Abstract
Expression of the type I transmembrane glycoprotein CD44 has recently been recognized as a signature for cancer stem cells. In this study, we demonstrate that CD44, once engaged, is internalized and translocated to the nucleus, where it binds to various promoters, including that of cyclin D1, leading to cell fate change through transcriptional reprogramming. In regulating cyclin D1 expression, the internalized CD44 forms a complex with STAT3 and p300 (acetyltransferase), eliciting STAT3 acetylation at lysine 685 and dimer formation in a cytokine- and growth factor–independent manner. A bipartite nuclear localization signal (NLS) was mapped to the cytoplasmic tail of CD44, which mediates its nuclear translocation. Expression of CD44(NLS) mutant sequesters STAT3 in cytosol. In the nucleus, the acetylated STAT3 dimer remains associated with CD44 and binds to the cyclin D1 promoter, leading to increased cyclin D1 expression and cell proliferation. This study describes a novel function for CD44 in transcriptional modulation through nuclear translocation of the internalized CD44 and complex formation with transcription factors.
Introduction
CD44 mediates the responses of cells to their microenvironment and participates in the regulation of growth, survival, differentiation, and motility (Al-Hajj et al., 2003; Ponta et al., 2003; Ponti et al., 2005). Several cell surface receptors, including EGF receptor family members, are known to migrate to the nucleus as an intact polypeptide or a proteolytic fragment with or without their corresponding ligands. These nuclear localized receptors have been shown to act as transcription factors (Lin et al., 2001; Ni et al., 2001; Wang et al., 2004) for genes like cyclin D1 (Lin et al., 2001), FGF2 (Peng et al., 2001), and COX-2 (Wang et al., 2004) or modulators for induction of c-jun and cyclin D1 (Reilly and Maher, 2001).
Although phosphorylation is a crucial posttranslational modification that regulates the activities of different proteins, there are many others, including acetylation (Yang and Seto, 2007), isgylation (Kim and Zhang, 2005), methylation (D'Alessio et al., 2007), sumoylation (Zhao, 2007), and ubiquitination (Giandomenico et al., 2003). Function-related posttranslational modifications of STAT3 in response to treatment with cytokine or growth factor include phosphorylation on tyrosine 705 and serine 727 residues. However, unphosphorylated or tyrosine-mutated STAT3 can still form dimers and induce transcription (Braunstein et al., 2003). Others have found that STAT3 dimerization is regulated by reversible acetylation of lysine at residue 685 in the SH2 domain of STAT3 (Yuan et al., 2005). IL-6–induced acetylation of the STAT3 N terminus is necessary for acute-phase induction of angiotensinogen (Ray et al., 2002). Together, these observations indicate that site-specific acetylation of STAT3 is an important regulatory modification that influences protein–protein interaction and transcriptional regulation.
The CD44 transmembrane glycoprotein family adds new aspects to these roles by participating in signal transduction processes. A previous study has shown that a fragment of CD44 can directly interact with the transcriptional machinery, resulting in the up-regulation of genes containing the TPA-responsive element, including CD44 itself (Kajita et al., 2001). However, the mechanism of their nuclear import and the function of nuclear CD44 are virtually unknown. In this study, we show for the first time that full-length CD44 is internalized and translocated into the nucleus, where, in complex with STAT3 and p300, it binds to the cyclin D1 promoter and enhances cell proliferation.
Results and discussion
Nuclear localization of full-length CD44
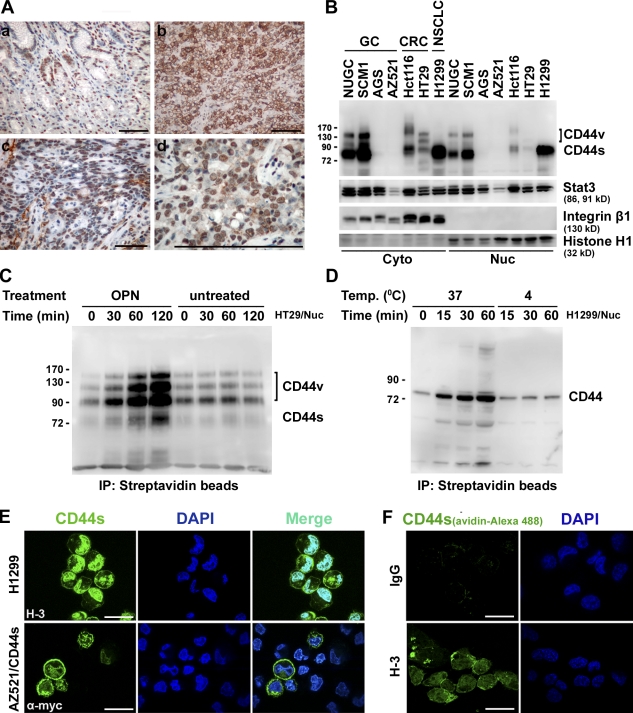
The CD44 gene contains 20 exons. 10 of the variant exons are alternatively spliced to encode a portion of the ectodomain, generating numerous CD44 spliced variant isoforms (CD44V) in some epithelial tissues and several cancers. The standard CD44 (CD44S), lacking all variant exons, is widely expressed in most cell types. We have previously shown that CD44V and its physiological ligand osteopontin (OPN) are frequently overexpressed in human gastric cancer and that OPN-engaged CD44V ligation confers cells an increased matrix-derived survival through inducing lipid raft coalescence to facilitate the CD44–Src–integrin signaling axis (Lee et al., 2007, 2008). In this study, we show that ligation of CD44 promotes its nuclear localization. Fig. 1 A shows the expression pattern of CD44 in primary gastric cancer. Immunohistochemistry revealed focal signals of CD44 in cells located at crypt regions in normal gastric mucosa (Fig. 1 A, a). In contrast, CD44 was expressed