Summary
Crystal structures of an aspartic proteinase from Trichoderma reesei (TrAsP) and its complex with a competitive inhibitor, pepstatin A, were solved and refined to crystallographic R-factors of 17.9% (Rfree=21.2%) at 1.70 Å and 15.8% (Rfree=19.2%) at 1.85 Å resolution, respectively. The three-dimensional structure of TrAsP is similar to structures of other members of the pepsin-like family of aspartic proteinases. Each molecule is folded in a predominantly β-sheet bilobal structure with the N-terminal and C-terminal domains of about the same size. Structural comparison of the native structure and the TrAsP:pepstatin complex reveals that the enzyme undergoes an induced fit, rigid body movement upon inhibitor binding, with the N- and C-lobes tightly enclosing the inhibitor. Upon recognition and binding of pepstatin A, amino acid residues of the enzyme active site form a number of short hydrogen bonds to the inhibitor that may play an important role in mechanism of catalysis and inhibition. The structures of TrAsP were used as a template for performing statistical coupling analysis (SCA) of the aspartic protease family. This approach permitted, for the first time, identification of a network of structurally linked residues putatively mediating conformational changes relevant to the function of this family of enzymes. SCA reveals co-evolved continuous clusters of amino acid residues which extend from the active site into the hydrophobic cores of each of the two domains and also include amino acid residues from the flap regions, highlighting the importance of these parts of the protein for its enzymatic activity.
Keywords: Trichoderma reesei, aspartic proteinase, crystal structure, pepsin-like fold, statistical coupling analysis
Introduction
Trichodermapepsin, abbreviated TrAsP, (EC 3.4.23.18) is a 329-residue aspartic proteinase isolated from the fungus Trichoderma reesei. T. reesei is an industrially important cellulolytic filamentous fungus, capable of secreting large amounts of several cellulose-degrading enzymes. The original isolate, QM6a, and its subsequent derivatives have been extensively studied with the aim to use T. reesei to produce low-cost enzymes for the conversion of plant biomass materials into industrially useful bioproducts, such as sugars and bioethanol.1,2 Fungal aspartic proteinases (APs) have been shown to participate in the processing of secreted enzymes and, as a rule, to act as regulatory enzymes. Thus, for example, the acid proteinase from Aspergillus awamori cleaves off a non-catalytic substrate-binding domain of glucoamylase, resulting in appearance of multiple glucoamylase forms during growth of the fungus in liquid medium.3 It was shown that the presence of multiple forms of cellobiohydrolase I and II in T. reesei is the result of a similar proteolytic modification.4
Much of the current understanding of aspartic proteinases structure came from the pioneering work of Andreeva et al.5 The molecules of APs from the fungal and mammalian sources contain two domains, each comprised mostly of β-sheets6–8. The active site contains two copies of an Asp-Thr/Ser-Gly motif, located at the bottom of a long and deep cleft located between the two lobes7. The substrate-binding site is able to accommodate roughly eight amino acid residues of an oligopeptide in an extended conformation. Each domain of a eukaryotic aspartic proteinase contributes one of the two catalytic aspartate residues. On the other hand, the smaller retroviral aspartic proteinases are dimeric proteins consisting of two identical subunits, and each subunit contributes one catalytic aspartate9. Consequently, it has been postulated that eukaryotic aspartic proteinases have evolved divergently by gene duplication and fusion from a primitive dimeric enzyme resembling retroviral proteinases.8
The aspartic proteinases are widely distributed among different organisms, with the exception of eubacteria. These enzymes are involved in a number of physiological processes such as digestion (pepsin), protein catabolism (cathepsin D) and blood pressure homeostasis (renin), and in pathological processes such as Alzheimer β-amyloid formation and metastasis of breast cancers (cathepsin D), retroviral infection (human immunodeficiency virus proteinase) and hemoglobin degradation in malaria (plasmepsins).9 Furthermore, some aspartic proteinases play an important role in food industry. For example, the milk-clotting enzymes chymosins, cardosins, and pepsins are utilized in fabrication of cheeses and soy sauces.10 Hence, detailed understanding of the structure and function of these enzymes may be useful in rational use of aspartic proteinases for industrial and therapeutic applications and for rational design of their inhibitors.
We present here the structure of native aspartic proteinase from T. reesei as well as the structure of its complex with the inhibitor, pepstatin A, refined to 1.70 Å and 1.85 Å, respectively. Comparison between the structures in the absence and presence of the inhibitor allowed us to describe the conformational changes of the enzyme upon inhibitor binding and recognition, and to reveal the residues that contribute to enzyme specificity. The structure of TrAsP was subsequently used as a template for performing statistical coupling analysis (SCA) of the aspartic protease family. This approach permitted, for the first time, the identification of a network of structurally linked residues putatively mediating conformational changes relevant to the function of this family of enzymes.
Results and discussion
Structure solution and the assessment of the quality of the models
The crystals of TrAsP and of its complex with pepstatin were isomorphous in the tetragonal space group P43212, with a single molecule in the asymmetric unit. The structures were solved at medium-high resolution by molecular replacement, using the coordinates of penicillopepsin, a related fungal enzyme, as the search model. The final model of uncomplexed TrAsP included all 329 residues of a single peptide chain, five ethylene glycol molecules, and 555 ordered water molecules. The stereochemical parameters of TrAsP, for both the main and side chains, had better than expected values for a 1.70 Å resolution model, with 91.7% of the residues in the most favored region of the Ramachandran plot, 8.3% in the additionally allowed regions, and with no residues in the disallowed regions, as evaluated using PROCHECK.11
The model of TrAsP:pepstatin complex was of comparable quality, including, in addition to the complete protein molecule, a molecule of pepstatin A and 621 ordered water molecules. The refinement statistics of both models are summarized in Table 1. The final (2mFobs–DFcalc, ϕcalc) electron density map is well defined and continuous throughout the proteinase molecules in both the apoenzyme and in the complex, allowing unambiguous assignment of all the amino acid residues.
Table 1.
Data collection and refinement statistics. Values for the highest resolution shell are given in parentheses.
| Native | Complex | ||
|---|---|---|---|
| Space group | P43212 | P43212 | |
| Cell dimensions at 90K (Å) | a=b=74.17, c=161.53 | a=b=74.28, c=160.03 | |
| Resolution range (Å) | 30.0-1.70 (1.79-1.70) | 30.0-1.85 (1.89-1.85) | |
| Total number of reflections | 196,637 (25,287) | 109,124 (6187) | |
| Number of unique reflections | 50,475 (7225) | 39,197 (3656) | |
| Redundancy | 3.9 (3.5) | 2.8 (1.7) | |
| Rmerge (%)a | 7.6 (47.3) | 6.1 (32.7) | |
| Completeness (%) | 98.4 (98.8) | 96.3 (89.2) | |
| <I/σ(I)> | 8.3 (2.0) | 9.6 (3.1) | |
| Refinement statistics | Proteinase | Proteinase | inhibitor |
| Number of non-hydrogen protein atoms | 2462 | 2445 | 47 |
| Number of waters molecules | 555 | 621 | |
| Number of reflections used in refinement (working/test data sets) | 49,801 (47,274/2527) | 37,690 (35,806/1884) | |
| Rfactor (%)b | 17.9 | 15.8 | |
| Rfree (%)c | 21.2 | 19.2 | |
| Average Bfactors | |||
| Main-chain (Å2) | 22.6 | 20.9 | 22.8 |
| Side-chains (Å2) | 25.9 | 23.4 | 23.6 |
| Water molecules (Å2) | 39.6 | 36.1 | |
| R.m.s. deviations from ideal geometry | |||
| Bonds (Å) | 0.017 | 0.019 | |
| Bonds angles (o) | 1.68 | 2.02 | |
| Ramachandran statistics | |||
| Most favoured regions | 91.7% | 90.7% | |
| Additional allowed regions | 8.3% | 9.3% | |
| Generously allowed/disallowed regions | 0% | 0.0% | |
Rmerge=Σhkl|I-<I>|/ΣhklI.
Rfactor=Σ(|Fobs|-|Fcalc|)/Σ|Fobs-Fcalc|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
Rfree was calculated using 5% reflections randomly selected in a test data set.
