceramide homeostasis in the ER. (A) Normally, only small amounts of newly synthesized ceramides can reach the active site of SMSr in the ER lumen as a result of their efficient removal from the cytosolic surface by CERT. (B) When CERT can't cope with the rate of ceramide production, more ceramides will reach the active site of SMSr, resulting in production of CPE. Because of its high affinity for the enzyme's active site, CPE remains bound to SMSr, thus blocking a further round of catalysis. CPE-trapped SMSr adopts a conformation that relays a signal inhibiting ceramide production or stimulating ceramide breakdown in the ER, thereby restoring ceramide homeostasis. Sph, sphingoid base.
Collectively, our data suggest that SMSr is a CPES with a dual role as ceramide sensor to control ceramide homeostasis in the ER. As its active site faces the ER lumen, SMSr would only sense ceramides present in the exoplasmic leaflet, i.e., molecules that escaped CERT-mediated extraction at the cytosolic surface and flipped to the lumenal side. Notably, this arrangement implies cooperativity between SMSr and CERT in keeping ER ceramide levels low. Thus, SMSr serves a biological role that is different from closely related SMS1 but analogous to the ensemble of membrane proteins that monitor sterol concentrations in the ER (Goldstein et al., 2006). In this respect, the SMS enzyme family displays an interesting parallel with the nutrient transporter family. Besides conventional transporters, the latter family also harbors transceptors that play a dual role as nutrient sensor because their conformational change during the transport process can trigger downstream signaling pathways (Holsbeeks et al., 2004).
Functional implications of SMSr-controlled ceramide homeostasis
Concurrent with the accumulation of ceramide in the ER, blocking SMSr activity causes a structural collapse of the early secretory pathway. The disruption of tER–Golgi organization completely parallels the rise in ER ceramide levels. Indeed, any condition that normalizes the ceramide concentration in the ER seems to restore Golgi stack morphology; i.e., the presence of enzymatically active SMSr, the consumption of ceramide by ER resident SMS, stimulation of ER export of ceramide by overexpression of CERT, or a block in ceramide production by ceramide synthase inhibitors. Conversely, blocking SM production to accumulate ceramides in the Golgi does not perturb its morphology.
How can an excess of ceramides in the ER affect Golgi organization? As the biogenesis of the Golgi is tightly linked to that of tER sites (Bevis et al., 2002), a disruption of tER sites likely contributes to the structural collapse of the Golgi. tER sites are thought to arise from patches of ER membrane proteins that have weak, cooperative affinity for one another (Bevis et al., 2002; Heinzer et al., 2008). These patches capture additional tER proteins until they grow large enough to produce COPII vesicles. A rise in ER ceramides may perturb the local physical environment required to stabilize patches of tER proteins. Alternatively, ceramides may affect tER site morphology by altering the membrane turnover of COPII components analogous to the situation in sterol-depleted cells (Runz et al., 2006). Moreover, ceramides serve as early mediators of apoptosis, an event accompanied by fragmentation of the Golgi complex. Apoptosis-induced Golgi fragmentation requires caspase 3–mediated cleavage of golgins, a family of coiled-coil proteins involved in maintenance of Golgi stack morphology (Lane et al., 2002). Interestingly, SMSr-depleted HeLa cells display hallmarks of apoptosis, including activation of caspase 3 (unpublished data). Whether blocking caspase activation prevents Golgi fragmentation in SMSr-deficient cells remains to be established.
A tight regulation of ceramide homeostasis in the ER likely has functional implications beyond sustaining the structural integrity of the early secretory pathway. Down-regulation of CERT sensitizes cancer cells to paclitaxel, suggesting that ER ceramides are integral to taxane-induced cell death (Swanton et al., 2007). Moreover, CERT−/− mouse embryos accumulate ceramide in the ER and display a chronic state of ER stress (Wang et al., 2009). As components of the unfolded protein response are up-regulated during animal development (Reimold et al., 2000; Iwakoshi et al., 2007), a defective transport of ceramide might potentiate ER stress in the embryo (Wang et al., 2009). Whether loss of SMSr sensitizes cells to ER stress–inducing conditions is currently under investigation.