abundantly in gastric tumor cells, with strong staining in the membrane and nucleus (Fig. 1 A, b–d). By Western blotting, CD44 was detected in the nuclear fractions of a variety of cell lines that expressed endogenous CD44 (Fig. 1 B). Internalization and nuclear translocation of CD44 were monitored in HT29 and H1299 cells. After biotinylation of cell surface proteins at 4°C, cells were removed to 37°C and incubated with OPN, a physiological ligand of CD44, or anti-CD44 mAb Hermes-3 (H-3). The nuclear fraction was immunoprecipitated with streptavidin and immunoblotted for CD44. As shown, ligation of CD44 by OPN or H-3 (Lee et al., 2008) promoted its nuclear translocation in a time-dependent manner (Fig. 1, C and D). Confocal microscopic examination further revealed nuclear localization of a prominent proportion of the endogenous CD44 in H1299 cells (Fig. 1 E, top) and ectopically expressed myc-tagged CD44 in AZ521/CD44 stable clones (Fig. 1 E, bottom), respectively. Furthermore, H1299 cells were incubated with biotin-conjugated H-3 at 4°C. After removal to 37°C for 60 min, cellular uptake and nuclear translocation of CD44 were readily detected by immunofluorescence (Fig. 1 F).
Figure 1.
Nuclear localization of full-length CD44. (A) Immunohistochemistry of nuclear CD44 in human gastric mucosal (a) and cancerous (b–d) tissues using anti-CD44v6 antibody, with nuclei counterstained in hematoxylin. (B) Western blot analyses of nuclear (Nuc) and cytosolic (Cyto) fractions of human gastric (GC), colon (CRC), and lung (NSCLC) cancer cell lines. NUGC, Nagoya University gastric cancer. (C and D) HT29 (C) and H1299 (D) cells were surface labeled with biotin at 4°C followed by incubation at 37°C in the presence of OPN (C) and H-3 (D). The nuclear fraction was incubated with streptavidin beads and subjected to Western blotting. (B–D) Molecular mass is shown in kilodaltons. (E) H1299 and AZ521/CD44 cells were suspended in medium for 30 min, replated on dishes for 1 h, and immunostained using H-3 (top) and anti-myc (bottom) antibodies. Representative images taken by confocal laser microscopy are shown. (F) H1299 cells were incubated with biotin-conjugated control IgG or H-3 at 4°C followed by further incubation at 37°C for 60 min. After removing surface-retained biotin, cells were fixed and stained by avidin. Bars: (A) 100 µm; (E) 10 µm; (F) 25 µm.
Nuclear translocation of CD44 by endosomal sorting
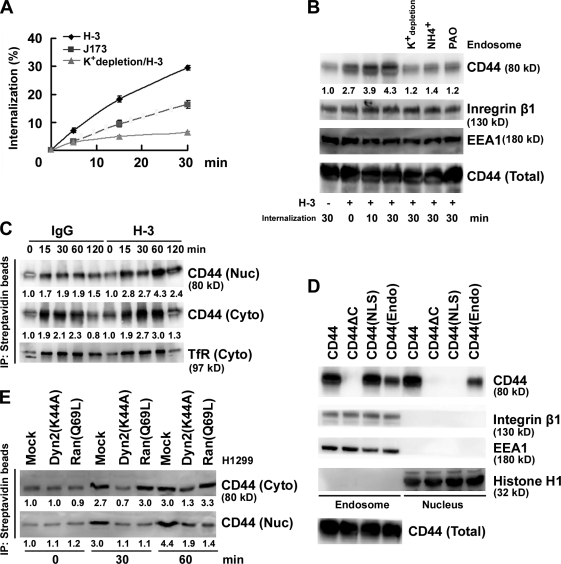
The kinetics of CD44 internalization were followed in H1299 cells incubated with biotin-conjugated anti-CD44 antibodies. Two anti-CD44 mAbs, J173, which does not induce CD44 ligation, and H-3, which can induce CD44 ligation, were applied to cells separately. Cellular intake of J173-bound CD44 increased as a function of time, and a greater intake of CD44 was observed upon H-3 treatment, suggesting that ligation of CD44 accelerated its internalization (Fig. 2 A). Consistent with this, increasing amounts of CD44 were recovered from the early endosome fraction isolated from H1299 cells after H-3 treatment, and pretreatment of cells with NH4+ or phenylarsine oxide or depletion of K+ to block endocytosis significantly inhibited the internalization and uptake of CD44 (Fig. 2 B). By biotin-labeled endocytosis assay, we showed that CD44 ligation enhanced not only the internalization of the biotin-labeled receptor but also its nuclear localization (Fig. 2 C). From the standard isoform of CD44, a CD44(NLS) mutant was generated by changing the putative NLS sequence 292RRRCGQKKK300 to 292AAACGQAAA300, a CD44(Endo) mutant was generated by changing sequences flanking the C-terminal dihydrophobic motif (L331/V332; Sheikh and Isacke, 1996) from 329VHLV332 to 329AHAA332, and a CD44ΔC mutant was generated by removing most of the C-terminal sequences. By subcellular fractionation, wild-type (WT) CD44 ectopically expressed in AZ521 cells was readily detected in the endosomal and nuclear fractions. In contrast, CD44ΔC failed to internalize, CD44(NLS) mutant internalized efficiently but failed to enter the nucleus, and CD44(Endo) mutant was internalized less efficiently (Fig. 2 D). To corroborate that endosomal sorting and the bipartite NLS are essential for the nuclear localization of CD44 protein, a dynamin mutant Dyn 2(K44A) (Cao et al., 1998) and a Ran GTPase mutant Ran(Q69L) (Macara, 2001) were ectopically expressed in H1299 cells, respectively. Fig. 2 E shows that overexpression of Dyn 2(K44A) significantly inhibited internalization of CD44, whereas overexpression of Ran(Q69L) has no effect on CD44 internalization but completely blocked its nuclear import. We conclude that CD44 is internalized through endosomal sorting and imported to the nucleus through the nuclear pore complex.