The structure of the TrAsP
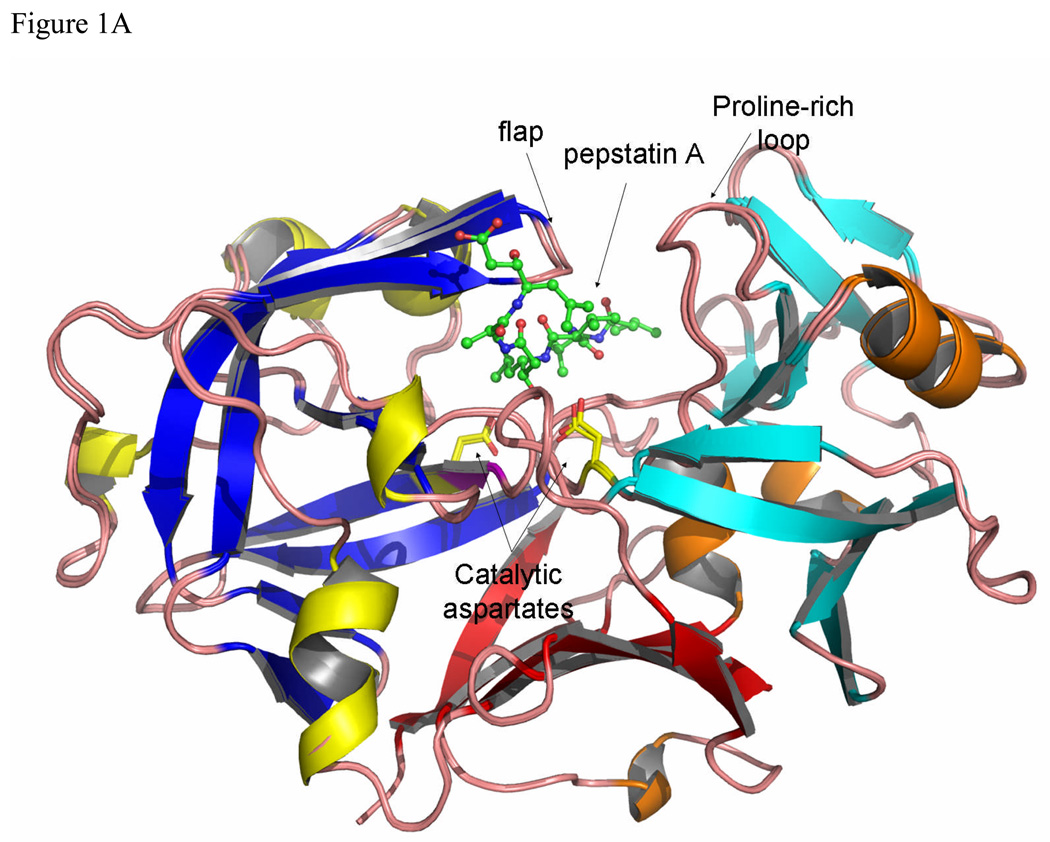
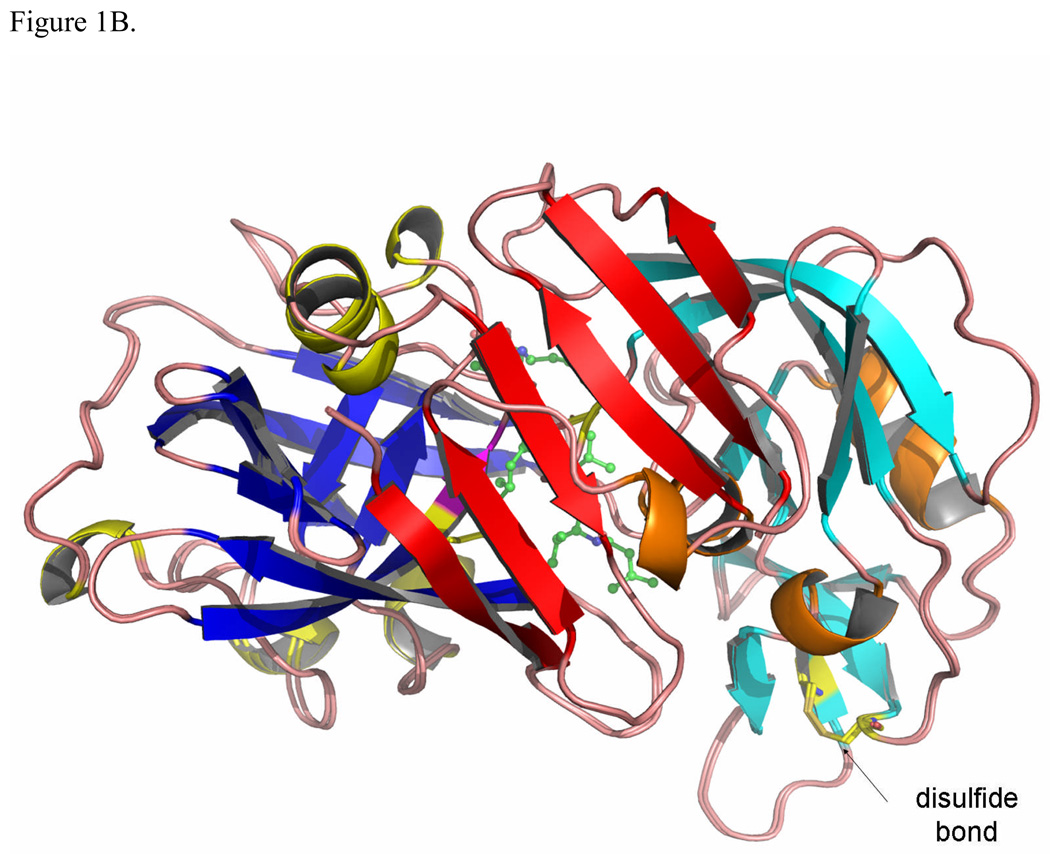
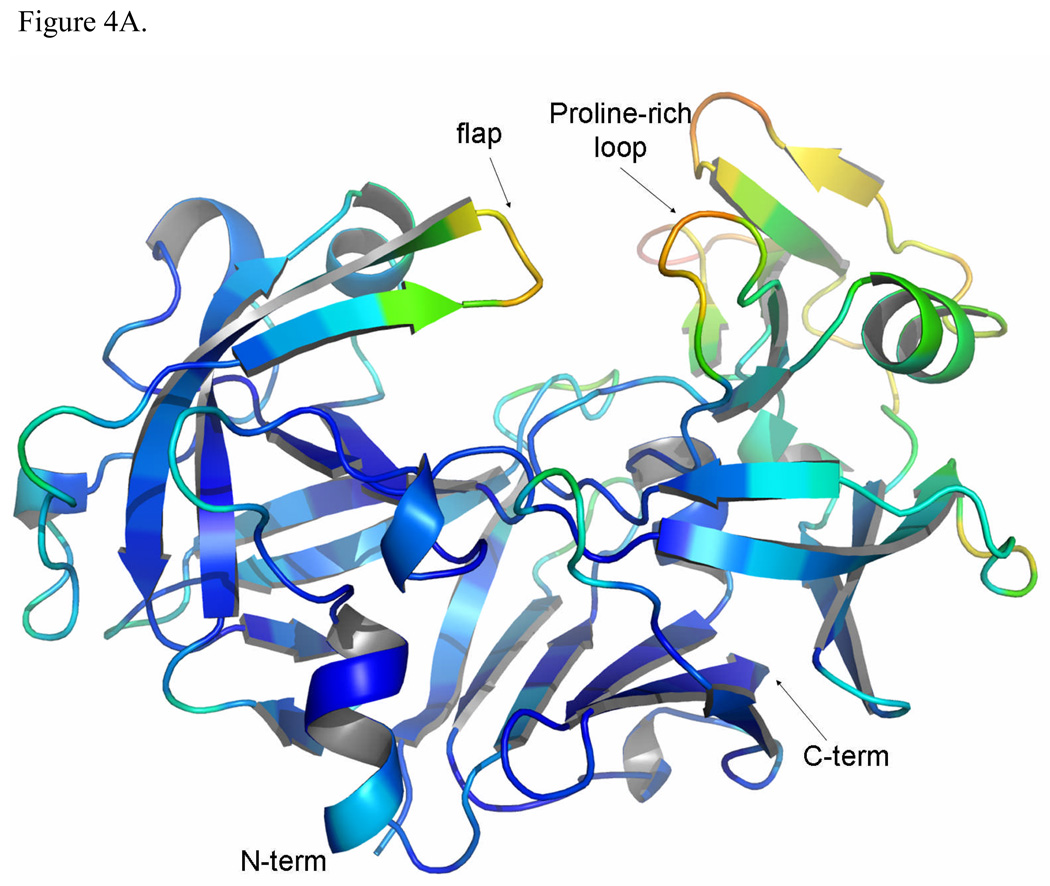
The overall structure of TrAsP is very similar to the structures of other pepsin-like aspartic proteinases and its description below will utilize the common system of numbering amino acids based on the sequence of porcine pepsin. The molecule contains twenty-five β-strands grouped into four β-sheets, connected by short α- and 310-helices, and by loop regions. The bilobal protein molecule has an approximate 2-fold symmetry, with the symmetry axis passing between the catalytic residues in the cleft between the two domains (Fig. 1). The N-terminal lobe includes residues from -2 to 171 and the C-terminal lobe, residues from 172 to 326, connected at the bottom of the active site cleft by a six-stranded, antiparallel β-sheet. Two catalytic residues, Asp32 and Asp215, are located in the middle of the cleft between the two lobes, at the ends of two ψ-like loops extending from each lobe (Fig. 1). Another loop forms a flap structure that protrudes from the N-terminal lobe and partially covers the catalytic site. It also makes several contacts with the pepstatin A molecule in the proteinase:inhibitor complex.