The ER is a highly efficient lipid distribution system that is intimately associated with other organelles, including mitochondria (Levine and Rabouille, 2005). Ceramides have been implicated in mitochondria-induced apoptosis and may reach these organelles via contact sites with the ER (Siskind et al., 2008; Stiban et al., 2008; Wang et al., 2009). The present identification of SMSr as a key regulator of ceramide homeostasis in the ER provides novel opportunities to explore the role of ceramides in mitochondria-induced cell death and for modulating taxane effects on tumors.
Materials and methods
Chemicals
NBD-Cer was obtained from Invitrogen, NBD-SM, POPE (2-oleoyl-1-palmitoyl-sn-glycerol-3-phosphoethanolamine), and POPC (2-oleoyl-1-palmitoyl-sn-glycerol-3-phosphocholine) were obtained from Avanti Polar Lipids, Inc. NBD-CPE was provided by P. Devaux (Institut de Biologie Physico-chimique, Paris, France). 2-[14C]ethan-1-ol-2-amine hydrochloride and 3-[14C]L-serine were obtained from GE Healthcare, and methyl-[14C]choline chloride and [3H]sphingosine were obtained from MP Biomedicals. D4-ethanolamine and D9-choline were obtained from Cambridge Isotope Laboratories. All other lipids and chemicals were obtained from Sigma-Aldrich.
Antibodies
The following antibodies were used: rabbit polyclonal anti-V5 and mouse monoclonal anti-Flag (Sigma-Aldrich), mouse monoclonal anti-V5 (Invitrogen), mouse monoclonal anti-GM130 (BD), mouse monoclonal anti-KDEL, rabbit polyclonal anti-calnexin and anti-GRP75 (Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-d120kd (EMD), rabbit polyclonal anti-dGOLGIN245 (provided by S. Munro, Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK), rabbit polyclonal anti-Na+/K+-ATPase (C356-M09; provided by J. Koenderink, Radboud University Nijmegen, Nijmegen, Netherlands), rabbit polyclonal antiprotein disulfide isomerase (provided by I. Braakman, Utrecht University, Utrecht, Netherlands), rabbit polyclonal anti-GFP (Invitrogen), mouse monoclonal anti-EEA1 (BD), rabbit polyclonal anti-dGMAP (Kondylis et al., 2007), rabbit polyclonal anti-dSec23p, and mouse monoclonal C594.9B (Kondylis and Rabouille, 2003). Horseradish peroxidase–conjugated secondary antibodies were obtained from PerBio, whereas antibodies conjugated to FITC and Texas red or Alexa dyes were obtained from Jackson Immunoresearch Laboratories and Invitrogen, respectively.
To raise an antibody against dSMSr, the N terminus of dSMSr (residues 1–199) was expressed as glutathione S-transferase fusion in Escherichia coli and purified by affinity chromatography on glutathione agarose (Sigma-Aldrich). The purified fusion protein was used by PickCell Laboratories for the immunization of mice, yielding monoclonal anti-dSMSr antibody B5-G7.