Figure 2.
Nuclear translocation of CD44 by endosomal sorting. (A) H1299 cells were incubated with biotin-conjugated anti-CD44 mAb H-3 and J173 at 4°C separately followed by further incubation at 37°C for 5, 15, and 30 min as indicated. Internalization was measured by flow cytometry. The percentage of internalization was calculated by setting the mean fluorescence intensity of cells after biotin labeling but without glutathione incubation as 100%. The value of each time point is shown as means ± SD from three independent experiments. (B) H1299 cells were cultured in complete medium with and without 50 mM NH4Cl or 20 µM phenylarsine oxide (PAO) or in K+ depletion medium at 37°C for 1 h followed by further incubation in the presence of H-3 for various times as indicated. Total cell lysates and endosomes purified by sucrose gradient centrifugation were subjected to Western blotting. (C) H1299 cells were surface labeled with biotin. Cytosolic (Cyto) and nuclear (Nuc) fractions were affinity purified with avidin-conjugated beads and analyzed by Western blotting. (D) Western blot analyses of endosomal and nuclear fractions in individual AZ521/CD44 cell clones. (E) Western blot analyses of cytosolic and nuclear fractions derived from H1299 cells transfected with and without plasmids encoding Dyn 2(K44A) or Ran(Q69L). (B, C, and E) The relative intensities of the bands, which were quantified by densitometry, are shown.
Nuclear CD44 accelerates cell proliferation and cell cycle progression through control of cyclin D1 expression
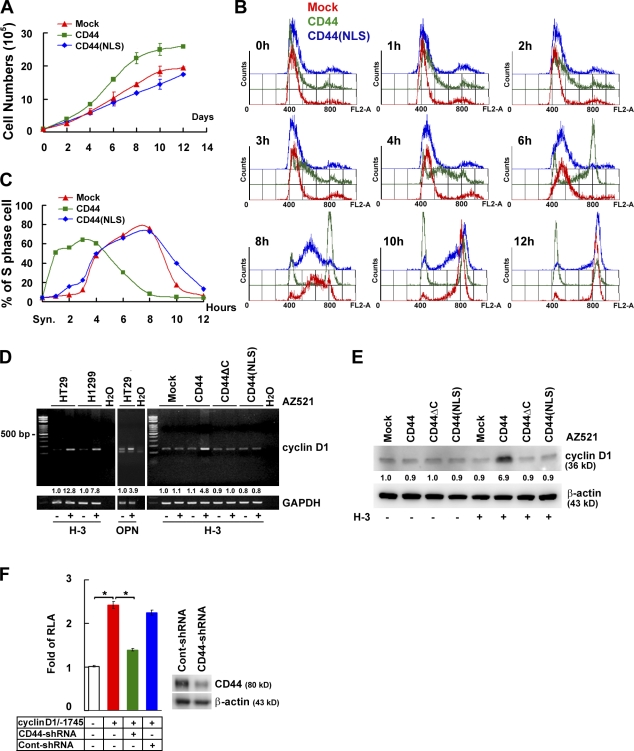
As shown in Fig. 3 A, ectopic expression of CD44 but not CD44(NLS) mutant conferred AZ521 cells increased proliferation, suggesting that localization of CD44 in the nucleus is an important aspect of its growth-stimulating function. AZ521 cell clones were synchronized at the G1–S boundary by double thymidine block. After release from the block, AZ521/CD44 cells exited G1 and entered S phase in 1–5 h, which is in contrast to the 4–8-h transition time taken by the AZ521/mock and AZ521/CD44(NLS) mutant cells (Fig. 3, B and C). Cyclin D1 is known to drive cell cycle progression at the G1/S transition. We showed that both cyclin D1 transcripts and proteins were expressed at elevated levels in CD44-expressing cells upon CD44 ligation by H-3 or OPN (Fig. 3, D and E). Consistent with this, knockdown of CD44 transcripts significantly reduced cyclin D1 promoter–mediated reporter gene expression (Fig. 3 F).