Figure 1.
The structure of TrAsP in cartoon representation, showing a superposition of the native and complexed structures. The characteristic structure elements are noted by different colors: blue β-strands and yellow helices define the N-terminal lobe and cyan β-strands and orange helixes define the C-terminal lobe. Each element in the N-terminal lobe is mirrored by a similar element in the C-terminal lobe. At the bottom of the molecule, represented in red, each lobe contributes three β-strands to form a six-stranded β-sheet that sometimes is called a third domain in addition to the N- and C-terminal lobes.7 (A) View of the structure looking “end-on” to the active site with pepstatin bound. (B) View from the “bottom” of the molecule with the structure rotated 90° front-to-back to illustrate the six-stranded β-sheet. The flap, polyproline loop and catalytic dyad are indicated.
The main chain of TrAsP contains two cis-peptide bonds, one located between Thr22 and Pro23 and another between Arg132 and Pro133. The first cis-Pro, on the tip of a conserved VIb β-turn, is a common feature of many proteins that belong to the AP family, whereas the second one seems to be specific only to fungal enzymes, such as TrAsP or endothiapepsin. There is a single disulfide bridge, between Cys250 and Cys283 in the C-terminal lobe of TrAsP molecule, also present in some others fungal APs. Another common feature12 of the AP:inhibitor complexes is an inverse γ-turn that occurs in pepstatin A, with a hydrogen bond between the CO and NH of the two statine residues occupying the positions P1 and P2’, respectively.
Conformational changes upon pepstatin A binding
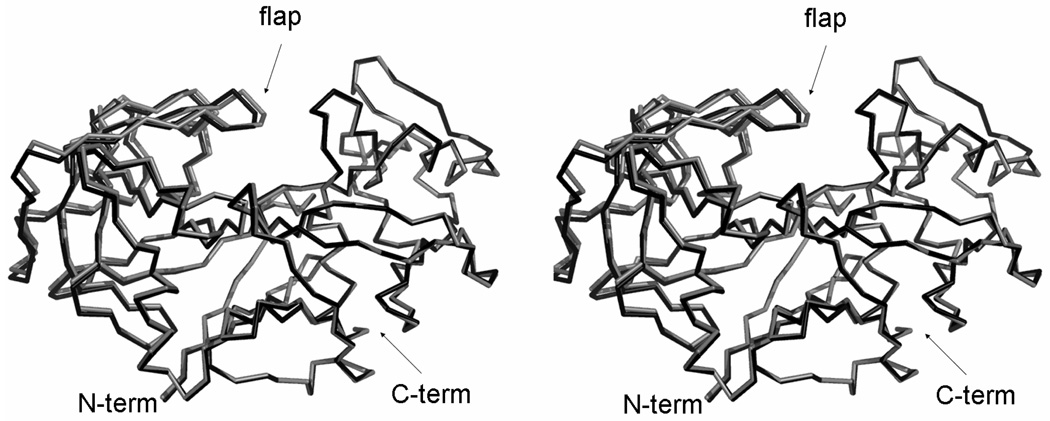
A comparison between the native form of TrAsP and the pepstatin A complex shows that the inhibitor binds in the active site cleft between the two lobes, and that the residues in the binding site shift away slightly to accommodate the inhibitor. The r.m.s. deviation between Cα positions in TrAsP and in the TrAsP:pepstatin complex, calculated with program SSM,13 is 0.32 Å for all 329 residues (Fig. 2).
Figure 2.
Superposition of the uncomplexed (black) and complexed (gray) traces of TrAsP, based on the alignment of the Cα atoms in the C-terminal domain (right) of the protein. Cα traces of both the apo- and holo- structures are shown in a stereoview. Superposition was performed with the program LSQKAB from the CCP4 suite.47
The enzyme embraces the inhibitor very tightly upon complex formation. This conformational change can be described in terms of a rigid body rotation of two parts of the enzyme defined by visual inspection, the first one formed by the residues -2–189 and 306–326, and the second comprising the residues 190–305. Therefore, the first rigid body is formed by both the N-terminal lobe and the central motif, whereas the second rigid body is formed by the C-terminal lobe. Superposition of the N-terminal lobes yields an r.m.s. deviation of 0.21 Å, whereas the corresponding r.m.s. deviation for the C-terminal lobe is 0.13 Å. Although small, these values are significantly lower than the value for the whole molecule, given above. The relative movement of the rigid bodies results in modest tightening of the binding cavity; the distance from Cα of Gly76, at the tip of the flap, to Cα of Ile297 lying opposite to Gly76 in a proline-rich loop (amino acid residues 294 to 304) decreases by about 1 Å. This rigid body movement becomes more evident when the residues from only one rigid body are superimposed, as shown in Fig. 2.
Apart from the conformational differences due to the rigid body movement, no other significant conformational changes occur upon inhibitor binding in TrAsP. This applies to the flap region and all other residues that are in contact with pepstatin A (Fig. 2, Fig. 3).
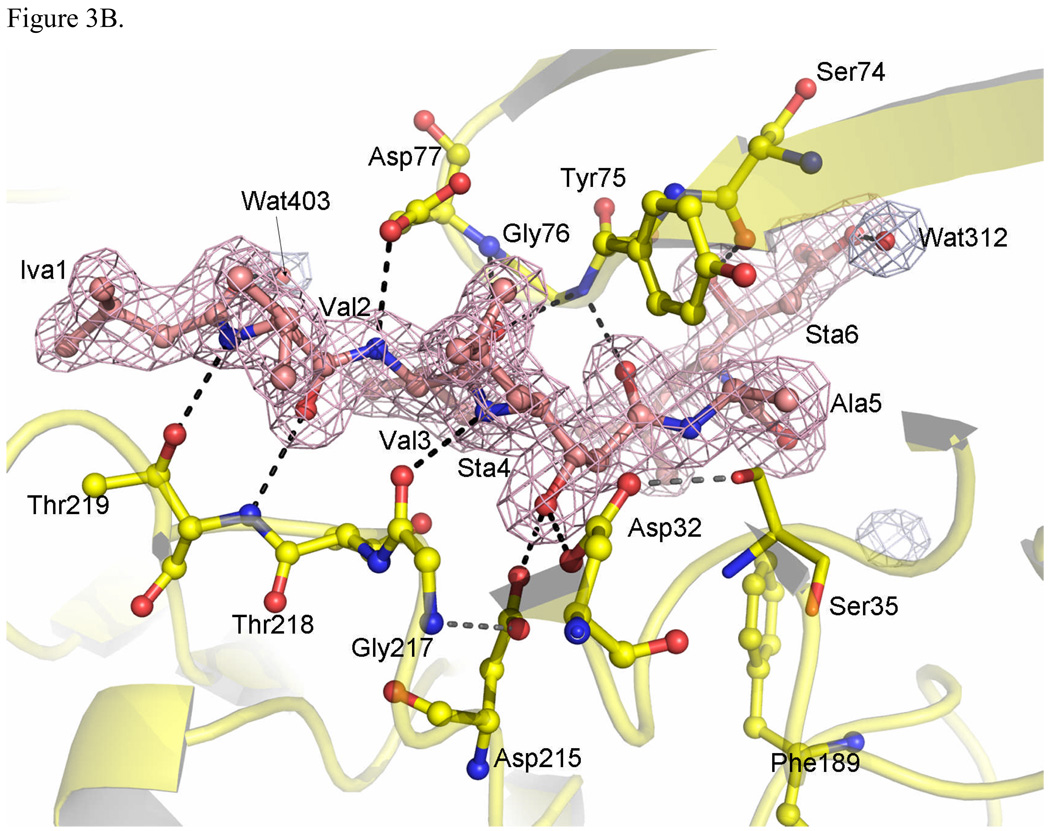
Figure 3.