DNA constructs
hSMSr, hSMS1, and dSMSr cDNAs were cloned into yeast expression vector pYES2.1/V5-His-TOPO and mammalian expression vector pcDNA3.1/V5-His-TOPO (Invitrogen) as described previously (Huitema et al., 2004). For expression studies in Drosophila S2 cells, dSMSr, hSMSr, and hSMS1 cDNAs were PCR amplified and ligated into the copper-inducible pMT/V5-His B vector (Invitrogen). Mutation of active site residues in dSMSr (H401A and D405E) and hSMSr (D348E) was performed using the megaprimer PCR method (Orr-Weaver et al., 1983). SAM-deficient mutants were obtained by truncation of the first 186 N-terminal residues in dSMSr or the first 68 N-terminal residues in hSMSr. An ER-resident hSMS1 swap mutant, hSMS1Nr, was made by swapping the cytosolic N terminus of hSMS1 (residues 1–133) with that of hSMSr (residues 1–150). The cDNA of human PEMT was purchased from RZPD (clone IRATp970D1078D6) and cloned into pMT/EGFP. Human CERT–Flag (Fugmann et al., 2007) and Drosophila CERT–Flag expression constructs (Rao et al., 2007) were provided by M. Olayioye (University of Stuttgart, Stuttgart, Germany) and J. Acharya (National Cancer Institute at Frederick, Frederick, MD), respectively.
Yeast culture
Yeast strain 4Δ.Lass5 (MATa ade2-101ochre his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 lag1::TRP1 lac1::LEU2 ydc1::natMX ypc1::kanMX4 p413MET25:Lass5; Cerantola et al., 2007) was transfected with hSMSr or dSMSr in pYES2.1/V5-His-TOPO and grown in selective synthetic medium containing 2% (wt/vol) galactose and 50 mg/l myo-inositol.
Animal cell culture and RNAi
CHO-K1 and LY-A cells were grown in F-12 medium with 10% FCS (Invitrogen), whereas LY-A/human CERT cells were grown in F-12 medium containing 0.8 mg/ml G-418 (Invitrogen). HepG2 cells were grown in DME with 10% FBS (Cambrex) and CaCo-2 cells in DME-GlutamaxI with 10% FCS. HeLa cells were grown in DME with 10% FCS. Stable transfections with hSMS1-V5, hSMSr-V5, or mouse α-mannosidase II-RFP in pcDNA3.1 and treatment with siRNA (QIAGEN) were performed as described previously (Tafesse et al., 2007). siRNA target sequences were: nonsense, 5′-AATTCTCCGAACGTGTCACGT-3′; LA, 5′-AACTGGACTTCCAGAAGAACA-3′; hSMSr #1, 5′-AATCTTCTTCATCTTGGCTGC-3′; hSMSr #2, 5′-CAAGAAGCTGGAATTTCTTGC-3′; hSMS1, 5′-AACTACACTCCCAGTACCTGG-3′. For rescue experiments, cells were transfected with siRNA-resistant hSMSr constructs 8 h after the start of siRNA treatment using Effectene (QIAGEN). Constructs were made siRNA resistant by introducing five silent point mutations in the siRNA target sequence.
Drosophila S2 cells were grown in Schneider's insect medium with 10% FBS. Treatment with double-stranded RNA (dsRNA) was performed as described previously (Clemens et al., 2000). On day 1, 106 cells were plated in a 35-mm dish and incubated with 30 µg dsRNA in 1 ml serum-free medium for 1 h at RT followed by addition of 2 ml complete medium. Treatment with dsRNA was repeated on day 4. For rescuing experiments, cells were transfected with SMS/pMT/V5-HisB constructs on day 5 using Effectene. Expression of recombinant SMS protein was induced by addition of 1 mM CuSO4 for 3 h followed by a 2-h chase in the presence of 150 µg/ml cycloheximide. dsRNA was synthesized by in vitro transcription of PCR products flanked by T7 RNA polymerase-binding sites (5′-TTAATACGACTCACTATAGGGAGA-3′) using the MEGASCRIPT T7 transcription kit (Applied Biosystems). dsRNA-targeting dSMSr was synthesized from two ∼800-bp PCR products that were generated from dSMSr cDNA using the following primer sets: 5′-T7-ATGTGCGACGGTGAAATTG-3′/5′-T7-GAGCCGGTTATTTCGCACAT-3′ (dsRNA-targeting SMSr [ds SMSr] #1) and 5′-T7-TTGGTCCTACTGAGGCGATTC-3′/5′-T7-CGACTGGTGAGGACTAAAGAAAGC-3′ (ds SMSr #2). dsRNA-targeting GFP (ds GFP) was synthesized from an ∼800-bp PCR product, which was generated from pMT/EGFP plasmid using the primer set 5′-T7-CTAGGCGGCCGCAAAAAATG-3′/5′-T7-CTGTTCGAAGGCGCGCCTTGT-3′.