Figure 3.
Nuclear CD44 accelerates cell proliferation and cell cycle progression through controlling cyclin D1 expression. (A) AZ521/CD44 cell clones were cultured in RPMI medium containing 1% FBS. Total viable cell numbers were determined. Data were derived from three independent experiments and are presented as means ± SD. (B and C) AZ521/CD44 cell clones were treated with H-3 and synchronized by double thymidine block. After being released into fresh medium containing 1% FBS, cellular DNA content was determined by FACS analysis after propidium iodide staining. The percentage of cells in S phase of the cell cycle is indicated in C. (D and E) Cells were cultured in serum-free medium for 24 h and incubated with or without H-3 for 1 (D) or 3 d (E). The expression of cyclin D1 was measured by RT-PCR (D) and Western blotting (E). The relative intensities of the bands, which were quantified by densitometry, are shown. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (F) Reporter assays were performed by transfection of a luciferase reporter plasmid driven by the 1.75-kb cyclin D1 promoter into H1299 cell clones stably harboring lentivirus-encoded control shRNA or shRNA targeting CD44. Data, presented as the means ± SD, were derived from at least three independent experiments. *, P < 0.05 by Student's t test. RLA, relative luciferase activity.
Nuclear CD44 associates with STAT3 and functions to modulate transcription
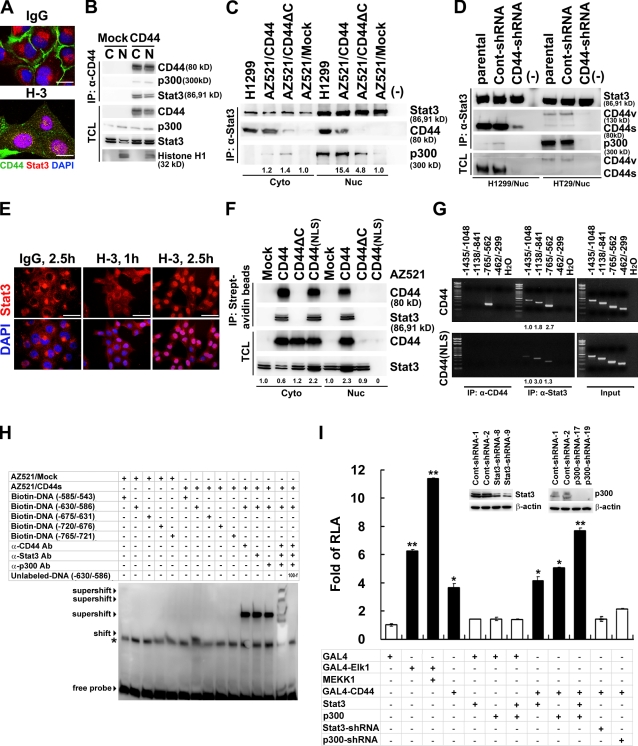
Chromatin immunoprecipitation (IP [ChIP]) was performed to search for DNA sequences bound by nuclear CD44 complexes. A total of 19 clones was obtained from the DNA fragments pulled down by anti-CD44 mAb. Through a National Center for Biotechnology Information BLAST (basic local alignment search tool) search, they contained sequences corresponding to the promoters of several genes, including cyclin D1 (Tables S1 and S2). Among them, 15 clones contained binding elements for signal transducer and activator of transcription (STAT) and p300. We tested whether nuclear CD44 exerts its transcriptional regulatory function through interacting with STAT3 and p300. By immunofluorescence, we showed that ligation of CD44 promoted nuclear colocalization of CD44 and STAT3 in H1299 cells (Fig. 4 A). Co-IP revealed that STAT3 and p300 were in complex with CD44 in AZ521/CD44 cells (Fig. 4 B). In-frame CD44 deletional mutants and STAT3 domain truncation mutants were generated for mapping the interacting domains in CD44 and STAT3 (Figs. S1 and S2). Co-IP showed that the N-terminal region encoded by constant exon 3 in CD44 and the N-terminal coiled-coil domain (aa 134–317) of STAT3 were important for their interaction. Our experiments using in vitro–translated CD44 and STAT3 suggested that additional factors, including posttranslational modification of CD44, were required for efficient interaction between CD44 and STAT3 (Figs. S1 and S2). Our data also suggested that p300 participates in the formation of the CD44–STAT3–p300 complex through its interaction with STAT3 (Fig. S3). We noted that little p300 was associated with STAT3 in AZ521/mock cells, whereas elevated p300 was found in the CD44-expressing H1299 and AZ521/CD44 cells, and removal of the CD44 C-terminal region precluded its interaction with STAT3 and significantly suppressed the association of p300 and STAT3 (Fig. 4 C). To corroborate that CD44 facilitates the association of STAT3 and p300, we knocked down the expression of endogenous CD44 in H1299 and HT29 cells and showed that the association of p300 and STAT3 was severely prohibited (Fig. 4 D). In the absence of cytokine- and growth factor–mediated signals, ligation of CD44 by H-3 (Fig. 4 E) or OPN (Fig. S4) promoted nuclear localization of STAT3 in a time-dependent manner. When AZ521 cell clones were incubated with biotin-labeled H-3 