Abstract
Genomic instability is a hallmark of human cancers. Spindle assembly checkpoint (SAC) is a critical cellular mechanism that prevents chromosome missegregation and therefore aneuploidy by blocking premature separation of sister chromatids. Thus, SAC, much like the DNA damage checkpoint, is essential for genome stability. In this study, we report the generation and analysis of mice carrying a Cdc20 allele in which three residues critical for the interaction with Mad2 were mutated to alanine. The mutant Cdc20 protein (AAA-Cdc20) is no longer inhibited by Mad2 in response to SAC activation, leading to the dysfunction of SAC and aneuploidy. The dysfunction could not be rescued by the additional expression of another Cdc20 inhibitor, BubR1. Furthermore, we found that Cdc20AAA/AAA mice died at late gestation, but Cdc20+/AAA mice were viable. Importantly, Cdc20+/AAA mice developed spontaneous tumors at highly accelerated rates, indicating that the SAC-mediated inhibition of Cdc20 is an important tumor-suppressing mechanism.
Introduction
Genomic instability is a hallmark of human cancers, a prominent form of which is chromosomal instability (CIN). CIN is most likely caused by errors in mitoses during which the duplicated genome is distributed into two daughter cells. Mitosis is composed of several phases: prophase (chromosomes start condensing), prometaphase (chromosomes condensed, removal of the bulk of sister cohesins, and establishment of bipolar spindles), metaphase (sister chromatids aligned in metaphase plate), anaphase/telophase (separation and pulling of sister chromatids), mitotic exit (loss of Cdk1 kinase activity and relaxation of the condensed chromosomes), and cytokinesis (end of mitosis and the formation of two new daughter cells). Genetic studies in yeasts have identified several important mitotic regulators. Key among them is the anaphase-promoting complex/cyclosome (APC/C), a multisubunit E3 ubiquitin ligase (Morgan, 1999; Page and Hieter, 1999). APC/C mediates ubiquitination of protein substrates including cyclin B1 and securin to drive the progression of mitosis. It recognizes its substrates through two adapter proteins, Cdc20 and Cdh1, which contain similar C-terminal substrate-interacting domains composed of seven WD-40 repeats (Hendrickson et al., 2001; Pfleger et al., 2001; Schwab et al., 2001; Harper et al., 2002; Kraft et al., 2005; Diaz-Martinez and Yu, 2007). Destruction boxes or KEN boxes are motifs frequently found in APC/C's substrates, but other motifs are also possible for the recognition (Harper et al., 2002).
Before anaphase, sister chromatids are held together by cohesin complexes that resist the pulling force generated by the microtubule spindle. It is the dissolution of sister cohesion that allows anaphase to happen. The cohesin complexes are composed of protein subunits encoded by Smc1, Smc3, Scc1/Mcd1, and Scc3 and are thought to form a ring structure that encloses sister chromosomes (Nasmyth, 2005). At the onset of anaphase, the Scc1 subunit of the cohesin complex is cleaved by separase, a CD clan protease of the caspase family (Uhlmann et al., 2000), leading to the opening of the ring and release of sister chromatids. The timing of anaphase is controlled by spindle assembly checkpoint (SAC), an elaborate biochemical mechanism that ensures that sister chromatids are held together by cohesion rings until all of the chromosomes have achieved bivalent spindle attachments. By doing that, SAC prevents chromosome missegregation and aneuploidy. Dysfunctional SAC likely underlies the CIN phenotype observed in cancer cells.
SAC is activated when the kinetochores are not occupied by microtubules or when there is no tension at the kinetochores (Lew and Burke, 2003; Pinsky and Biggins, 2005). A single lagging chromosome is sufficient to activate SAC and cause an arrest in metaphase (Rieder et al., 1995). The same arrest is induced upon treating cells with spindle microtubule-disrupting agents such as nocodazole or colcemid. SAC activation (Diaz-Martinez and Yu, 2007) results in the inhibition of APC–Cdc20 by Mad2 and BubR1, and thus, the stabilization of securin and cyclin B1. Securin is an inhibitor of separase, and cyclin B1–Cdk1 kinase can phosphorylate separase (Stemmann et al., 2001). Phosphorylation of separase opens up the site for the binding and inhibition by the Cdk1–cyclin B1 complex (Gorr et al., 2005; Boos et al., 2008). Therefore, separase is dually inhibited by securin and phosphorylation when the checkpoint is activated, preventing premature separation of sister chromatids. These two inhibitory mechanisms are redundant in somatic cell lineages (Mei et al., 2001; Huang et al., 2005, 2008), but the phosphorylation is uniquely required in mouse embryonic germ cells (Huang et al., 2008). Furthermore, the stabilization of cyclin B1 prevents other events necessary for mitotic exit, leading to cell cycle arrest at prometaphase (Morgan, 1999).
Genetic analyses in budding yeasts have clearly demonstrated that the SAC is essential in preventing CIN (Li and Murray, 1991; Yamamoto et al., 1996). The discovery of mutations in BUBR1 and BUB1 in a subset of colon cancer cell lines (Cahill et al., 1998) suggests a weakened spindle checkpoint as the cause of CIN that contributed to the oncogenic process, which was further substantiated by the finding that BUBR1 is mutated in mosaic variegated aneuploidy, a rare human disorder characterized by increased percentage of aneuploid cells (usually >25%) and predisposition to childhood cancers (Hanks et al., 2004). To determine the role of spindle checkpoint in tumorigenesis, a large amount of efforts have gone to the generation and analysis of mice with targeted deletions in various SAC components. Because the spindle checkpoint is essential in mice, the analyses were restricted to heterozygous mice or mice carrying hypomorphic alleles. However, despite the compromises in the checkpoint, these mice did not display the expected large increases in the rate of spontaneous tumor development (Michel et al., 2001; Babu et al., 2003; Baker et al., 2004; Dai et al., 2004; Iwanaga et al., 2007). Furthermore, these spindle checkpoint components may have functions outside the checkpoint, and it might be the noncheckpoint-related functions that contribute to tumorigenesis when disrupted. Thus, whether the SAC is an as important tumor-suppressing mechanism as the DNA damage checkpoint is in question.
In this study, we report the generation and analysis of mice carrying a mutant Cdc20 allele (Cdc20AAA) in which the Mad2-binding sites in Cdc20 were mutated. The mutant Cdc20 protein can no longer be inhibited by Mad2 and renders the spindle checkpoint dysfunctional. These mice contain a high percentage of aneuploid cells and develop spontaneous neoplasms at a much increased rate, indicating that the SAC-mediated inhibition of Cdc20 is an important tumor-suppressing mechanism. Furthermore, analysis of the mutant cells revealed that the timing of anaphase depended on Mad2–Cdc20 interaction, and additional expression of BubR1 could not rescue the spindle checkpoint defects caused by AAA-Cdc20.