In vitro enzyme assays
100 ODs of yeast cells was lysed by bead bashing in 10 ml ice-cold reaction buffer (0.3 M sucrose, 15 mM KCl, 5 mM NaCl, 1 mM EDTA, 20 mM Hepes-KOH, pH 7.0) containing freshly added protease inhibitors. HeLa and S2 cells were lysed in ice-cold reaction buffer by passing 20 times through a 26-G 0.75 needle. 200 µl postnuclear supernatant (PNS; 700 g for 10 min at 4°C) was combined with 200 µl reaction buffer containing 0.002% Triton X-100, 40 nmol POPE and/or POPC, and 50 µM NBD-Cer (Avanti Polar Lipids, Inc.), and incubated at 37°C (HeLa/yeast) or 27°C (S2 cells) for 2 h. Reactions were stopped by adding 1 ml MeOH and 0.5 ml CHCl3, and lipids were extracted according to Bligh and Dyer (1959). The lower phase was evaporated under N2 and the reaction products analyzed by TLC, which was developed first in acetone and then in CHCl3/MeOH/25% NH4OH (50/25/6 [vol/vol/vol]; Figs. 1 and 2) or CHCl3/acetone/MeOH/acetic acid/H2O (50/20/10/10/5 [vol/vol/vol/vol/vol]; Fig. 4). Fluorescent lipids were visualized on an image analysis system (STORM 860; GE Healthcare) and quantified with Quantity One software (Bio-Rad Laboratories).
Lipid MS
100 ODs of yeast cells was washed with water, treated with trichloroacetic acid to inactivate lipases, and finally boiled for 10 min before lipid extraction by bead bashing in CHCl3/MeOH/H2O (10/10/3 [vol/vol/vol]) according to Folch et al. (1957). HeLa and S2 lipid extracts were prepared according to Folch et al. (1957), but omitting the salt. The lower phase was evaporated under N2. Glycerolipids were removed by mild alkaline hydrolysis in 0.5 M sodium methoxide in MeOH for 1 h at RT. The lipids were analyzed with a triple quadrupole mass spectrometer (PE Sciex API-365; Applied Biosystems). Positive ions were generated by a turbo ion spray ionization source operating at +5.5 kV ionization potential. N2 was used as drying gas at 350°C. The declustering potential (cone voltage) was set to 50 V, the collision energy to 36 V, the entrance potential to −10 V, and the focus potential to 160 V. The lipid molecular species and classes were quantified as described previously in Tafesse et al. (2007).
Metabolic labeling
S2 cells (2–5 × 106) were labeled in 0.5 ml complete Schneider's insect medium with 10 nmol NBD-Cer, 1 µCi [14C]ethanolamine, 1 µCi [14C]choline, or 1 µCi [14C]serine at 27°C for the indicated time. Mammalian cells (10-cm dish; 50% confluency) were labeled in complete medium with 1 µCi [14C]ethanolamine or 1 µCi [3H]sphingosine for the indicated time. Lipids were extracted in CHCl3/MeOH/10 mM acetic acid (1/4.4/0.2 [vol/vol/vol]) and processed according to Bligh and Dyer (1959). Half of the extract was subjected to mild alkaline hydrolysis. Radiolabeled lipids were analyzed by TLC in CHCl3/MeOH/25% NH4OH (50/25/6 [vol/vol/vol]; [14C]ethanolamine/choline), CHCl3/MeOH/2 M NH4OH (40/10/1 [vol/vol/vol]; [14C]serine), CHCl3/MeOH/0.2% CaCl2 (60/40/9 [vol/vol/vol]; [3H]sphingosine), detected by exposure to imaging screens (BAS-MS; FujiFilm), scanned on a Personal Molecular Imager (Bio-Rad Laboratories), and quantified with Quantity One software.