in an endocytosis assay, the internalized CD44 formed a complex with STAT3 in both the cytosol and nucleus, whereas the CD44(NLS) mutant only formed a complex with STAT3 in the cytoplasm, and CD44ΔC was not internalized (Fig. 4 F). STAT3 was sequestered from the nucleus in cells overexpressing the CD44(NLS) mutant (Fig. 4 F). These data suggested that internalized CD44 formed a complex with STAT3 in the cytosol and that CD44–STAT3 co-migrated to the nucleus in a CD44-dependent manner. In agreement, ChIP assays showed that STAT3 was bound weakly but mainly to the fragment of −1138/−841 bp in AZ521/CD44(NLS) cells, whereas a robust binding activity was observed for STAT3 as well as CD44 to the DNA fragment of −765/−562 bp in AZ521/CD44 cells (Fig. 4 G). These data suggest that, in the presence of CD44, STAT3-mediated transactivation of cyclin D1 was up-regulated from basal activity to CD44-regulated activity via redirecting STAT3 to a preferred binding site. By electrophoretic mobility shift assay (EMSA), we showed that CD44, STAT3, and p300 were associated in a complex and bound predominantly to the cis-element located within −630/−586 bp (Fig. 4 H). Next, we examined whether CD44 acts as a transcription-related coactivator by GAL4-CD44 transactivation assays. In AZ521 cells, GAL4-CD44 induced a 3.5-fold expression of a reporter driven by a GAL4-dependent promoter. Overexpression of STAT3 and p300 greatly enhanced, whereas knockdown of the expression abolished, the reporter activity (Fig. 4 I).
Figure 4.
Nuclear CD44 associates with STAT3 and functions to modulate transcription. (A) Confocal microscopy of H1299 cells cultured in serum-free medium for 24 h and treated with control IgG or H-3 for 1 h. (B) Nuclear (N) and cytosolic (C) fractions were prepared from AZ521/mock and AZ521/CD44 cells and immunoprecipitated followed by Western blotting. (C) Nuclear (Nuc) and cytosolic (Cyto) fractions were immunoprecipitated followed by Western blotting. (D) Nuclear extracts were prepared from the parental H1299 and HT29 cells or cell clones stably harboring lentivirus-encoded control (Cont) shRNA or shRNA targeting CD44 and immunoprecipitated followed by Western blotting. (E) Confocal microscopy of AZ521/CD44 cells with anti-STAT3 (in red) after incubation with control IgG or H-3 for 1 and 2.5 h. (F) AZ521/CD44 cells were incubated with biotin-conjugated H-3 at 4°C. After removal to 37°C for 1 h, cytosolic and nuclear fractions and immunoprecipitates from streptavidin beads were analyzed by Western blotting. (G) Nuclear extracts were prepared from AZ521/CD44 (top) and AZ521/CD44(NLS) mutant (bottom) cells after cross-linking with 1% formaldehyde. ChIP was performed using anti-CD44 and anti-STAT3. PCR amplification of designated regions within cyclin D1 promoter was performed. (H) Nuclear extracts were prepared from AZ521/mock and AZ521/CD44s cells cultured in serum-free medium for 24 h. EMSA was performed with biotin-labeled double-stranded oligonucleotide probes corresponding to various regions of cyclin D1 promoter in the presence and absence of anti-CD44, anti-STAT3, or anti-p300 antibodies. Shifted and supershifted complexes are indicated by arrowheads. A nonspecific band is indicated by an asterisk. Ab, antibody. (I) Reporter assays were performed in AZ521 cells and AZ521 cell clones stably harboring lentivirus-encoded control shRNA or shRNA targeting STAT3 or p300 using the pFR-Luc reporter plasmid containing five copies of GAL4 DNA–binding sites. Data are presented as the means ± SD and were derived from at least three independent experiments. *, P < 0.05; and **, P < 0.01 by Student's t test. RLA, relative luciferase activity; TCL, total cell lysate. (C, F, and G) The relative intensities of the bands, which were quantified by densitometry, are shown. Bars: (A) 10 µm; (E) 25 µm.