Results
Genetic disruption of Mad2-mediated inhibition of Cdc20
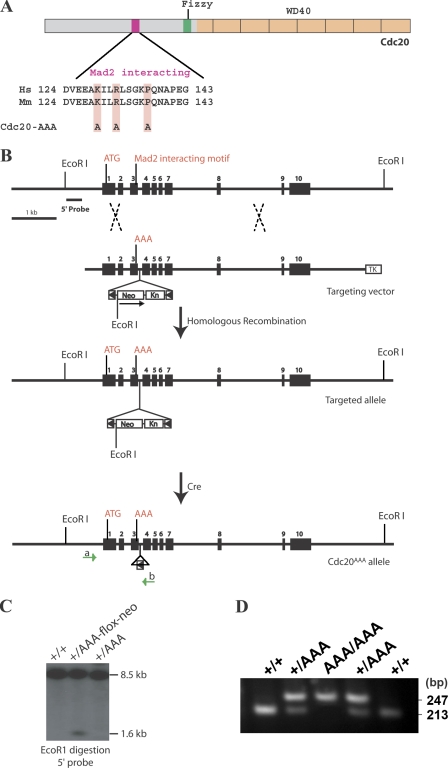
The motif for Mad2 binding on Cdc20 (Fig. 1 A) has been identified and is homologous to the one used by other Mad2-interacting proteins such as Mad1 (Zhang and Lees, 2001; Luo et al., 2002). We substituted the two charged residues (K129 and R132) and the proline (P137) within the motif with Ala to generate the Cdc20AAA allele in mice via homologous recombination in mouse embryonic stem (ES) cells. The strategy is shown in Fig. 1 B. The targeting construct brought in the mutations along with a floxed neo cassette. The targeted allele, Cdc20AAA-flox-neo, is likely a null allele caused by the presence of the neo cassette (in fact, a truncated protein is expressed from the targeted allele; unpublished data). The point mutant only becomes active upon Cre-mediated loop out of the neo cassette. Thus, the mutation is conditional. We transiently expressed Cre in one of the targeted clones to activate the mutant allele. Sequencing of the Cdc20 cDNA derived from the resulting Cdc20+/AAA cells demonstrated the expression of the mutant allele.
Figure 1.
Generation of Cdc20AAA allele. (A) A diagram depicting the domain structure of Cdc20 protein. (B) Schematic presentation of the strategy for the generation of Cdc20AAA. Dashed X's indicate crossing over in homologous recombination. Green arrows indicate PCR genotyping primers. Kn, kanamycin-resistant gene; TK, thymidine kinase. (C) Southern blot analysis of the mutant alleles. (D) PCR genotyping of Cdc20AAA using primers a and b.
We injected two Cdc20+/AAA-flox-neo ES clones into mouse blastocysts for the production of chimeric mice. Both clones gave germline transmission of the AAA-flox-neo allele. Cdc20+/AAA-flox-neo mice were crossed to a Cre deleter strain to generate Cdc20+/AAA mice. Fig. 1 C shows a Southern blot analysis of AAA-flox-neo and AAA alleles, and Fig. 1 D shows PCR genotyping of a litter derived from the Cdc20+/AAA to Cdc20+/AAA cross. Cdc20+/AAA mice were healthy and fertile. They did not show obvious signs of premature aging. However, intercrossing of Cdc20+/AAA mice failed to produce Cdc20AAA/AAA mice (Table I). Examination of embryos showed that Cdc20AAA/AAA mice could survive up to embryonic day (E) 12.5. The mutant embryos were smaller than wild-type controls from early stages on (Fig. S1 A). Analysis of apoptosis indicated that the mutants suffered massive apoptotic cell death (Fig. S1 B), which likely caused the lethality.
Table I.
Intercrosses of Cdc20+/AAA mice
| Age | Cdc20+/+ | Cdc20+/AAA | Cdc20AAA/AAA |
| Neonate | 73 (40.3%) | 108 (59.7%) | 0 (0%) |
| E14.5–18.5 | 12 (40.0%) | 18 (60.0%) | 0 (0%) |
| E10.5–12.5 | 13 (19.7%) | 38 (57.6%) | 15 (22.7%) |
| E9.5 | 8 (19.0%) | 22 (52.4%) | 12 (28.6%) |
| E8.5 | 3 (21.4%) | 7 (50.0%) | 4 (28.6%) |
Numbers on the left indicate number of animals/embryos.
The point mutation in Cdc20 causes functional loss of SAC
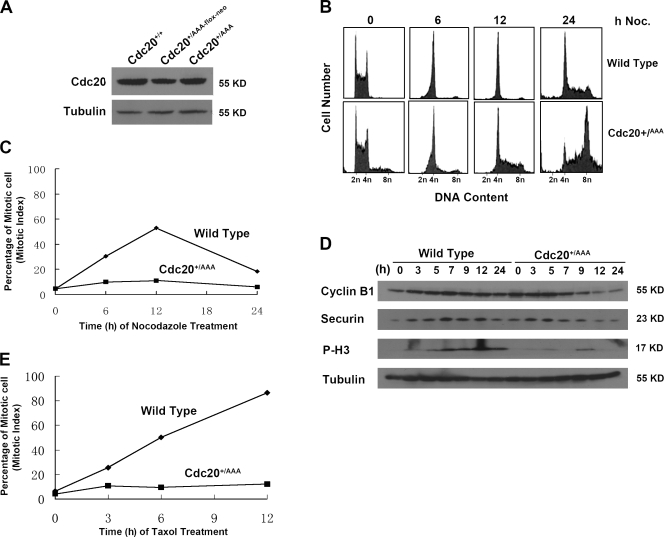
To determine the effect of the mutation we introduced into Cdc20 on mitosis at cellular level, we first isolated two independent Cdc20+/AAA clones through transient Cre expression in Cdc20+/AAA-flox-neo ES cells. Western blot analysis indicated that the mutation did not interfere with the expression of Cdc20 protein (Fig. 2 A), although the level in Cdc20+/AAA-flox-neo cells showed a slight decrease, most likely because the AAA-flox-neo allele is a null allele. Wild-type and Cdc20+/AAA ES cells were treated with 50 ng/ml nocodazole and harvested at different time points for analysis. We found that the mutant cells could not arrest in response to spindle microtubule disruption. By 12 h, although the majority of the control cells were found in G2/M with 4 N DNA content, Cdc20+/AAA cells continued cell cycle and increased their DNA content to >4 N (Fig. 2 B). By 24 h, most of the mutant cells had reached an 8-N DNA content. At this point, some of the wild-type cells also adapted to the spindle checkpoint and entered cell cycle. The continued cycling by the mutant cells (and the wild-type cells at a later time point) was likely a result of the lack of p53 function in ES cells (Hong and Stambrook, 2004). When the number of mitotic cells was counted, we found that the mutant cells did not accumulate in mitosis, whereas the control cells did (Fig. 2 C). Moreover, when the levels of cyclin B1 and securin were analyzed, we found that the mutant cells could not maintain the levels of these two mitotic regulators as the wild-type cells in response to the disruption of microtubules (Fig. 2 D). However, these two proteins did accumulate to some extent before being degraded, suggesting that there is either residual spindle checkpoint function in the mutant cells, that the Chfr-mediated prophase checkpoint was at work (Scolnick and Halazonetis, 2000), or both. In agreement with the observation that nocodazole treatment did not cause an increase in mitotic indices in the mutant cells, there was no accumulation of phosphorylated histone H3 in the mutants as in wild-type cells (Fig. 2 D). These data demonstrate that the mutation we had generated in Cdc20 disrupts SAC and is dominant.