Isolation of ER membranes
HeLa cells were washed twice and harvested by scraping in ice-cold homogenization buffer (HB; 10 mM Hepes, pH 7.8, 1 mM EGTA, 25 mM KCl, 1 mM protease inhibitor cocktail, and 1 mM PMSF) supplemented with 0.25 M sucrose. Cells were pelleted by centrifugation at 600 g and gently disrupted by a dounce homogenizer in 11 ml HB at 4°C. The homogenate was centrifuged twice at 500 average g (gav; 5 min at 4°C) to generate a PNS. The PNS was centrifuged at 10,000 gav (10 min at 4°C) to pellet mitochondria. The postmitochondrial supernatant was loaded onto a 1 ml 68% (wt/wt) sucrose cushion (prepared in HB) and centrifuged at 100,000 gav. (1 h at 4°C) to collect crude microsomes. The microsomes were resuspended in 1 ml HB and loaded on top of a 25–60% (wt/wt) sucrose step gradient that was generated in HB using the following steps: 2 ml 25%, 2 ml 30%, 2 ml 37%, 2 ml 39%, 2 ml 42%, and 1 ml 60%. After centrifugation at 100,000 gav (18 h at 4°C) in a rotor (SW41 Ti; Beckman Coulter), 20 × 0.6 ml fractions were collected from the top. PM and ER membranes peaked in fractions 7–9 and 17 and18, respectively. Equal amounts of protein from the PNS, the crude mitochondrial membrane pellet, and the collected PM and ER membrane peaks were subjected to immunoblot analysis. This revealed that the collected ER membranes were virtually devoid of other organelles except for a minor contamination with PM (<10%).
Microscopy and image analysis
Cells were fixed in 4% paraformaldehyde/PBS, processed for immunofluorescence as described previously for S2 (Kondylis and Rabouille, 2003) and HeLa cells (Tafesse et al., 2007), and mounted in Vectashield medium containing DAPI (Vector Laboratories). Images were captured at RT using a confocal microscope (LSM 510 Meta; Carl Zeiss, Inc.) with a 63× 1.40 NA Plan Apo oil objective (Carl Zeiss, Inc.) or with a confocal microscope (D-eclipse C1; Nikon) with 60× 1.40 NA Plan Apo oil objective (Nikon). The fluorochromes used were DAPI, λex = 360 nm and λem= 460 nm; FITC/Alexa Fluor 488, λex = 488 nm and λem = 515 nm; Texas red/Alexa Fluor 568, λex = 568 nm and λem = 585 nm. Projections were obtained by collecting series of 0.4-µm sections that were acquired and combined using LSM software (Carl Zeiss, Inc.) or EZ-C1 software (Nikon). Images were further processed using Photoshop software (version 7.0.1; Adobe).
The tER–Golgi fragmentation phenotype in S2 cells was assessed as described previously in Kondylis and Rabouille (2003). In brief, all cells from each projection were assigned to either one of two categories: cells with a wild-type Golgi morphology, which corresponds to 17–25 large, dGMAP, or d120kd-labeled spots, and cells with a fragmented Golgi morphology, corresponding to numerous smaller labeled spots. Results were expressed as a percentage of cells displaying a fragmented Golgi morphology. The averages were derived from three independent experiments, each analyzing at least 400 (or ≥75 transfected) cells per condition. The statistical significance of all data obtained was assessed by two-tailed unpaired Student's t tests. P ≤ 0.001 was considered significant and is marked by an asterisk, whereas P > 0.001 is indicated (Fig. 7, C and D).