Active site of TrAsP. (A) Active site region in the apoenzyme, with the residues shown in blue, and the electron density (2mFobs-DFcalc, ϕcalc) of the bound water molecules in blue mesh, contoured at 1.0σ. (B) Residues that make contacts with pepstatin A in the complex are shown in yellow and the electron density omit map of pepstatin in orange. The electron density omit maps (2mFobs-DFcalc) were calculated using the program OMIT from the CCP4 suite,47 and were contoured at 1.5 σ.
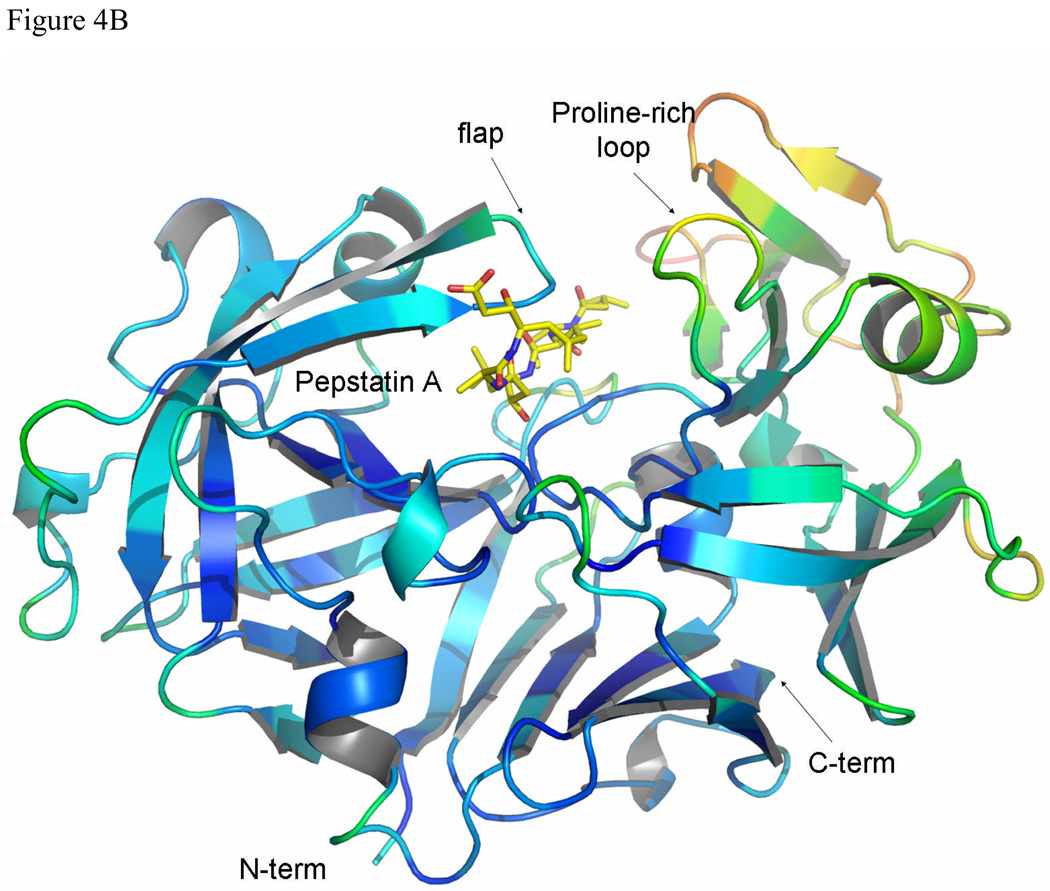
Temperature factors
The average temperature factors of TrAsP do not change significantly upon inhibitor binding (Table 1 and Fig. 4), with the exception of pronounced decrease in the mobility of the flap residues in the complex. The average temperature factors for the atoms of the flap (residues 74 to 80) are 31.3 Å2 and 19.9 Å2 for the free and complexed TrAsP, respectively. Mobility of the flap is reduced upon inhibitor binding by extensive hydrogen bonding and van der Waals contacts with pepstatin. Asp77, at the tip of the flap, shows the largest mobility changes. The neighboring residues, Gly76 and Gly78, were proposed to serve as hinges for the motion of the tip in other APs.14 Mobility of the flap in the apoenzyme is needed for enhancing substrate/inhibitor binding, allowing positioning of the active side residues in the geometry optimal for catalysis.
Figure 4.
A comparison between the temperature factors in uncomplexed (A) and complexed (B) TrAsP, colored using the program PYMOL.50 The color range of the protein is from dark-blue for low temperature factor values in the more ordered regions to red for high temperature factor values in the less ordered regions. Pepstatin molecule is shown as sticks and is not color-coded according to its B-factors.
The average B-factors of the C-terminal lobe are significantly higher that those of the N-terminal lobe in both the native structure and in the enzyme:inhibitor complex (Fig. 4). The residues of the C-terminal lobe are more distant from the enzyme’s active site, and are also more exposed to the solvent, which probably explains their higher flexibility, considered to be a common feature of the AP family.15 This flexibility, among other factors, explains the range of conformational variability of the C-terminal domains observed in three-dimensional structures of the members of AP family.
In addition, the structural flexibility of the C-terminal domain probably facilitates a rigid body movement involved in the catalytic function of APs. Data from kinetic studies involving aspartic proteinases16,17 have demonstrated that several steps during catalysis involve conformational changes of the protein, such as opening of the binding cleft to the entry and positioning of inhibitor/substrate. It has been suggested15 that the structural flexibility of the C-terminal domain plays an important role in the function of aspartic proteinases. Since part of the binding energy should be spent on distorting the enzyme, the structural flexibility would reduce the required binding energy, thus facilitating the process.
Active site
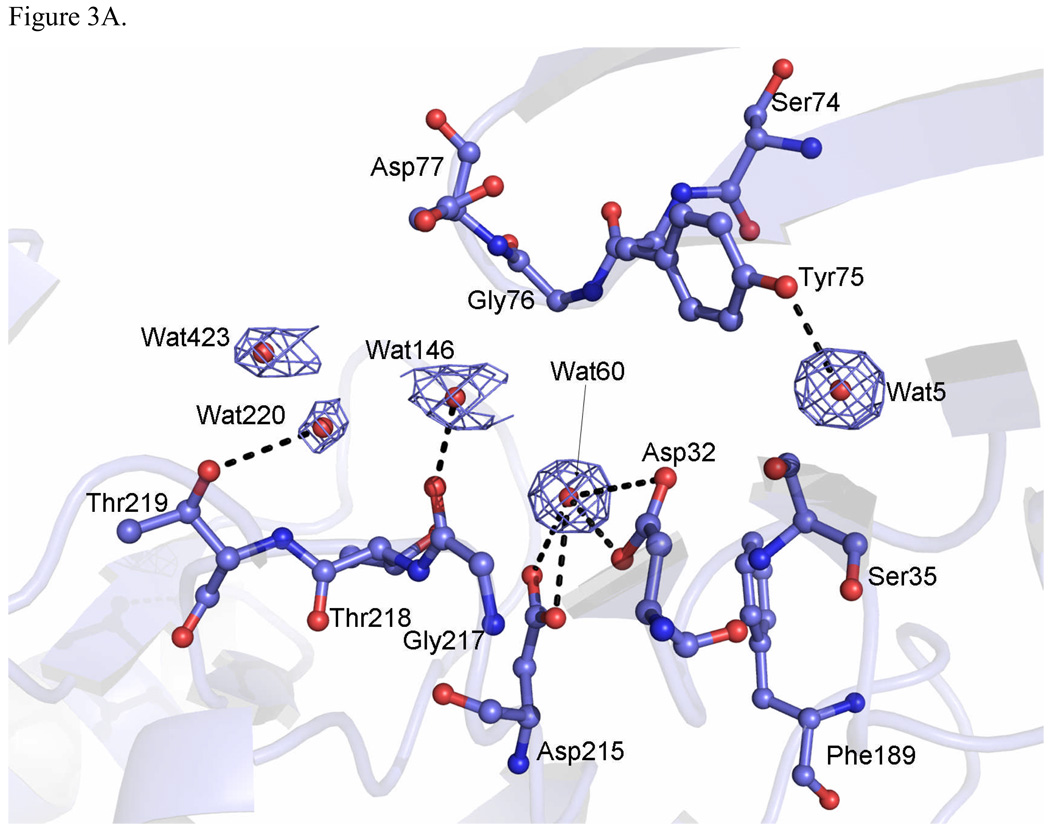
The active site, located between the two lobes of the molecule at the bottom of a large cleft, is one of the most highly conserved regions among the AP family.8 The carboxyl groups of two catalytic aspartates, Asp32 and Asp215, are held almost coplanar by a hydrogen bonding network which involves main-chain and conserved side-chain groups.