Figure 2.
Cdc20AAA disrupts SAC. (A) Western blot analysis of Cdc20 expression. (B) FACS profiles of the cells treated with 80 nM nocodazole (noc). (C) Mitotic indices of the cells in A. (D) Western blot analysis of the cells treated with nocodazole for various lengths of time. P-H3, phosphorylated histone 3. (E) Mitotic indices of the cells treated with 100 ng/ml taxol.
It has been suggested that Mad2 and BubR1 are responsive to different kinetochore abnormalities; e.g., unoccupied kinetochores were suggested to activate Mad2 and lack of tension was proposed to activate BubR1 (Zhou et al., 2002a,b; Logarinho et al., 2004). With our AAA-Cdc20 cells, we tested this possibility experimentally. Cdc20+/AAA and wild-type ES cells were treated with taxol, a microtubule stabilizer that activates SAC by preventing the dynamics of microtubules and therefore the generation of tension. Taxol caused marked increases in mitotic indices over time in the control cells but little increases in the mutant cells (Fig. 2 E), indicating that Mad2-mediated inhibition of Cdc20 is also required for the execution of SAC in response to lack of tension across kinetochores.
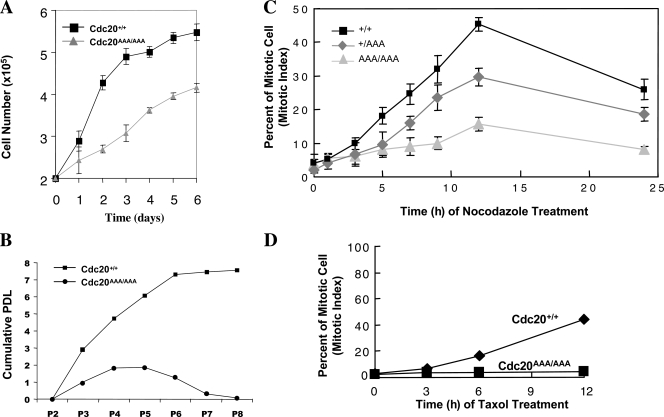
We derived mouse embryonic fibroblasts (MEFs) from E12.5 embryos. Cdc20AAA/AAA MEFs grew noticeably slower than the wild-type control even at early passages (<3; Fig. 3 A), and the growth succumbed to a complete stop as a result of massive cell death as passage number increased (Fig. 3 B). However, the culture could be rescued with immortalization by the expression of SV40 large T antigen, suggesting that the cell death was mediated by the p53, Rb, or both pathways. When immortalized MEFs (iMEFs) were treated with nocodazole, wild-type cells accumulated in mitosis, but Cdc20AAA/AAA cells failed to do so (Fig. 3 C), indicating that the mutation disrupts SAC in MEFs as well. Similar to the ES cells, Cdc20AAA/AAA iMEFs did not respond to taxol treatment (Fig. 3 D). Cdc20+/AAA iMEFs displayed a milder defect in the checkpoint function than the homozygous mutant cells (Fig. 3 C).
Figure 3.
Analysis of Cdc20AAA/AAA MEFs. (A) Growth curve analysis. (B) Population doubling (PDL) analysis under the 3T9 protocol. (C) Mitotic indices of the iMEFs treated with 100 nM nocodazole. (D) Mitotic indices of the iMEFs treated with 100 ng/ml taxol. Error bars indicate SD.
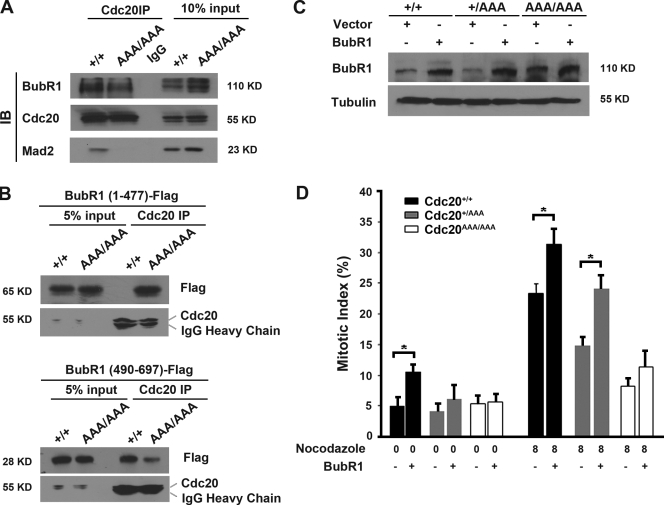
To demonstrate that the mutation abolishes the interaction between Cdc20 and Mad2, we treated the iMEFs with nocodazole (to activate the spindle checkpoint) and MG132 (to prevent the mutant cells from leaving mitosis). Cdc20 was immunoprecipitated, and Western blotting was performed to determine whether Mad2 was in the immune complexes. As shown in Fig. 4 A, although Mad2 was clearly present in the immunoprecipitation from wild-type cell lysates, it was absent from the mutant, demonstrating that the mutation we generated indeed disrupted the interaction between Cdc20 and Mad2. However, BubR1 could still interact with the mutant Cdc20, albeit less efficiently, suggesting that the interaction between BubR1 and Cdc20 does not strictly depend on Mad2–Cdc20 interaction.
Figure 4.