S2 cells were fixed and processed for immunoelectron microscopy as described previously in Kondylis and Rabouille (2003). Ultrathin cryosections were examined using an electron microscope (1200 EX; JEOL). All antibodies used for immunolabeling are described in Online supplemental material.
Delta and VSV-045G transport assays
The Delta transport assay was performed using the cell line Delta-WTNdeMYC as described previously in Kondylis and Rabouille (2003). In brief, S2 cells were treated with different dsRNA for the indicated time. The Delta protein was induced for 1 h with 1 mM CuSO4 and chased in the absence of CuSO4 for different time points. Subsequently, cells were fixed, immunostained, and Delta arrival at PM was monitored by immunofluorescence microscopy. For VSV-045G transport assay, HeLa cells were treated with different siRNA constructs for 7 d, transfected with VSV-045G-GFP, kept at 39.5°C for 16 h, and chased at 32°C for different time points as indicated.
Online supplemental material
Fig. S1 shows the effect of externally added phospholipid on CPE production levels in lysates of normal and hSMSr-overexpressing HeLa cells. Fig. S2 shows that loss of the SAM domain causes a redistribution of SMSr from the ER to the Golgi. Fig. S3 shows that SMSr-depleted cells accumulate all major species of ceramide but no sphingosine. Fig. S4 shows the single channels of images presented in Fig. 6. Fig. S5 shows that anterograde protein transport to the PM is not impaired in SMSr-depleted cells. Table S1 shows a quantitative analysis of ceramide species in control, hSMSr-depleted, and hSMS1-depleted HeLa cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903152/DC1.
Acknowledgments
We thank Kenneth Hanada, Sean Munro, Andreas Conzelmann, Philippe Devaux, Jairaj Acharya, and Monilola Olayioye for gifts of reagents and cell lines, Tarja Grundström, Elleke Bosma, and Martin Harterink for technical assistance, and our colleagues at the department of Membrane Enzymology for stimulating discussions and critical reading of the manuscript.
A.M. Vacaru was supported by the European Union Framework V Program (the Flippase Project), P. Ternes was supported by a Marie Curie Intra-European Fellowship and a European Molecular Biology Organization Long-Term Fellowship, V. Kondylis was supported by a Horizon Programme from National Regie-Orgaan Genomics (grant 050-71-029), and J.C.M. Holthuis was supported by grants from the Dutch Organization of Sciences (NWO-CW) and the Utrecht University High Potential Program.
Footnotes
Abbreviations used in this paper: CERT, ceramide transfer protein; CPE, ceramide phosphoethanolamine; CPES, CPE synthase; ds GFP, dsRNA-targeting GFP; ds SMSr, dsRNA-targeting SMSr; dSMSr, Drosophila SMSr; dsRNA, double-stranded RNA; gav, average g; GlcCer, glucosylceramide; HB, homogenization buffer; hSMSr, human SMSr; LA, lamin A; LCB, long-chain base; MS, mass spectrometry; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PEMT, PE N-methyltransferase; PM, plasma membrane; PNS, postnuclear supernatant; SAM, sterile α motif; si hSMSr, siRNA-targeting hSMSr; si LA, siRNA-targeting LA; SM, sphingomyelin; SMS, SM synthase; SMSr, SMS related; tER, transitional ER.