In uncomplexed TrAsP, a water molecule (Wat60) is tightly bound to both aspartate carboxyl moieties by several hydrogen bonds (Fig. 3A). The distances between the solvent molecule and these Asp residues are 3.45 Å to Asp32Oδ1, 2.94 Å to Asp32Oδ2, 3.04 Å to Asp215Oδ1, and 2.87 Å to Asp215Oδ2. This is a conserved water molecule observed in other native aspartic proteinases that has been implicated in catalysis. Upon substrate binding, this water molecule is partially displaced and polarized by one of the aspartate carboxyls and may then be involved in a nucleophilic attack on the carbonyl carbon atom of the peptidic scissile bond (P1-P1’) to form a tetrahedral intermediate, which is bound non-covalently to the enzyme.18 In the proposed mechanism, the tetrahedral intermediate is stabilized by hydrogen bonds to the negatively charged carboxyl of Asp32. Fission of the scissile main-chain C-N bond is accompanied by transfer of a proton to the leaving amino group, either from Asp215 or from bulk solvent.
Pepstatin A is a peptide-like inhibitor containing six amino acid residues (isovaleryl-L-valyl-L-valyl-L-statine-L-alanyl-L-statine or Iva1-Val2-Val3-Sta4-Ala5-Sta6). It is produced by Streptomyces and contains two residues of statine (an unusual amino acid (3S,4S)-4-amino-3-hydroxy-6-methylheptanoic acid).19 The inhibitory potency of pepstatin A toward aspartic proteinases has been attributed to the presence of the central Sta4 residue which contains a hydroxyethylene analog (-CHOH-CH2-) that could mimic the tetrahedral transition state in place of the scissile peptide bond (Fig. 3B). In the TrAsP:pepstatin complex this hydroxyl replaces the catalytic water molecule (Wat60) found at the active center of the native enzyme, and forms short hydrogen bonds with the inner carboxyl oxygen of Asp32 and the outer carboxyl oxygen of Asp215. The other two hydrogen bonds to the Sta4 hydroxyl group involve the outer carboxyl oxygen of Asp32 and the inner oxygen of Asp215, but both have unfavorable geometry with donor-acceptor distances too long and are very weak. Furthermore, other neighboring residues also are involved in short hydrogen bonds with the catalytic residues, as depicted in Fig. 3B.
The TrAsP-pepstatin A interactions
The inhibitor subsites have a generally hydrophobic character (Fig. 3), thus hydrophobic contacts are expected: P1–Sta4 forms hydrophobic interactions with Tyr75, Leu120, and Phe111, the inhibitor P2–Val3 side chain interacts with Leu222 and Ile297, and the P2’–Sta6 side chain interacts with Phe189, Ile297, and Ile299. The inhibitor P3–Val2 and P1–Sta4 side chains are closely packed against each other. The inverse γ-turn involving both the pepstatin Sta residues changes the direction of the inhibitor chain, leading the P3’–Sta6 toward the protein surface. As a result, the backbone of the P2’–Ala5 and P3’–Sta6 residues deviates from the regular extended conformation. The side chain of P4–Iva1 also points toward the molecular surface. Both the P4–Iva1 and P3’–Sta6 sites at the inhibitor extremities have more contacts with solvent molecules.
Pepstatin A forms thirteen hydrogen bonds to the enzyme and six to water molecules (some of them marked in Fig. 3B). In the N-terminal part of the inhibitor, the carbonyl oxygen of the P4-Iva1 is hydrogen bonded to two water molecules Wat331 and Wat357. The P3-Val2 participates in two hydrogen bonds to Thr219 residue: the amide nitrogen atom of P3-Val2 is hydrogen bonded to the side-chain oxygen atom of Thr219 and the carbonyl oxygen atom of the P3-Val2 interacts with the amide nitrogen of Thr219. The carbonyl oxygen atom of the P3-Val2 is also hydrogen bonded to the water molecule Wat201. The P2-Val3 is involved in three hydrogen bonds to the flap residues: the side-chain oxygen Oδ2 of Asp77 is hydrogen bonded to the amide nitrogen atom of P2-Val3, the amide nitrogen from this same residue participates in a rather weak hydrogen bond interaction with the carbonyl oxygen atom of P2-Val3 and the same carbonyl oxygen is hydrogen bonded to the amide nitrogen atom of Gly76.
The flap is an important region defining the specificity of APs. For example, most aspartic proteinases have a preference for cleavage of covalent bonds between two hydrophobic amino acids, but fungal peptidases have the ability to activate trypsinogen by cleaving a bond after a lysine residue in the P1 position. Site-directed mutagenesis has been used to prove that the presence of a conserved aspartic acid residue in the flap (Asp77 in TrAsP) is essential for cleavage of the Lys-containing substrates.19
In the central part of the inhibitor, the residue P1–Sta4 is involved in four hydrogen bonds: the amide nitrogen of P1–Sta4 is hydrogen bonded to the carbonyl oxygen of Gly217, the central hydroxyl of P1–Sta4 makes one hydrogen bond with each catalytic aspartate (Asp32Oδ2 and Asp215Oδ1), and the carbonyl oxygen of P1–Sta4 is hydrogen bonded to the amide nitrogen of Gly76. Finally, in the C-terminal part of the inhibitor, the amide nitrogen of P2’–Ala5 is hydrogen bonded to the carbonyl oxygen of Gly34 and the residue P3’–Sta6 participates in four hydrogen bonds: statine hydroxyl is hydrogen bonded to the carbonyl oxygen of Ser74 and to the water Wat420, while the carboxylate is hydrogen bonded to waters Wat141 and Wat312.
Among all the observed hydrogen bonds, those formed by the catalytic aspartates in the TrAsP:pepstatin complex structure are of particular interest. As shown in Table 2, the donor-acceptor distance for one of them is short (less than 2.6 Å), two are typical hydrogen bond distances, and one is long. The presence of short hydrogen bonds in the APs:inhibitor complexes has been supported by X-ray studies at atomic resolution and by NMR and neutron diffraction studies.20,21 Short hydrogen bonds (2.4–2.6 Å) are also known as low-barrier hydrogen bonds (LBHB), since the proximity of the donor and acceptor atoms reduces the energy barrier which normally prevents proton transfer from the donor to the acceptor group.22 Thus, rapid exchange of a proton between the donor and acceptor atoms can occur, and this has been proposed as an important effect in the catalytic mechanisms of a number of enzymes. The studies indicate that low-barrier hydrogen bond formation is due to steric compression upon inhibitor binding. This is corroborated by the observations that: (1) the short hydrogen bonds are absent in the native structure where a water molecule is bound to both carboxylates and (2) after inhibitor binding, the two lobes of the enzyme undergo a relative rotation that compresses the substrate-binding pocket.
Table 2.
Interaction distances between the catalytic aspartates in TrAsP and pepstatin A
| TrAsP | Distance | Pepstatin A |
|---|---|---|
| Asp32 Oδ2 | 3.40 Å | STA4 OH |
| Asp32 Oδ1 | 2.69 Å | STA4 OH |
| Asp215 Oδ2 | 2.53 Å | STA4 OH |
| Asp215 Oδ1 | 3.03 Å | STA4 OH |
The hydrogen-bonding pattern present in the TrAsP:pepstatin A complex is consistent with the network of active site hydrogen bonds found in the previously reported complexes of aspartic proteinases with statine-based inhibitors.21 This implies that the outer oxygen atom of Asp215 is protonated when the inhibitor is bound at the active site of TrAsP and, consequently, Asp32 is negatively charged, which is in agreement with the transition state of the catalytic mechanism proposed for aspartic proteinases.
Comparisons with other fungal aspartic proteinases
A fungal aspartic proteinase with known structure that is closest in its amino acid sequence to TrAsp is endothiapepsin. The primary structures of these two APs share 61% identity, and most of the remaining differences in their sequences are quite conservative. A number of structures of endothiapepsin, both as apoenzyme and complexed with different inhibitors, have been published. For the purpose of a comparison of the apoenzymes, we have selected the highest-resolution structure, refined at 0.9 Å (PDB code 1OEW20). The structure of a complex of endothiapepsin with pepstatin has also been published, at the resolution of 2.0 Å (PDB code 4ER223). Although these structures were refined with different protocols and at vastly different resolution, they are very similar, with an r.m.s. deviation between Cα positions of only 0.23 Å. Despite this very low deviation, a subtle motion of the domains can still be detected when the N-terminal and C-terminal lobes of the apo- and inhibited enzyme are superimposed, as discussed above for TrAsP.