The interaction between BubR1 and AAA-Cdc20. (A) Coimmunoprecipitation analysis of the interaction between Mad2 and Cdc20. Cdc20 was immunoprecipitated from nocodazole- and MG132-treated (8 h) iMEF cells, and the immune complexes were analyzed with Western blotting. Note that Cdc20 was immunoprecipitated with mouse monoclonal antibodies and detected with rabbit polyclonal antibodies. IB, immunoblotting. (B) The two Cdc20-binding domains of BubR1 interact with AAA-Cdc20 differentially. Flag-tagged BubR1 (1–477) or BubR1 (490–679) was transiently expressed in iMEFs from which Cdc20 was immunoprecipitated. The immune complexes were analyzed with Western blotting. Note that the same mouse monoclonal anti-Cdc20 antibodies were used for immunoprecipitation (IP) and Western blotting. (C) Exogenous expression of BubR1 in iMEFs. (D) Mitotic index analysis of iMEFs transfected with a BubR1-expressing plasmid. The asterisk indicates statistical significance (P < 0.05). Error bars indicate SD.
A recent study demonstrated the presence of two Cdc20-interacting motifs in BubR1 (Davenport et al., 2006). One motif is located at the N terminus (aa 1–477) and the other in the middle (aa 490–679) of BubR1. It was suggested that the N-terminal motif bound Cdc20 in a Mad2-dependent manner, whereas the C-terminal motif bound in a Mad2-independent fashion. With the availability of cells expressing mutant Cdc20 that could not interact with Mad2, we tested the interaction between Cdc20 and BubR1 further. The two Cdc20-interacting fragments (Flag tagged at the C termini) of BubR1 were transiently expressed in iMEFs and immunoprecipitated with anti-Flag antibodies. Coprecipitated Cdc20 protein was detected with Western blotting. As shown in Fig. 4 B (1–477), BubR1 could not interact with AAA-Cdc20, whereas 490–679 BubR1 could, supporting the notion that the binding of the N terminus of BubR1 to Cdc20 requires Mad2–Cdc20 interaction. However, the mutant Cdc20 bound 490–679 BubR1 much less efficiently than wild-type Cdc20 (Fig. 4 B), suggesting that Mad2–Cdc20 interaction also plays a role in promoting the binding of BubR1's C-terminal part to Cdc20.
Next, we tested whether overexpression of BubR1 could compensate for the lost interaction between Mad2 and Cdc20 in the mutant cells. Empty vector or BubR1-expressing vector together with a GFP-expressing plasmid were transfected into iMEFs. Mitotic indices of the transfected (GFP positive) cells were determined before and after 12-h nocodazole treatment. Although additional expression of BubR1 (Fig. 4 C) significantly increased the mitotic indices of wild-type cells even in the asynchronously growing population, it had little impact on the homozygous mutant cells either treated with nocodazole or untreated (Fig. 4 D), suggesting that the inhibition of Cdc20 by BubR1 requires the function of Mad2. In fact, the endogenous level of BubR1 was already noticeably elevated in Cdc20AAA/AAA cells (Fig. 4, A and C), but the SAC was still ineffective. It is unclear at the moment what causes the elevation in BubR1 levels in AAA-Cdc20 homozygous cells. In the heterozygous cells, additional BubR1 expression was able to improve the checkpoint function to some extent (Fig. 4 D), most likely working through the wild-type Cdc20 protein.
Abnormal mitoses in MEFs harboring Cdc20AAA
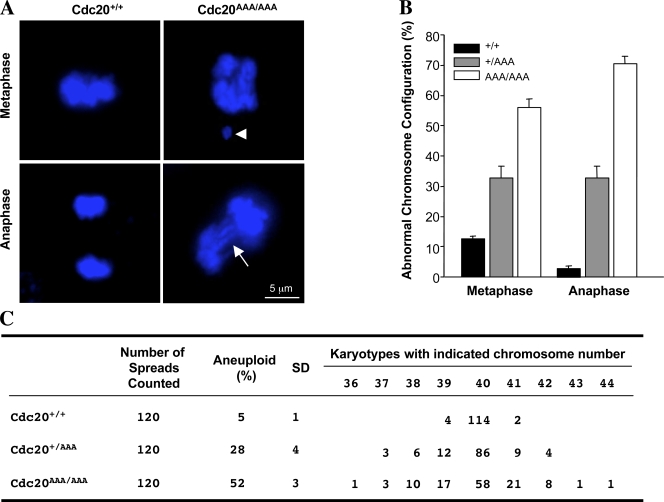
During normal mitotic divisions, SAC is essential in delaying anaphase until all sister chromatids are aligned at metaphase plate. We analyzed mitoses in asynchronously growing MEFs undisturbed by microtubule-disrupting agents. Microscopic observation indicated that the mitoses were highly abnormal in the mutants (Fig. 5, A and B). In metaphase (when a metaphase plate was clearly visible), the chromosomes in mutant cells were misaligned (Fig. 5 A), and in anaphase, there were lagging chromosomes or chromosome bridges in the mutant cells (Fig. 5 A). Quantitation of the abnormal mitoses are shown in Fig. 5 B. The abnormal mitoses in the mutant MEFs suggested that these cells should become aneuploid. Indeed, as early as passage 3, both heterozygous and homozygous mutant MEFs showed high percentages of cells with abnormal karyotypes, reaching 28% and 52%, respectively, whereas the wild-type cells only showed 5% aneuploidy (Fig. 5 C).
Figure 5.
Aberrant mitoses and aneuploidy in the mutant MEFs. (A) Representative images of mitotic cells. DNA was visualized with DAPI staining. The arrowhead indicates a DNA mass drifted far away from the bulk, and the arrow indicates anaphase bridges. (B) Quantitation of abnormal mitoses. (C) Karyotype analysis is shown. Error bars indicate SD.
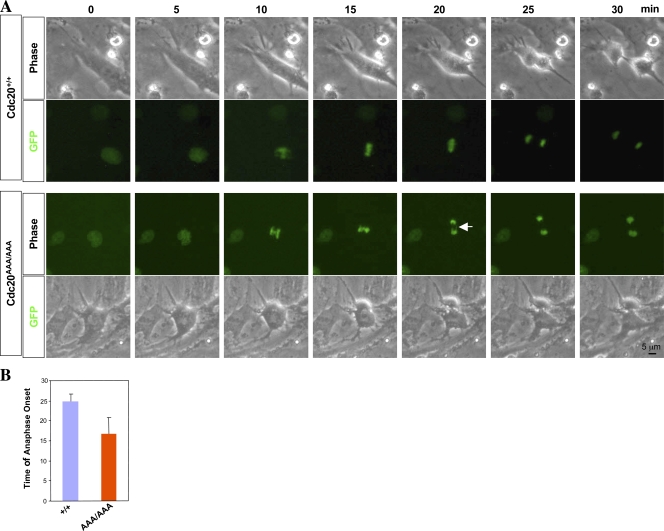
The abnormal mitoses in the mutant cells also suggested that anaphases began prematurely in the mutant. To demonstrate that, we marked the chromosomes of the cells with H2B-GFP carried on a retroviral vector. Unperturbed mitoses starting from nuclear envelop breakdown were recorded with a live cell imaging system. As shown in Fig. 6 A, the mutant cell entered anaphase much quicker than the wild type. 30 mitoses were analyzed, and the result is shown in Fig. 6 B. On average, the mutant entered anaphase ∼8–10 min earlier than the control (Fig. 6 B), indicating that Mad2-mediated inhibition of Cdc20 is critical in the timing of anaphase. In agreement with the advancement of anaphase in mutant cells, Cdc20AAA/AAA MEFs contained reduced levels of securin and cyclin B1 (Fig. S2).