Ligation of CD44 promotes cyclin D1 expression via stimulation of STAT3 acetylation
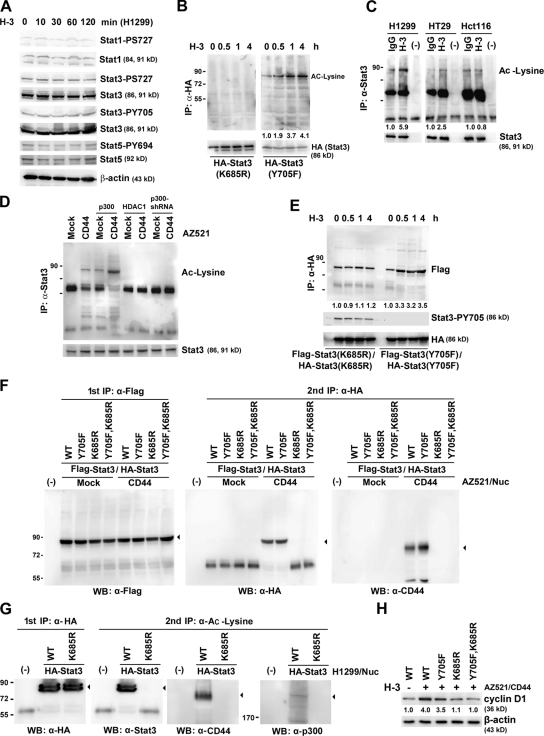
The major mechanism of STAT3 activation in tumor cells is mediated through the phosphorylation of STAT3 on tyrosine residue 705, leading to its dimerization followed by translocation into the nucleus for transcriptional regulation. In our study, ligation of CD44 by H-3 did not induce phosphorylation of STAT3 (Fig. 5 A). Recent studies have demonstrated that unphosphorylated or tyrosine-mutated STAT proteins can form stable dimers through acetylation of lysine 685 and enhance transcription (Braunstein et al., 2003; Yuan et al., 2005). We examined whether CD44 ligation promotes STAT3 acetylation. Fig. 5 B shows that ligation of CD44 enhanced the acetylation of STAT3(Y705F) in H1299 cells in a time-dependent manner, indicating that the status of phosphorylation was not a prerequisite for STAT3 acetylation. In agreement, ligation of CD44 facilitated acetylation of STAT3 in H1299 and HT29 cells but had much less effect in the p300-hemizygous HCT116 cells (Fig. 5 C). In AZ521 cells, ectopic expression of CD44 greatly enhanced STAT3 acetylation, which was further enhanced by coexpression of p300 and blocked by HDAC1 expression or knockdown of p300 expression (Fig. 5 D). Co-IP showed that CD44 ligation greatly facilitated the association of HA-tagged and Flag-tagged STAT3(Y705F) but not STAT3(K685R) (Fig. 5 E). Nuclear extracts were prepared from AZ521/CD44 cells expressing both HA-STAT3 and Flag-STAT3 and subjected to tandem IP using anti-Flag mAb first followed by anti-HA mAb (Fig. 5 F). Consistent with this, Western blotting showed dimer formation of STAT3(WT) and STAT3(Y705F) but not STAT3(K685R) (Fig. 5 F, middle). Most importantly, CD44 was found to be associated with these STAT3 dimers (Fig. 5 F, right). By tandem IP, we demonstrated that STAT3 in complex with CD44 and p300 was acetylated (Fig. 5 G). In AZ521 cells, ectopic expression of the acetylation-proficient STAT3, WT and Y705F, but not the acetylation-defective STAT3, K685R and K685R/Y705F, enhanced cyclin D1 expression in response to H-3 (Fig. 5 H), suggesting that CD44-elicited cyclin D1 expression is mediated through acetylated STAT3. In addition to cyclin D1, other STAT3-responsive genes, including BCL-XL, VEGF, and MMP2, were also found to be up-regulated through CD44 ligation–mediated STAT3 activation (Fig. S5).
Figure 5.
Ligation of CD44 promotes cyclin D1 expression via stimulation of STAT3 acetylation. (A) Western blot analyses of H1299 cells treated with H-3 for various times as indicated. (B) H1299/HA-STAT3 cells were treated with H-3 for the time points indicated and subjected to IP followed by Western blotting. (C) H1299, HT29, and HCT116 cells were treated with or without H-3 for 4 h and subjected to IP followed by Western blotting. (D) AZ521/mock and AZ521/CD44 cells were transfected with or without plasmids encoding p300 or HDAC1 or infected by lentivirus-encoding shRNA targeting p300. Total cell lysates were prepared, immunoprecipitated with anti-STAT3, and analyzed for acetylated lysine (Ac-Lysine) by Western blotting. (E) H1299 cells ectopically expressing the Flag-tagged and HA-tagged STAT3 mutants were treated with H-3 mAb. At the time points indicated, lysates were immunoprecipitated and subjected to Western blotting. (F) AZ521/mock and AZ521/CD44 cells were transfected with plasmids encoding the Flag-tagged and HA-tagged STAT3 mutants as indicated. Nuclear extracts were subjected to tandem IP. Proteins in the first and second immunoprecipitates were analyzed by Western blotting as indicated. (G) H1299 cells were transfected with plasmids encoding HA-tagged STAT3(WT) or STAT3(K685R). Nuclear extracts were subjected to tandem IP. Proteins in the first and second immunoprecipitates were analyzed by Western blotting as indicated. Arrowheads indicate the target proteins with expected sizes. (H) AZ521/CD44 cells were transfected with plasmids encoding individual STAT3. After treatment without or with H-3 for 3 d, Western blot analysis was performed. (B, C, E, and H) The relative intensities of the bands, which were quantified by densitometry, are shown. Molecular mass is shown in kilodaltons. WB, Western blot.
More than 40 different polypeptide ligands induce STAT phosphorylation. Although unphosphorylated STAT is known to drive the expression of certain genes, limited information is available on how unphosphorylated STAT3 drives gene expression (Yang et al., 2005, 2007; Yuan et al., 2005). In this study, we demonstrate that STAT3 is activated upon engagement of the cell surface receptor CD44, in the absence of cytokine and growth factors. This is initiated by accelerated internalization and nuclear localization of CD44 followed by complex formation of CD44–acetylated STAT3–p300, leading to enhanced expression of downstream target genes, including cyclin D1.