References
- Andrieu-Abadie N., Levade T. 2002. Sphingomyelin hydrolysis during apoptosis.Biochim. Biophys. Acta. 1585:126–134 [DOI] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Aronov P.A., Fontes K., Ramos K., Hammock B.D., Powers T. 2008. Regulation of ceramide biosynthesis by TOR complex 2.Cell Metab. 7:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis B.J., Hammond A.T., Reinke C.A., Glick B.S. 2002. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris.Nat. Cell Biol. 4:750–756 [DOI] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. 1959. A rapid method of total lipid extraction and purification.Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- Bretscher M.S., Munro S. 1993. Cholesterol and the Golgi apparatus.Science. 261:1280–1281 [DOI] [PubMed] [Google Scholar]
- Cerantola V., Vionnet C., Aebischer O.F., Jenny T., Knudsen J., Conzelmann A. 2007. Yeast sphingolipids do not need to contain very long chain fatty acids.Biochem. J. 401:205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways.Proc. Natl. Acad. Sci. USA. 97:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G.H. 1957. A simple method for the isolation and purification of total lipides from animal tissues.J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- Fugmann T., Hausser A., Schoffler P., Schmid S., Pfizenmaier K., Olayioye M.A. 2007. Regulation of secretory transport by protein kinase D–mediated phosphorylation of the ceramide transfer protein.J. Cell Biol. 178:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J.L., DeBose-Boyd R.A., Brown M.S. 2006. Protein sensors for membrane sterols.Cell. 124:35–46 [DOI] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide.Nature. 426:803–809 [DOI] [PubMed] [Google Scholar]
- Hannun Y.A., Obeid L.M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids.Nat. Rev. Mol. Cell Biol. 9:139–150 [DOI] [PubMed] [Google Scholar]
- Heinzer S., Worz S., Kalla C., Rohr K., Weiss M. 2008. A model for the self-organization of exit sites in the endoplasmic reticulum.J. Cell Sci. 121:55–64 [DOI] [PubMed] [Google Scholar]
- Hirschberg K., Rodger J., Futerman A.H. 1993. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver.Biochem. J. 290:751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsbeeks I., Lagatie O., Van Nuland A., Van de Velde A., Thevelein J.M. 2004. The eukaryotic plasma membrane as a nutrient-sensing device.Trends Biochem. Sci. 29:556–564 [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., Pomorski T., Raggers R.J., Sprong H., van Meer G. 2001. The organizing potential of sphingolipids in intracellular membrane transport.Physiol. Rev. 81:1689–1723 [DOI] [PubMed] [Google Scholar]
- Huitema K., van den Dikkenberg J., Brouwers J.F., Holthuis J.C. 2004. Identification of a family of animal sphingomyelin synthases.EMBO J. 23:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi N.N., Pypaert M., Glimcher L.H. 2007. The transcription factor XBP-1 is essential for the development and survival of dendritic cells.J. Exp. Med. 204:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S.D., Nagiec M.M. 2003. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p.Eukaryot. Cell. 2:284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Rabouille C. 2003. A novel role for dp115 in the organization of tER sites in Drosophila.J. Cell Biol. 162:185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., van Nispen tot Pannerden H.E., Herpers B., Friggi-Grelin F., Rabouille C. 2007. The Golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE.Dev. Cell. 12:901–915 [DOI] [PubMed] [Google Scholar]
- Kumagai K., Kawano M., Shinkai-Ouchi F., Nishijima M., Hanada K. 2007. Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT.J. Biol. Chem. 282:17758–17766 [DOI] [PubMed] [Google Scholar]
- Lane J.D., Lucock J., Pryde J., Barr F.A., Woodman P.G., Allan V.J., Lowe M. 2002. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis.J. Cell Biol. 156:495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T., Rabouille C. 2005. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues.Curr. Opin. Cell Biol. 17:362–368 [DOI] [PubMed] [Google Scholar]
- Malgat M., Maurice A., Baraud J. 1986. Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain.J. Lipid Res. 27:251–260 [PubMed] [Google Scholar]