Figure 6.
Advanced timing of anaphase in mutant MEFs. (A) Clips from time-lapse video recordings. The arrow indicates lagging chromosomes. (B) Quantitation of the timing of anaphase onset given in minutes. Error bars indicate SD.
Chromosome instability and spontaneous tumorigenesis in Cdc20+/AAA mice
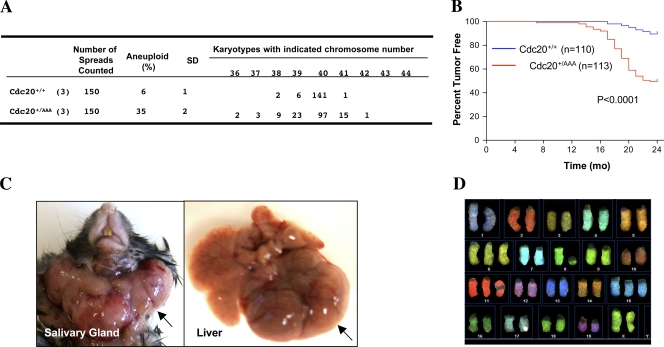
To determine whether the increased aneuploidy in the mutant MEFs was also true in adult mice, we karyotyped splenocytes isolated from 5-mo-old wild-type and Cdc20+/AAA mice (three animals per genotype). The isolated cells were cultured for 48 h and enriched for mitotic fraction with nocodazole and MG132 treatment and processed for chromosome spreading. As shown in Fig. 7 A, Cdc20+/AAA splenocytes contained ∼35% aneuploid cells, whereas the wild type contained only 6%. To determine whether such a high percentage of aneuploidy has any impact on spontaneous tumorigenesis, we subjected cohorts of wild-type and Cdc20+/AAA mice to long-term (24 mo) observation of tumor development. These mice were under a mixed genetic background of C57BL/6 and 129SV. As shown in Fig. 7 B, Cdc20+/AAA mice had a significantly increased rate of tumor formation. Palpable tumors could be detected as early as 6.8 mo in the mutant mice. By 24 mo, 50% of the mutant mice developed tumors, whereas only 10% of the wild-type animals did. 38% of the mice that had tumors developed tumors at multiple organ sites. Fig. 7 C shows a couple of large images of the tumors. Logrank test indicated that the tumor-free curves of wild-type and mutant mice were significantly different with P < 0.0001. Among 29 tumor samples analyzed histopathologically, four were hepatomas (13.8%) and the rest were lymphomas. Spectrum karyotyping (SKY) analysis of one lymphoma sample demonstrated aneuploidy (2 N + 3) in this tumor (Fig. 7 D). The tumors developed in wild-type animals were all lymphomas.
Figure 7.
Spontaneous tumorigenesis in Cdc20+/AAA mice. (A) Karyotype analysis of the splenocytes. (B) Tumor-free survival analysis of wild-type and Cdc20+/AAA mice. (C) Large images of tumors. (D) SKY analysis of one lymphoma.
Discussion
Cancer is thought to be caused by multistep genetic changes that had started in a single cell. Epidemiological and genetic analyses have led to an estimation of the number of mutations needed for malignant transformation to be about six (Armitage and Doll, 1954, 2004; Kinzler and Vogelstein, 1996). Given the mutation rate in humans, it is extremely rare for a single cell to acquire all the necessary mutations. Thus, it has been postulated that destabilizing the genome is necessary for cancer development. A prominent form of genetic instability in cancer is chromosomal. CIN can be gains or losses of whole chromosomes or translocation of chromosome segments. Alterations in chromosome number or aneuploidy are found in nearly all major human tumor types (Mertens et al., 1994). Examples include the loss of chromosome 10 in glioblastomas, often reflecting the inactivation of the tumor suppressor gene PTEN (phosphatase and tensin homologue; Wang et al., 1997) and the gain of chromosome 7 in papillary renal carcinomas, reflecting a duplication of a mutant Met oncogene (Zhuang et al., 1998). However, most of the time, no specific molecular advantages can be associated with a particular gain or loss of a chromosome. In fact, cancer cells display gross abnormalities in their chromosome numbers, most likely as a result of defects in the quality control of sister chromatid separation, including defects in the SAC (for reviews see Chi and Jeang, 2007; Ganem et al., 2007; Weaver and Cleveland, 2007; Storchova and Kuffer, 2008).
A complete lack of SAC is incompatible with viability in higher eukaryotes. Mad2-deficient mice show early embryonic lethality around the blastocyst stage (Dobles et al., 2000). Loss of Mad2 in Caenorhabditis elegans is also incompatible with the viability (Kitagawa and Rose, 1999). Other central components of the SAC, Bub1, BubR1, and Mad1, are all essential as well (Basu et al., 1999; Kitagawa and Rose, 1999; Babu et al., 2003; Wang et al., 2004). Although Mad2-deficient cells are nonviable (Michel et al., 2001), the nonviability could be rescued with the deletion of p53 (Burds et al., 2005). Thus, a complete lack of SAC function does not cause cell lethality by itself. Rather, the cells are eliminated by other protecting mechanisms such as p53-induced apoptosis, perhaps because of the severe aneuploid nature of these cells. However, the SAC is not “all or none.” It can be compromised to certain degrees as indicated by the fact that the mice heterozygous for the essential components are viable despite clear defects in the checkpoint; e.g., not only do the cells derived from these mice arrest less efficiently than wild-type cells in response to microtubule disruption, there were also certain percentages of aneuploid cells present in the adult animals. The percentage of the aneuploid cells could be tolerated as high as >30% (BubR1 hypomorphic mice and our AAA-Cdc20 heterozygous mice). It is unclear at present whether such a high percentage of aneuploidy has any impact on normal physiology of the animals.