Materials and methods
Reagents and constructs
The CD44 mutants with C-terminal deletion (CD44ΔC) have been previously described (Lee et al., 2008). NLS and Endo mutants of CD44 were constructed by site-directed mutagenesis using the WT CD44S cDNA template. The vectors encoding Dyn 2(K44A) and Ran(Q69L) were gifts from M.-C. Hung (University of Texas M.D. Anderson Cancer Center, Houston, TX; Giri et al., 2005). The vectors encoding STAT3(Y705F) and STAT3(K685R) were gifts from Y.-E. Chin (Brown University School of Medicine; Providence, RI; Yuan et al., 2005). The hybridoma for H-3 was obtained from American Type Culture Collection. Preparation and purification of OPN have been described previously (Lee et al., 2007).
Cell culture and molecular and protein techniques
Most of the experiments were performed in HT29 cells that expressed various isoforms of CD44, H1299 cells that expressed only the standard form of CD44, and the CD44-null AZ521 cells. Transient transfection was performed using the Lipofectamine 2000 reagent (Invitrogen). Ligation of CD44 was performed by incubating cells precultured in serum-free medium for 24 h in the same medium containing 20 µg/ml H-3 at 37°C for 1 h or as indicated. Reporter assay was performed as previously described using the pRL-SV40 vector (Promega) containing the reporter Renilla luciferase gene as an internal control (Liu et al., 2004). The lentivirus-based knockdown approach has been previously described (Lee et al., 2007, 2008). The pLKO.1–short hairpin RNA (shRNA) plasmids encoding an shRNA with scramble sequences or sequences targeting human CD44, STAT3, or p300 were purchased from the National RNAi Core Facility. Western blotting was performed as previously described (Lee et al., 2004). Images were recorded using the luminescent image analyzer (LAS-3000; Fujifilm), and the intensities of the bands were quantitated by densitometry using Multi Gauge version 3.0 software (Fujifilm). For immunofluorescence, cells were fixed in 4% paraformaldehyde, incubated with primary antibody and Alexa Fluor 594– or Alexa Fluor 488–conjugated secondary antibody, counterstained with DAPI, and examined under a laser-scanning confocal system (Radiance 2100; Bio-Rad Laboratories) with Plan-Apochromat oil immersion 60× or 100× NA 1.4 objective lenses (Nikon) using the standard analysis software (Lasersharp 2000; Bio-Rad Laboratories). The images were arranged and labeled using Photoshop software (Adobe).
Receptor internalization assay
Cells (70–80% confluence) were briefly washed with ice-cold PBS and treated with sulfo-NHS-SS-biotin at 1.0 mg/10 ml/10-cm dish (Thermo Fisher Scientific) with gentle agitation for 60 min at 4°C. Cells were then incubated with control IgG or H-3 at 37°C in complete medium for the designated times as indicated. Cells were incubated with 0.1 M glycine in PBS for 30 min at 4°C to quench the unreacted biotin. Surface-retained biotin was removed using reduced glutathione (50 mM glutathione, 75 mM NaCl, 1 mM EDTA, 1% BSA, and 0.75% [vol/vol] of 10 N NaOH). After lysis, biotinylated proteins were precipitated using streptavidin beads from equal amounts of cell lysates. The amounts of receptor bound to beads were determined by immunoblotting.
EMSA
Nuclear extracts were incubated with 5′-biotin–labeled double-stranded oligonucleotides for 30 min at room temperature in the presence or absence of anti-CD44, anti-STAT3, or anti-p300 antibodies. In some experiments, an excess of unlabeled oligonucleotide (cold oligonucleotide) was added as competitor. DNA–protein complexes were fractionated by PAGE and visualized by horseradish peroxidase–conjugated streptavidin using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific).
Statistical analysis
Statistical analysis of data was performed by a Student's t test using SigmaPlot software (SSPS). Difference was considered to be statistically significant at P < 0.05.
Online supplemental material
Fig. S1 shows that the STAT3 N-terminal region bears a CD44 docking site. Fig. S2 shows that the N-terminal region and posttranslational modification of CD44 are necessary for the association with STAT3. Fig. S3 shows that p300 may not directly interact with CD44, and it remains in the CD44–STAT3–p300 complex through its interaction with STAT3. Fig. S4 shows the time course study of STAT3 nuclear localization after CD44 ligation by OPN. Fig. S5 shows that the expression of STAT3-responsive genes is increased after ligation of CD44. Table S1 shows cis-elements bound by nuclear CD44. Table S2 shows DNA sequences bound by nuclear CD44 complexes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812060/DC1.
Acknowledgments
We thank Dr. Fen-Yau Li, Dr. Chin-Wen Chi, and Hwa-Li Kao for assistance in immunohistochemistry.
This work was supported by the Genomics and Proteomics program project grant (AS94M009-3 to J.-Y. Chen) and a postdoctoral training grant (to J.-L. Lee) from Academia Sinica.
Footnotes
Abbreviations used in this paper: ChIP, chromatin IP; EMSA, electrophoretic mobility shift assay; H-3, Hermes-3; IP, immunoprecipitation; OPN, osteopontin; shRNA, short hairpin RNA; STAT, signal transducer and activator of transcription; WT, wild type.