- Malgat M., Maurice A., Baraud J. 1987. Sidedness of ceramide-phosphoethanolamine synthesis on rat liver and brain microsomal membranes.J. Lipid Res. 28:138–143 [PubMed] [Google Scholar]
- Mandala S.M., Thornton R., Tu Z., Kurtz M.B., Nickels J., Broach J., Menzeleev R., Spiegel S. 1998. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response.Proc. Natl. Acad. Sci. USA. 95:150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon E.C., Ehses I., Rother J., van Echten G., Sandhoff K. 1992. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver.J. Biol. Chem. 267:11144–11148 [PubMed] [Google Scholar]
- Morales A., Lee H., Goni F.M., Kolesnick R., Fernandez-Checa J.C. 2007. Sphingolipids and cell death.Apoptosis. 12:923–939 [DOI] [PubMed] [Google Scholar]
- Muehlenberg B.A., Sribney M., Duffe M.K. 1972. Occurrence and biosynthesis of ceramide phosphorylethanolamine in chicken and rat liver.Can. J. Biochem. 50:166–173 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak J.W., Rothstein R.J. 1983. Genetic applications of yeast transformation with linear and gapped plasmids.Methods Enzymol. 101:228–245 [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Hirschberg K., Polishchuk R.S., Gerlich D., Phair R.D., Lippincott-Schwartz J. 2008. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system.Cell. 133:1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R.P., Yuan C., Allegood J.C., Rawat S.S., Edwards M.B., Wang X., Merrill A.H., Jr., Acharya U., Acharya J.K. 2007. Ceramide transfer protein function is essential for normal oxidative stress response and lifespan.Proc. Natl. Acad. Sci. USA. 104:11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Etkin A., Clauss I., Perkins A., Friend D.S., Zhang J., Horton H.F., Scott A., Orkin S.H., Byrne M.C., et al. 2000. An essential role in liver development for transcription factor XBP-1.Genes Dev. 14:152–157 [PMC free article] [PubMed] [Google Scholar]
- Runz H., Miura K., Weiss M., Pepperkok R. 2006. Sterols regulate ER-export dynamics of secretory cargo protein ts-O45-G.EMBO J. 25:2953–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siskind L.J., Feinstein L., Yu T., Davis J.S., Jones D., Choi J., Zuckerman J.E., Tan W., Hill R.B., Hardwick J.M., Colombini M. 2008. Anti-apoptotic Bcl-2 family proteins disassemble ceramide channels.J. Biol. Chem. 283:6622–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel S., Milstien S. 2003. Sphingosine-1-phosphate: an enigmatic signalling lipid.Nat. Rev. Mol. Cell Biol. 4:397–407 [DOI] [PubMed] [Google Scholar]
- Stiban J., Caputo L., Colombini M. 2008. Ceramde synthesis in the endoplasmic reticulum can permeabilize mitochondria to pro-apoptotic proteins.J. Lipid Res. 49:625–634 [DOI] [PubMed] [Google Scholar]
- Swanton C., Marani M., Pardo O., Warne P.H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A.A., et al. 2007. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs.Cancer Cell. 11:498–512 [DOI] [PubMed] [Google Scholar]
- Tafesse F.G., Ternes P., Holthuis J.C. 2006. The multigenic sphingomyelin synthase family.J. Biol. Chem. 281:29421–29425 [DOI] [PubMed] [Google Scholar]
- Tafesse F.G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J.C. 2007. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells.J. Biol. Chem. 282:17537–17547 [DOI] [PubMed] [Google Scholar]
- Terova B., Heczko R., Slotte J.P. 2005. On the importance of the phosphocholine methyl groups for sphingomyelin/cholesterol interactions in membranes: a study with ceramide phosphoethanolamine.Biophys. J. 88:2661–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.E. 2008. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids.J. Lipid Res. 49:1377–1387 [DOI] [PubMed] [Google Scholar]
- Wang X., Rao R.P., Kosakowska-Cholody T., Masood M.A., Southon E., Zhang H., Berthet C., Nagashim K., Veenstra T.K., Tessarollo L., et al. 2009. Mitochondrial degeneration and not apoptosis is the primary cause of embryonic lethality in ceramide transfer protein mutant mice.J. Cell Biol. 184:143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S.N., Lupashin V.V. 2005. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells.J. Cell Biol. 168:747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]