Superposition of the apoenzyme structures of TrAsP and endothiapepsin results in an r.m.s. deviation of 0.91 Å for 327 Cα pairs, whereas the comparison of the pepstatin complexes results in an r.m.s. deviation of 0.89 Å. The largest differences between the structures are found in the loop that contains residue 318, where endothiapepsin has a single-residue insertion compared to pepsin and TrAsp. Other differences between the structures are seen in the loops 195–205 and 239–246, both distant from the active site. The conformation of pepstatin is virtually identical in the area that interacts with the catalytic aspartates, and differs only at both termini. A significant rearrangement of the isovaleryl side chain on the N terminus may be due to the presence of a much smaller Leu222 in TrAsP, compared to Tyr222 in endothiapepsin.
It has been postulated that water molecules play an important role in pepsin and pepsin-like enzymes activity, as reviewed by Andreeva and Rumsh.7 The similarity between the two fungal enzymes also extends to some of the bound solvent. For example, five buried water molecules (numbered 190, 211, 200, 194, and 234 in the TrAsP:pepstain complex) interacting with the conserved Tyr165, as well as with strands 12–15, 29–31, and 216–219 are present in almost identical positions in all four compared structures. However, some other comparatively inaccessible water molecules are different, for example in the vicinity of Leu316 in TrAsP, where this larger residue replaced a glycine present in endothiapepsin. It is thus not surprising that the enzymatic properties of these two highly homologous enzymes are not identical.
Another aspartic proteinase closely related to TrAsP is penicillopepsin, which shares with it 53% sequence identity. Not surprisingly, superposition of the Cα coordinates of the TrAsP:pepstatin complex and inhibited penicillopepsin refined at 0.89 Å resolution (PDB code 1BXO24) resulted in slightly higher r.m.s. deviation of 1.05 Å. The largest differences were observed for the loops 7–12 and 278–282B, due to a deletion in the penicillopepsin sequence. Other loops exhibiting significant differences were 195–205 and 239–246. All of these loops are distant from the substrate-binding site, the area where the two structures are most similar, including virtually identical conformation of the flap. Structural similarities also extend to the solvent structure, including the presence of the five water molecules mentioned above in approximately the same positions.
The amino acid sequences of fungal aspartic proteinases are more distant from those of the mammalian enzymes, with TrAsP sharing only 30% identity with human pepsin. This evolutionary distance is reflected in much larger r.m.s. deviation between the coordinates of TrAsP:pepstatin and of the inhibited human pepsin refined at 1.93 Å resolution (PDB code 1QRP25). These two sets of coordinates superimpose with an r.m.s. deviation of 1.38 Å for only 219 Cα pairs, with comparatively large rearrangement of the surface areas, but with much less deviation in the areas of the active site. Interestingly, of the five water molecules strictly conserved in fungal aspartic proteinases, only an equivalent of Wat200 is found in human pepsin, most likely due to the presence of a valine rather than a tyrosine in position 165.
Statistical coupling analysis of the aspartic proteinase family
It is clear that pairwise comparisons of the structures of aspartic proteinases are capable of yielding only a limited picture of the global conservation and evolutionary differences within the family. A recently introduced method termed Statistical Coupling Analysis (SCA)26 has been successfully used to delineate the similarities in several other large families of proteins, but has not been applied as yet to aspartic proteinases. Using TrAsP as a structural template, we performed SCA in order to gain better insight into structural and functional interactions between amino acid residues within the family of aspartic proteinases. SCA is a sequence-based analysis that assesses evolutionary conservation and mutual correlations in a multiple sequence alignment for a given protein family. It assumes that protein structure and function evolve over a long period of time in a large-scale, random mutagenesis process constrained by natural selection.26 In a multiple sequence alignment, if the sequence space for the protein family is large enough to be representative of the amino acid distribution found in Nature, one would expect the positions restrained by both structure and/or function to reveal increased conservation. Furthermore, functional and/or structural coupling between two positions in a protein sequence would lead to significant correlations in the distribution of amino acids for these positions in the multiple sequence alignment (MSA).
The approach developed to evaluate site conservation (ΔGstat) is based on the sum of individual amino acids binomial probability, given their “natural probabilities”, computed assuming a Boltzmann distribution.26,27 The natural probabilities are estimated for each protein family taking the amino acid frequencies found in the entire MSA to account for the family-specific amino acid frequencies (e.g., a protein family that exhibits various conserved disulfide bridges is expected to have an increased content of cysteines). The couplings between two positions, i and j, are evaluated using the same concepts, upon which a perturbation is introduced in position i, i.e., a subset of the alignment is chosen so that all sequences have a given amino acid in the i position. The probabilities for all other j positions are then re-evaluated for the subset and compared to the probabilities found in the entire dataset. Large changes in the amino acid probabilities in all j positions caused by the perturbation in i position