Despite the aneuploidy displayed by SAC mutants, only small increases in cancer susceptibility have been reported. For example, tumor incidence was increased (to 6%) in mice with severely reduced BubR1 levels (Baker et al., 2004). Also, 28% of mice heterozygous for Mad2 develop small, self-limiting, late onset (18–19 mo) papillary lung adenocarcinomas (Michel et al., 2001), and mice heterozygous for functional BubR1 or Bub3 are more prone to the development of colorectal (Dai et al., 2004) or lung (Babu et al., 2003; Dai et al., 2004) tumors after treatment with azoxymethane or DMBA (9,10-dimethyl-1,2-benzanthracene), respectively. More recently, mice heterozygous for Mad1 were reported to display a small increase (from 9 to 19%) in the incidence of spontaneous tumors at old ages (Iwanaga et al., 2007), and mice heterozygous for centromere protein E similarly displayed a small increase in tumor incidence (Weaver et al., 2007). These minor tumor phenotypes put the significance of the SAC in preventing the tumorigenesis in question. However, results from Cdc20+/AAA (this study) and Bub1-deficient mice (Jeganathan et al., 2007) suggest that the loss of SAC function can be highly tumorigenic. What could be the reason for the differences in the rates of tumor development among various mouse strains with SAC defects? One obvious possibility is the severity of the checkpoint defects and thus the rates of aneuploidy. Indeed, both Cdc20+/AAA and Bub1 hypomorphic mice contain high percentages of aneuploid cells (>30% when splenocytes were analyzed). However, mice deficient in BubR1 or doubly heterozygous for Bub3 and Rae1 contain a similarly high percentage of aneuploid cells, and yet, these mice are not tumor prone (Baker et al., 2004, 2006). One might argue that the premature aging process in BubR1-deficient and Bub3+/− Rae1+/− mice prevented these animals from developing tumors. However, such an argument could not be applied to centromere protein E+/− mice, which also display similarly high percentages of aneuploid cells as Cdc20+/AAA mice but only have a mild increase in tumor incidence (Weaver et al., 2007). Thus, the rate of aneuploidy cannot be the only determinant on the likelihood of tumorigenesis in SAC mutants. Another possibility is that Cdc20 and Bub1 may have functions outside mitosis. Loss (in the case of Bub1) or gain (in the case of AAA-Cdc20) of these functions in combination with aneuploidy may therefore drive the robust tumor development in these mice and potentially explain the difference in the tumor spectra between these two strains. Indeed, Cdc20 was suggested to function in regulating gene expression in mammalian cells (Yoon et al., 2004), and budding yeast CDC20 was found to be able to override G2/M DNA damage checkpoint when overexpressed (Lim and Surana, 1996) and needs to be repressed in S phase to prevent premature mitotic entry (Clarke et al., 2003). Given the fact that AAA-Cdc20 is no longer inhibited by Mad2, this mutant Cdc20 protein might have gained additional functions that help oncogenic transformation. In the case of Bub1, it seems to be required for the induction of cell death when chromosome missegregates (Jeganathan et al., 2007) and may have other unspecified functions that prevent tumorigenesis.
Both Mad2 and BubR1 can bind to Cdc20 directly. The Mad2-binding sites in Cdc20 are conserved in other Mad2-interacting proteins such as Mad1 (Zhang and Lees, 2001; Luo et al., 2002). However, the BubR1-interacting domain in Cdc20 remains ill defined (Luo et al., 2002). Recent studies in budding yeasts suggested that MAD3 (BubR1 in mammals) uses its KEN box to interact with CDC20 and inhibits APCCdc20 as a pseudosubstrate (Burton and Solomon, 2007), and it seems that a similar mechanism is used by BubR1 in mammals (Malureanu et al., 2009). One possibility for why both Mad2 and BubR1 are essential SAC components is stoichiometry. In other words, the number of Mad2 or BubR1 molecules is perhaps by themselves insufficient to inhibit all APCCdc20 complexes. However, our results (Fig. 4 E) indicate that this is unlikely the case. It is more likely that the mitotic checkpoint complex (MCC) containing Mad2, BubR1, Bub3, and Cdc20 blocks the E3 activity of APCCdc20 (Sudakin et al., 2001) instead of Mad2 and BubR1 acting separately. Indeed, MCC was much more potent in inhibiting APCCdc20 than Mad2 alone or BubR1 in complex with Bub3 (Sudakin et al., 2001), suggesting a synergy between Mad2 and BubR1 in enforcing the checkpoint. Recent biochemical and genetic complementation analyses suggest that MCC might be transient (Kulukian et al., 2009; Malureanu et al., 2009). The interaction between Mad2 and Cdc20 is to make Cdc20 susceptible to BubR1 binding and inhibition. In other words, activated Mad2 acts as a catalyst for the formation of BubR1–Bub3–Cdc20 or BubR1–Bub3–APCCdc20 complexes. Therefore, Mad2 deletion, BubR1 deletion, or loss of Mad2–Cdc20 interaction all lead to active APCCdc20 and thus no SAC. However, Cdc20AAA is certainly less disruptive than Mad2 or BubR1 deletion because Cdc20AAA/AAA mice could survive up to midgestation, whereas Mad2- and BubR1-deficient mice died much earlier. This discrepancy is likely caused by the incomplete disruption of the Mad2–Cdc20 interaction by the mutation. There might still be some residual interaction between Mad2 and the mutant Cdc20 that could not be detected with coimmunoprecipitation.
Materials and methods
Generation of Cdc20 mutant mice
We isolated Cdc20 genomic clones using the method based on homologous recombination (Zhang et al., 2002). A knockin construct was made (Fig. 1 B) and introduced to mouse ES cells (AB2.2). Recombinant ES clones were identified through Southern blot analysis. Two such clones were injected into blastocysts derived from C57BL/6 mice to produce chimeras. Subsequent breeding of the chimeric mice resulted in germline transmission of the knockin allele (AAA-flox-neo). To activate the AAA mutation, the mice were crossed with Meox2+/Cre to delete the neo cassette. A PCR-based protocol was developed to genotype Cdc20AAA allele with the following primer pair: forward, 5′-AGCCTGGTCTCTCAACCTGA-3′; reverse, 5′-GGTAGCCTGTGGCAAGAGAG-3′.
Isolation and analysis of MEFs
MEFs were isolated from 12.5-d-postcoitum embryos and cultured in DME supplemented with 2 mM glutamine, 1% penicillin/streptomycin, and 15% FBS. Immortalization of MEF cells were realized via retroviral expression of SV40 large T antigen. For live image analysis, MEF cells (passage 2) were infected with retroviruses carrying H2B-YFP (provided by J. van Deursen, Mayo Clinic, Rochester, MN) and were recovered for 24 h after the infection. The cell culture dish containing the infected cells was placed on the temperature-controlled warm stage of a microscope (Axiovert 200; Carl Zeiss, Inc.) equipped with an environmental chamber. The temperature was maintained at 37°C and the CO2 level at 10% with the CTI3700 controller (Carl Zeiss, Inc.). The cells were imaged every 5 min for 4 h. Imaging software (WS/20A; Carl Zeiss, Inc.) was used to analyze the progression of mitosis. At least 30 mitotic cells were analyzed. To express BubR1, iMEFs were transfected with pCDNA3-BubR1 together with a GFP-expressing plasmid at a 4:1 ratio. 24 h after the transfection, the cells were treated with nocodazole for 8 h, fixed with 4% PFA, and stained with DAPI. The number of mitotic cells was counted in GFP-positive populations.