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. 2003. Prospective identification of tumorigenic breast cancer cells.Proc. Natl. Acad. Sci. USA. 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein J., Brutsaert S., Olson R., Schindler C. 2003. STATs dimerize in the absence of phosphorylation.J. Biol. Chem. 278:34133–34140 [DOI] [PubMed] [Google Scholar]
- Cao H., Garcia F., McNiven M.A. 1998. Differential distribution of dynamin isoforms in mammalian cells.Mol. Biol. Cell. 9:2595–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio A.C., Weaver I.C., Szyf M. 2007. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes.Mol. Cell. Biol. 27:7462–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico V., Simonsson M., Gronroos E., Ericsson J. 2003. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors.Mol. Cell. Biol. 23:2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri D.K., Ali-Seyed M., Li L.Y., Lee D.F., Ling P., Bartholomeusz G., Wang S.C., Hung M.C. 2005. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor.Mol. Cell. Biol. 25:11005–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. 2001. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration.J. Cell Biol. 153:893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.I., Zhang D.E. 2005. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays.Methods Enzymol. 398:491–499 [DOI] [PubMed] [Google Scholar]
- Lee J.L., Lin C.T., Chueh L.L., Chang C.J. 2004. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis.J. Biol. Chem. 279:14602–14609 [DOI] [PubMed] [Google Scholar]
- Lee J.L., Wang M.J., Sudhir P.R., Chen G.D., Chi C.W., Chen J.Y. 2007. Osteopontin promotes integrin activation through outside-in and inside-out mechanisms: OPN-CD44V interaction enhances survival in gastrointestinal cancer cells.Cancer Res. 67:2089–2097 [DOI] [PubMed] [Google Scholar]
- Lee J.L., Wang M.J., Sudhir P.R., Chen J.Y. 2008. CD44 engagement promotes matrix-derived survival through the CD44-SRC-integrin axis in lipid rafts.Mol. Cell. Biol. 28:5710–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Makino K., Xia W., Matin A., Wen Y., Kwong K.Y., Bourguignon L., Hung M.C. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor.Nat. Cell Biol. 3:802–808 [DOI] [PubMed] [Google Scholar]
- Liu C.A., Wang M.J., Chi C.W., Wu C.W., Chen J.Y. 2004. Rho/Rhotekin-mediated NF-kappaB activation confers resistance to apoptosis.Oncogene. 23:8731–8742 [DOI] [PubMed] [Google Scholar]
- Macara I.G. 2001. Transport into and out of the nucleus.Microbiol. Mol. Biol. Rev. 65:570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C.Y., Murphy M.P., Golde T.E., Carpenter G. 2001. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase.Science. 294:2179–2181 [DOI] [PubMed] [Google Scholar]
- Peng H., Moffett J., Myers J., Fang X., Stachowiak E.K., Maher P., Kratz E., Hines J., Fluharty S.J., Mizukoshi E., et al. 2001. Novel nuclear signaling pathway mediates activation of fibroblast growth factor-2 gene by type 1 and type 2 angiotensin II receptors.Mol. Biol. Cell. 12:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponta H., Sherman L., Herrlich P.A. 2003. CD44: from adhesion molecules to signalling regulators.Nat. Rev. Mol. Cell Biol. 4:33–45 [DOI] [PubMed] [Google Scholar]
- Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M.A., Daidone M.G. 2005. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties.Cancer Res. 65:5506–5511 [DOI] [PubMed] [Google Scholar]
- Ray S., Sherman C.T., Lu M., Brasier A.R. 2002. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity.Mol. Endocrinol. 16:824–836 [DOI] [PubMed] [Google Scholar]
- Reilly J.F., Maher P.A. 2001. Importin β-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation.J. Cell Biol. 152:1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh H., Isacke C.M. 1996. A di-hydrophobic Leu-Val motif regulates the basolateral localization of CD44 in polarized Madin-Darby canine kidney epithelial cells.J. Biol. Chem. 271:12185–12190 [DOI] [PubMed] [Google Scholar]
- Wang S.C., Lien H.C., Xia W., Chen I.F., Lo H.W., Wang Z., Ali-Seyed M., Lee D.F., Bartholomeusz G., Ou-Yang F., et al. 2004. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2.Cancer Cell. 6:251–261 [DOI] [PubMed] [Google Scholar]
- Yang J., Chatterjee-Kishore M., Staugaitis S.M., Nguyen H., Schlessinger K., Levy D.E., Stark G.R. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation.Cancer Res. 65:939–947 [PubMed] [Google Scholar]
- Yang J., Liao X., Agarwal M.K., Barnes L., Auron P.E., Stark G.R. 2007. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB.Genes Dev. 21:1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Seto E. 2007. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention.Oncogene. 26:5310–5318 [DOI] [PubMed] [Google Scholar]
- Yuan Z.L., Guan Y.J., Chatterjee D., Chin Y.E. 2005. Stat3 dimerization regulated by reversible acetylation of a single lysine residue.Science. 307:269–273 [DOI] [PubMed] [Google Scholar]
- Zhao J. 2007. Sumoylation regulates diverse biological processes.Cell. Mol. Life Sci. 64:3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]