are indicative of an evolutionary coupling between the corresponding amino acid residues. Importantly, the perturbations cannot be applied in either the completely random, or fully conserved positions of MSA. In the first case, the subset of MSA corresponding to a given perturbation in the non-conserved position i would not result in a statistically significant number of sequences and therefore will not be useful in the analysis. Perturbation in the fully conserved position will return the initial MSA, and, hence, would also be uninformative for the SCA study. In practice, only partly conserved positions that would permit perturbations leading to statistically significant subsets of sequences could result in significant statistical variations of the frequency distributions in other amino acid positions and would be able to produce strong ΔΔGstat signals. This does not mean that strictly conserved positions, with a very high ΔGstat values, are not correlated with any other positions within the given MSA. The significance of non-permissiveness of perturbation in the completely conserved positions is in the correlation of the latter positions with all the other amino acid residues of the protein through strict requirement of these completely conserved amino acid residues for protein folding and/or function.
SCA technique has its limitations. For example, it does not reveal the physical reasons for coupling between amino acids that should be inferred and comprehended on the basis of site-directed mutagenesis experiments and functional studies. It also depends on the quality and completeness of the protein sequence alignment used for the particular study, i.e. on the statistical robustness of the initial alignment. Data that are poorly distributed in a sequence space, incomplete, or badly sampled can seriously skew the results of the SCA and bias the ΔGstat and ΔΔGstat calculations. For that reason, absolute values of these variables should be taken with certain degree of caution and the metrics and methods used in SCA for clustering of the amino acid couplings undergo constant improvements. However, given well-defined MSAs, statistical coupling calculations performed previously for several other protein families yielded very interesting results and conclusions about clusters of functionally important amino acid residues.26–30
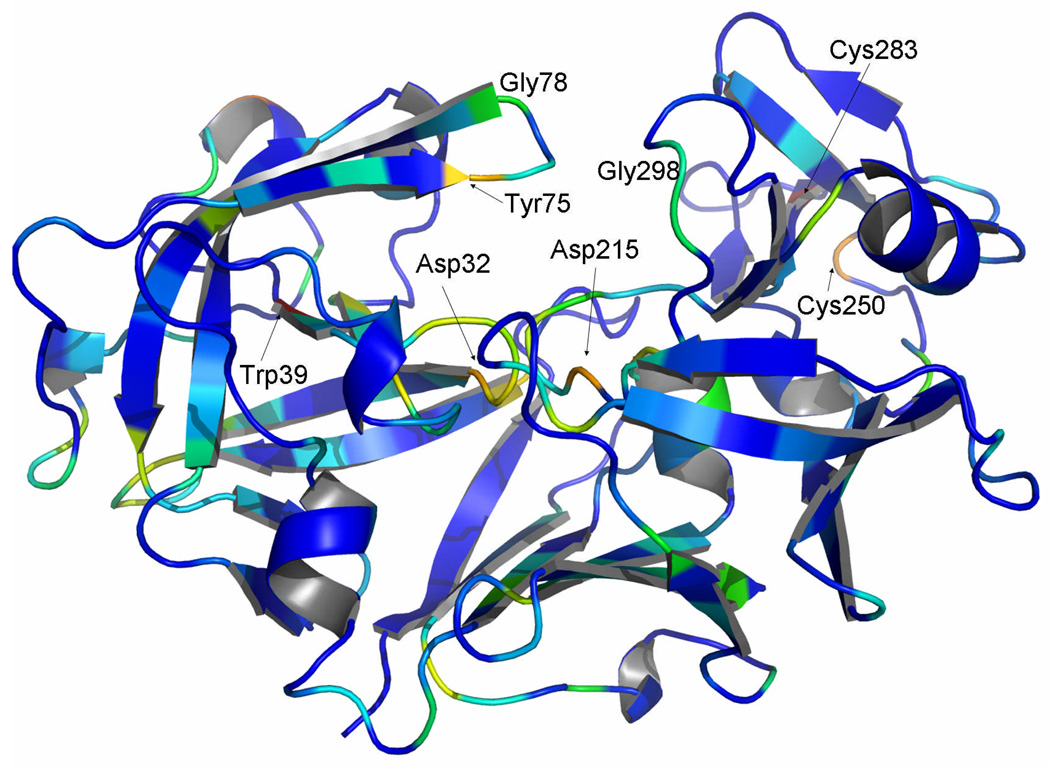
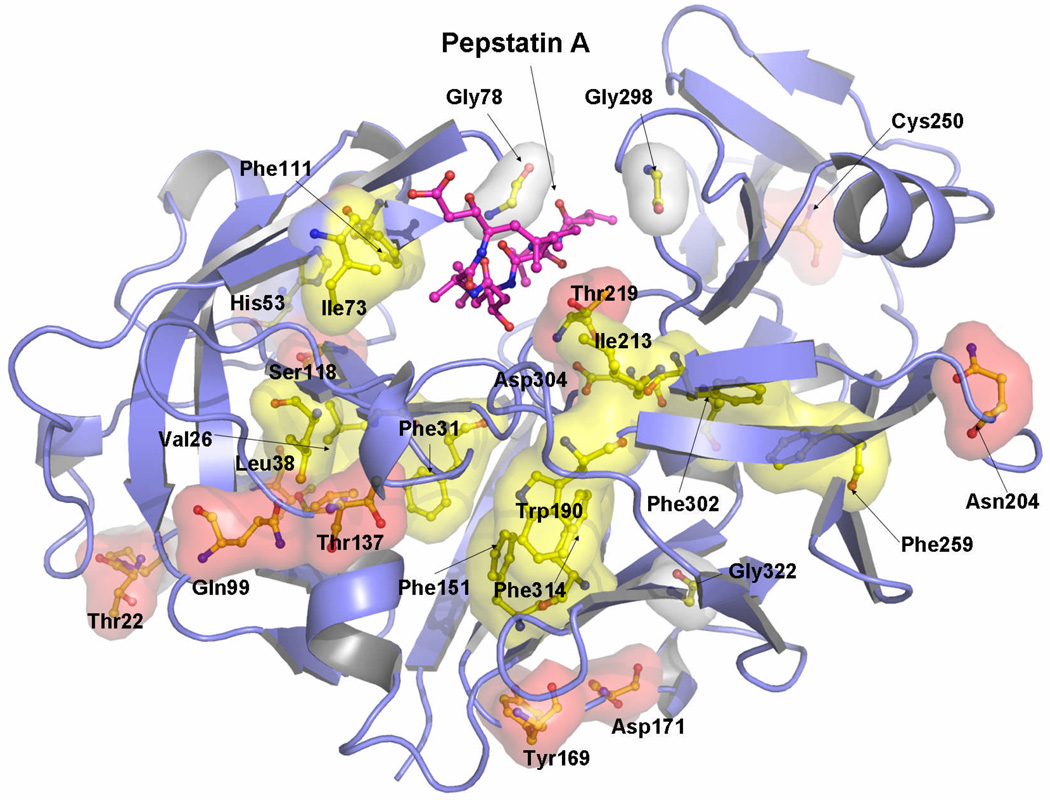
Following these principles, statistical energy of conservation (ΔGstat)26 has been calculated for the pepsin family, revealing evolutionarily conserved sites, some of them remote from the enzyme active site (Fig. 5). Several amino acid positions within the β-sheet region of both domains display almost total conservation, such as Cys283 and Cys250, which form a disulfide bond that stabilizes the protein31,32 and, in addition, is likely to be an important factor in the process of protein folding33. Another amino acid residue exhibiting very high ΔGstat values is Trp39. In addition, the two active site aspartic acids, Asp32 and Asp215, and the amino acid residues located in two loop regions (Tyr75, Gly298, and Gly78, located in a hinge of the flap region) are also highly conserved and have high ΔGstat values (Fig. 5). The hydroxyl group of Tyr75 and Trp39Nε1 are at 2.78 Å distance in TrAsP and involved in forming a hydrogen bond. Interaction between Trp39 and Tyr75 side chains stabilizes the flap conformation in the presence and in the absence of pepstatin, forming a cap above the hydrophobic core of the N-terminal lobe. The hydrophobic core of the N-terminal domain of TrAsP is a driving force for folding of proteins34,35 and, as alluded to below, is essential for the conformational flexibility of APs. Gly78 forms a hinge of the flap region and is crucial for protein mobility and function while Gly298 is adjacent to the proline-rich loop and is also important for protein conformational dynamics. High conservation of these glycine residues, together with the total conservation of catalytic aspartates in the proteolytically active members of the family, are indicative that the dynamics and the conformational flexibility of the flap region and of the proline-rich loop are essential for the AP catalytic activity and function.
Figure 5.
Crystal structure of TrAsP shown in a cartoon representation, colored by ΔGstat as computed from SCA.26 Evolutionary conservation is represented on a scale ranging from dark blue (poor conservation) to red (conserved positions). Some evolutionarily highly conserved positions are labeled.
Although ΔGstat is a very good indicator of the amino acid conservation at a given position within the alignment, the absolute values of ΔGstat should be interpreted with caution, since they are influenced by the general statistics of the sequence data set and the presence of the mutated and partially sequenced proteinases in the PFAM server. Furthermore, SCA relies on the “natural distribution” of amino acids in the ΔGstat computation, which increase the likelihood that the least frequently found amino acids (Cys, Trp, Tyr) will score relatively high in ΔGstat anaysis.
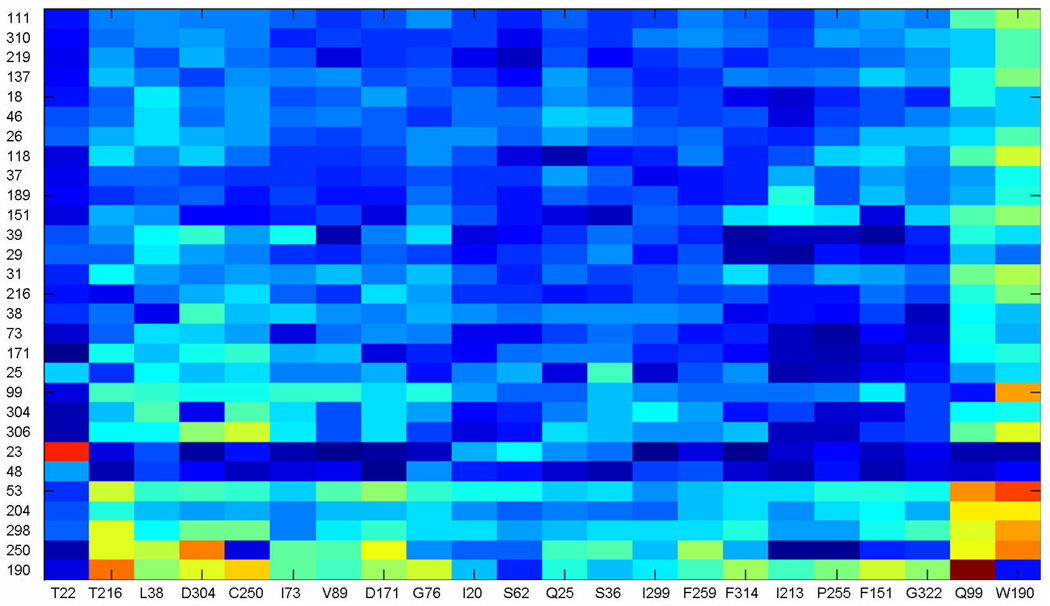
To define statistical correlations between amino acid appearances in particular positions within the aspartic proteinase family, we computed co-variations of these positions described by ΔΔGstat.26 An analysis of ΔΔGstat computed from our sequence alignment revealed a number of coupled positions, shown in a matrix form in Fig. 6. Strikingly, mapping of the statistically coupled amino acid residues onto 3D structure of TrAsP revealed two major clusters, one in each lobe of the protein, which form a continuous network within the protein molecule (Fig. 7). Each cluster emerges from a separate protein domain and merges with the other one in the area of the active site.
Figure 6.