For Western blotting, the cells were harvested and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail [Roche]). 50 µg total protein was separated in SDS-PAGE and immunoblotted with antibodies against cyclin B1 (Santa Cruz Biotechnology, Inc.), securin (NeoMarkers), or Cdc20 (Santa Cruz Biotechnology, Inc. and Millipore).
For coimmunoprecipitation, the iMEFs were lysed with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40), and the lysates were incubated with anti-Cdc20 antibody (Millipore) or anti-Flag antibodies (Sigma-Aldrich) for 90 min at 4°C. The immune complexes were precipitated with protein G agarose beads (GE Healthcare), washed four times with NETN buffer, and eluted with 2× SDS loading buffer. The eluted proteins were separated by SDS-PAGE and probed with antibodies against Cdc20, BurbR1, and Mad2 (Santa Cruz Biotechnology, Inc.).
Karyotype analysis
To isolate splenocytes, spleens were collected and minced between two microscope slides. The released the cells were cultured for 48 h in RPMI 1640 supplemented with 10% FBS, 5 µg/ml lipopolysaccharide, 1 µg/ml anti–mouse CD28, and 1 µg/ml anti–mouse CD3e (BD).
To prepare chromosome spreads, MEFs (at passage 3) and splenocytes were treated with 10 µM MG132 (Calchemico) for 5 h at 37°C, harvested and resuspended in 5 ml 0.075 M KCl, and incubated in the hypotonic solution at 37°C for 10 min. The cells were fixed in Carnoy's solution (methanol/acetic acid [3:1]), washed with PBS, and resuspended in 0.5 ml Carnoy's solution. The cell suspension was dropped onto prewetted microscope slides and air dried. Chromosomes were visualized by 10-min staining in 5% Giemsa solution.
For SKY, lymphoma cells were isolated and cultured. Chromosome spreads were prepared from asynchronously growing population of cells and stained with chromosome-specific probes in our cytogenetic core laboratory.
Histology analysis
Embryos or tumor tissues were fixed overnight in 4% PFA/PBS, pH 7.4, and embedded in paraffin. 4-mm sections were prepared and stained with hematoxylin and eosin according to standard protocols. For immunostaining of activated caspase 3, tissue sections were boiled for 10 min in citrate buffer (10 mM sodium citrate and 0.05% Tween 20, pH 6.0) in a microwave oven to retrieve antigens and were stained with antiactive caspase 3 antibodies (Cell Signaling Technology).
Imaging
For regular fluorescence imaging, we used a microscope (E800; Nikon) with a Plan Fluor 100×/1.30 oil objective (Nikon). Images were captured with a digital camera (SPOT-RT model 2.3.1; Diagnostic Instruments, Inc.). Fluochromes used are DAPI, Texas red, and FITC. For live imaging of YFP-labeled cells, we used a microscope (Axiovert 200; Carl Zeiss, Inc.) with a Plan Fluor 20×/0.30 objective (Carl Zeiss, Inc.). Images were taken with a digital camera (AxioCam; Carl Zeiss, Inc.).
Online supplemental material
Fig. S1 shows the embryonic phenotype of Cdc20AAA/AAA mice, and Fig. S2 shows immunostaining of securin and cyclin B1 in the mutant MEF cells in mitosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904020/DC1.
Acknowledgments
We thank the transgenic core laboratory of Baylor College of Medicine for the production of Cdc20 mutant mice, Dr. A. Heron (Baylor College of Medicine, Houston, TX) for pathological analysis of tumors, and the Cytogenetic Core Laboratory of Texas Children's Hospital for SKY analysis. We thank Dr. J. van Deursen for providing us reagents.
M. Li is supported by a postdoctoral training grant from the National Institutes of Health. This work was funded by research grants from the National Cancer Institute (CA122623 and CA116097 to P. Zhang).
Footnotes
Abbreviations used in this paper: APC/C, anaphase-promoting complex/cyclosome; CIN, chromosomal instability; ES, embryonic stem; iMEF, immortalized MEF; MCC, mitotic checkpoint complex; MEF, mouse embryonic fibroblast; SAC, spindle assembly checkpoint; SKY, spectrum karyotyping.
References
- Armitage P., Doll R. 1954. The age distribution of cancer and a multi-stage theory of carcinogenesis.Br. J. Cancer. 8:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage P., Doll R. 2004. The age distribution of cancer and a multi-stage theory of carcinogenesis. 1954.Int. J. Epidemiol. 33:1174–1179 [DOI] [PubMed] [Google Scholar]
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation.J. Cell Biol. 160:341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice.Nat. Genet. 36:744–749 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Malureanu L., Perez-Terzic C., Terzic A., van Deursen J.M. 2006. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice.J. Cell Biol. 172:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B.C., Lopes C., Sunkel C.E., Goldberg M.L. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila.J. Cell Biol. 146:13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D., Kuffer C., Lenobel R., Korner R., Stemmann O. 2008. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase.J. Biol. Chem. 283:816–823 [DOI] [PubMed] [Google Scholar]
- Burds A.A., Lutum A.S., Sorger P.K. 2005. Generating chromosome instability through the simultaneous deletion of Mad2 and p53.Proc. Natl. Acad. Sci. USA. 102:11296–11301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.L., Solomon M.J. 2007. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint.Genes Dev. 21:655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B. 1998. Mutations of mitotic checkpoint genes in human cancers.Nature. 392:300–303 [DOI] [PubMed] [Google Scholar]
- Chi Y.H., Jeang K.T. 2007. Aneuploidy and cancer.J. Cell. Biochem. 102:531–538 [DOI] [PubMed] [Google Scholar]
- Clarke D.J., Segal M., Andrews C.A., Rudyak S.G., Jensen S., Smith K., Reed S.I. 2003. S-phase checkpoint controls mitosis via an APC-independent Cdc20p function.Nat. Cell Biol. 5:928–935 [DOI] [PubMed] [Google Scholar]
- Dai W., Wang Q., Liu T., Swamy M., Fang Y., Xie S., Mahmood R., Yang Y.M., Xu M., Rao C.V. 2004. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency.Cancer Res. 64:440–445 [DOI] [PubMed] [Google Scholar]
- Davenport J., Harris L.D., Goorha R. 2006. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1.Exp. Cell Res. 312:1831–1842 [DOI] [PubMed] [Google Scholar]
- Diaz-Martinez L.A., Yu H. 2007. Running on a treadmill: dynamic inhibition of APC/C by the spindle checkpoint.Cell Div. 2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M.L., Benezra R., Sorger P.K. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2.Cell. 101:635–645 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D. 2007. Tetraploidy, aneuploidy and cancer.Curr. Opin. Genet. Dev. 17:157–162 [DOI] [PubMed] [Google Scholar]
- Gorr I.H., Boos D., Stemmann O. 2005. Mutual inhibition of separase and Cdk1 by two-step complex formation.Mol. Cell. 19:135–141 [DOI] [PubMed] [Google Scholar]
- Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Mehes K., Nash R., Robin N., et al. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B.Nat. Genet. 36:1159–1161 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Burton J.L., Solomon M.J. 2002. The anaphase-promoting complex: it's not just for mitosis any more.Genes Dev. 16:2179–2206 [DOI] [PubMed] [Google Scholar]
- Hendrickson C., Meyn M.A., III, Morabito L., Holloway S.L. 2001. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase-to-anaphase transition.Curr. Biol. 11:1781–1787 [DOI] [PubMed] [Google Scholar]
- Hong Y., Stambrook P.J. 2004. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation.Proc. Natl. Acad. Sci. USA. 101:14443–14448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Hatcher R., York J.P., Zhang P. 2005. Securin and separase phosphorylation act redundantly to maintain sister chromatid cohesion in mammalian cells.Mol. Biol. Cell. 16:4725–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Andreu-Vieyra C.V., York J.P., Hatcher R., Lu T., Matzuk M.M., Zhang P. 2008. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells.PLoS Biol. 6:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y., Chi Y.H., Miyazato A., Sheleg S., Haller K., Peloponese J.M., Jr., Li Y., Ward J.M., Benezra R., Jeang K.T. 2007. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice.Cancer Res. 67:160–166 [DOI] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis.J. Cell Biol. 179:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler K.W., Vogelstein B. 1996. Lessons from hereditary colorectal cancer.Cell. 87:159–170 [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Rose A.M. 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans.Nat. Cell Biol. 1:514–521 [DOI] [PubMed] [Google Scholar]
- Kraft C., Vodermaier H.C., Maurer-Stroh S., Eisenhaber F., Peters J.M. 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates.Mol. Cell. 18:543–553 [DOI] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding.Dev. Cell. 16:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Burke D.J. 2003. The spindle assembly and spindle position checkpoints.Annu. Rev. Genet. 37:251–282 [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W. 1991. Feedback control of mitosis in budding yeast.Cell. 66:519–531 [DOI] [PubMed] [Google Scholar]
- Lim H.H., Surana U. 1996. Cdc20, a beta-transducin homologue, links RAD9-mediated G2/M checkpoint control to mitosis in Saccharomyces cerevisiae.Mol. Gen. Genet. 253:138–148 [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E. 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells.J. Cell Sci. 117:1757–1771 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.Mol. Cell. 9:59–71 [DOI] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase.Dev. Cell. 16:118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J., Huang X., Zhang P. 2001. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts.Curr. Biol. 11:1197–1201 [DOI] [PubMed] [Google Scholar]
- Mertens F., Johansson B., Mitelman F. 1994. Isochromosomes in neoplasia.Genes Chromosomes Cancer. 10:221–230 [DOI] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., Benezra R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells.Nature. 409:355–359 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. 1999. Regulation of the APC and the exit from mitosis.Nat. Cell Biol. 1:E47–E53 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2005. How do so few control so many? Cell. 120:739–746 [DOI] [PubMed] [Google Scholar]
- Page A.M., Hieter P. 1999. The anaphase-promoting complex: new subunits and regulators.Annu. Rev. Biochem. 68:583–609 [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Lee E., Kirschner M.W. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex.Genes Dev. 15:2396–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B.A., Biggins S. 2005. The spindle checkpoint: tension versus attachment.Trends Cell Biol. 15:486–493 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores.J. Cell Biol. 130:941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Mocker D., Seufert W. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC.EMBO J. 20:5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick D.M., Halazonetis T.D. 2000. Chfr defines a mitotic stress checkpoint that delays entry into metaphase.Nature. 406:430–435 [DOI] [PubMed] [Google Scholar]
- Stemmann O., Zou H., Gerber S.A., Gygi S.P., Kirschner M.W. 2001. Dual inhibition of sister chromatid separation at metaphase.Cell. 107:715–726 [DOI] [PubMed] [Google Scholar]
- Storchova Z., Kuffer C. 2008. The consequences of tetraploidy and aneuploidy.J. Cell Sci. 121:3859–3866 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2.J. Cell Biol. 154:925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast.Cell. 103:375–386 [DOI] [PubMed] [Google Scholar]
- Wang Q., Liu T., Fang Y., Xie S., Huang X., Mahmood R., Ramaswamy G., Sakamoto K.M., Darzynkiewicz Z., Xu M., Dai W. 2004. BUBR1 deficiency results in abnormal megakaryopoiesis.Blood. 103:1278–1285 [DOI] [PubMed] [Google Scholar]
- Wang S.I., Puc J., Li J., Bruce J.N., Cairns P., Sidransky D., Parsons R. 1997. Somatic mutations of PTEN in glioblastoma multiforme.Cancer Res. 57:4183–4186 [PubMed] [Google Scholar]
- Weaver B.A., Cleveland D.W. 2007. Aneuploidy: instigator and inhibitor of tumorigenesis.Cancer Res. 67:10103–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor.Cancer Cell. 11:25–36 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s).J. Cell Biol. 133:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y.M., Baek K.H., Jeong S.J., Shin H.J., Ha G.H., Jeon A.H., Hwang S.G., Chun J.S., Lee C.W. 2004. WD repeat-containing mitotic checkpoint proteins act as transcriptional repressors during interphase.FEBS Lett. 575:23–29 [DOI] [PubMed] [Google Scholar]
- Zhang P., Li M.Z., Elledge S.J. 2002. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step.Nat. Genet. 30:31–39 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lees E. 2001. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation.Mol. Cell. Biol. 21:5190–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Panda D., Landen J.W., Wilson L., Joshi H.C. 2002a. Minor alteration of microtubule dynamics causes loss of tension across kinetochore pairs and activates the spindle checkpoint.J. Biol. Chem. 277:17200–17208 [DOI] [PubMed] [Google Scholar]
- Zhou J., Yao J., Joshi H.C. 2002b. Attachment and tension in the spindle assembly checkpoint.J. Cell Sci. 115:3547–3555 [DOI] [PubMed] [Google Scholar]
- Zhuang Z., Park W.S., Pack S., Schmidt L., Vortmeyer A.O., Pak E., Pham T., Weil R.J., Candidus S., Lubensky I.A., et al. 1998. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas.Nat. Genet. 20:66–69 [DOI] [PubMed] [Google Scholar]