SCA matrix after cluster analysis. For each possible perturbation ΔΔGstat were calculated in each position in the MSA, according to the defined criteria (see Materials and Methods), which generated a NxM matrix, where the N are the number of perturbations and M the number of positions in the MSA. The initial matrix was used in iterative rounds of cluster analysis in MATLAB. In each round, the positions in the matrix with significant ΔΔGstat values were kept and the ones with small signal, discarded. The final matrix is shown, with the perturbations given as columns and positions of the MSA as rows. The color scale range from dark blue for small values to bright red for high ΔΔGstat values.
Figure 7.
Coupled positions maped onto the crystal structure of TrAsP. The residue surfaces were computed and showed using the program PYMOL.50 Hydrophobic residues are shown in yellow surface, polar residues in red, a histidine (His53) in blue, and glycines and prolines in white surface.
A closer inspection of the coupled locations indicated by SCA calculations reveals a number of positions known to play a role in protein activation. The strong coupling between positions 22 and 23 (Thr22 and Pro23 in TrAsp) pinpoints a cis-Pro bond, highly conserved in the aspartic protease family. It is known that cis-Pro bonds, found mostly in β-turns, are important for protein folding and stability, and their removal strongly affects protein stability36 and function.37 A comparative analysis of the cis-peptides within the PDB shows that the presence of a cis-bond implies certain restrictions on the chances of occurrence of the particular amino acid in the preceding position38 and thus turns itself detectable by SCA. In agreement with the method of analysis, the second cis-bond (Arg132-Pro133), specific for TrAsP and other fungal proteinases, did not appear in the final SCA matrix (Fig. 5). Interestingly, Ser36, another residue which appeared in the final SCA matrix (Fig. 5), interacts via its carbonyl oxygen atom with the amide nitrogen of Thr22, whereas the carbonyl oxygen of Pro46 makes a hydrogen bond with the amide nitrogens of Ser 48, Ser49, and Ala50.
In the flap of the N-terminal lobe, a hydrophobic isoleucine residue (Ile73) is strongly coupled with several hydrophobic and aromatic positions in the active site pocket (Leu38 and Trp39). A similar position in the C-terminal lobe (occupied by Ile299), sometimes called the second flap, co-evolved with a polar aspartic acid (Asp304) in the active site. Importantly, Gly298 and Ile299 of the second flap region reveal a significant degree of evolutionary coupling with the catalytic site. Moreover, two hinge residues of the flaps, Gly76 (N-terminal) and Gly298 (C-terminal), which are important to the flap movement, also appear in our analysis. Gly298, in particular, shows an evolution profile coupled to Thr216 and Asp304, two residues that play a direct role during the process of catalysis by orienting and positioning the substrate in the catalytic cleft in a conformation appropriate for the enzymatic reaction. Furthermore, the side chains of Phe189, Ile299, and Phe111 which appear in SCA (Fig. 6) form direct interactions with P3′-Sta6, P1-Sta4, and P3-Val2 of pepstatin A, respectively. The fact that the active site residues show strong statistical coupling with the hinge region is consistent with the “induced fit” mechanism of enzymatic catalysis and implies a necessity of simultaneous conservation of these parts of the protein. Co-evolution of the residues in the binding site and in the hinges of the flaps that is suggested by SCA makes it tempting to speculate that binding sites for the substrate and conformational adjustments of the flap regions have probably developed simultaneously during the process of evolution of the aspartic proteinase family. Consistently with the functional and structural studies, this result suggests that the catalytic mechanism of this class of proteinases employs induced fit.
Another interesting feature resulting from the ΔΔGstat analysis is the presence of hydrophobic cores consisting of coupled residues in both protein lobes. In the N-terminal lobe, the residues Val18, Ile20, Val26, Leu29, Phe31, Leu38, and Val89 form a hydrophobic cluster covered by the β-sheets, whereas Phe151, Trp190, Ile213, Phe259, Ala306, and Phe314 form a larger cluster in the C-terminal lobe. Both clusters are buried and somewhat distant from the active site, i.e., none of the residues directly interact with pepstatin A in our crystal structure (Fig. 7). The same feature was observed in the related HIV-1 aspartic proteinase (HIV-1 PR), studied by molecular dynamics simulation39 and NMR spin relaxation.40 These studies of HIV-1 PR revealed that a number of buried hydrophobic residues could slide one over the other and that this “hydrophobic sliding” was necessary for the flap movements and, as a consequence, for the catalytic activity.39 Moreover, these authors demonstrated that conservative mutations in this core, i.e., mutations that still preserve its hydrophobic characteristics, maintained the enzymatic activity. This evidence provided further support to the relevance of the physicochemical properties of the residues found in the hydrophobic clusters for AP function. Ishima et al.40 observed that hydrophobic cores are present not only in HIV-1 PR but also in related simian immunodeficiency, Rous sarcoma, and equine infectious anemia virus proteinases, and proposed a similar feature for the human T-cell leukemia virus proteinase, the structure of which was not yet determined when the study was performed. The authors, however, failed to identify a similar hydrophobic core in eukaryotic aspartic proteinases, presumably because of their low sequence homology to viral proteinases32. In line with the results of Foulkes-Murzycki and coworkers,39 they also observed, using NMR spin relaxation technique, that the dynamic properties of the hydrophobic clusters should be necessary to accommodate structural perturbations caused by substrate binding.40
Moreover, the two clusters are involved in several interactions. For example, the side-chain oxygen Oδ1 of Thr216 is hydrogen bonded to both the amide nitrogen of Thr33 and the carbonyl oxygen of Phe31 from the N-terminal lobe hydrophobic cluster, whereas the amide nitrogen of Thr216 interacts with the side-chain oxygen Oδ1 of Asp304, which is involved in orientation of the pepstatin A in the active-site cleft. At the same time, the side-chain oxygen Oδ1 of Thr33 is at a hydrogen-bonding distance from both the amide nitrogen of Thr216 and the carbonyl oxygen of Ala214. Thr33, in its turn, forms a water-mediated contact with the carbonyl oxygen of Trp190 from the second, C-terminal, hydrophobic cluster. Trp190 also co-evolved with His53; the side chain of the latter residue is engaged in hydrogen bond interactions with the carbonyl groups of Phe111, Val112, Asp114, and Ile117. As mentioned before, Phe111 is involved in hydrophobic interactions with P3-Val2 of pepstatin A. All three amino acid residues, Thr216, Trp190 and His53, are strongly coupled (Fig. 6). Furthermore, Trp190 of the second hydrophobic cluster and Cys250, which participates in a highly conserved disulfide bond with Cys283, display significant coupling with the N-terminal flap residue Ile73. Statistical coupling of the amino acid residues involved in inhibitor binding and recognition, protein mobility, and hydrophobic clusters required for induced fit movements of the enzyme domains, are all consisted with the AP function.
In addition, all the residues that appear in Figure 6 and Figure 7 are implicitly coupled with the highly conserved residues essential for folding (Tyr75, Trp39, Cys250, and Cys283) and activity (Asp32, Asp215, Gly29, and Gly78), shown in Figure 5, most of which do not appear in the final SCA cluster (Fig. 6) due to their almost total conservation. Given the fact that APs cannot fold or function in the absence of these amino acid residues at the respective positions, they are necessarily correlated with all the rest of the statistically coupled positions. Some of the highly conserved amino acid residues, such as Cys250 and Trp39, are slightly less frequent in the MSA, and therefore appear in the final SCA (Figure 6), whereas others (Tyr75, Cys283, Asp32, Asp215, Gly29 and Gly78) are so highly conserved that the perturbations at the correspondent positions of MSA do not result in significant ΔΔGstat values.
The sequence-based analysis used in this work was proven to be very sensitive to evolutionary conservation patterns in protein families24–30 and was used to provide a better comprehension of the tertiary structure of TrAsP by means of primary structure analysis of the whole family, highlighting the importance of a concerted conformational movement of the flaps and the importance of hydrophobic clusters present in TrAsP structure for AP activity.
The results shown here indicate that the hydrophobic sliding mechanism may be a general dynamic mechanism utilized by the aspartic proteinase family and not limited to only retroviral proteinases. Since SCA is based on the relationships between distributions of probabilities of finding a given residue in an MSA position, it is likely that the chemical characteristics, rather than specific residues, are the main feature of the hydrophobic sliding, i.e., conservative mutations that keep the hydrophobicity and the internal van der Waals interactions are likely to preserve the dynamic properties of the system.
A number of other positions that appear in our coupling analysis within the range of ΔΔGstat values observed in the final SCA cluster, might be relevant to aspartic proteinases biology (Fig. 6). For some of them (such as, for example, Gln99, Gln204 or Ala310) the role in enzyme activation and function still is to be elucidated. The statistical analysis performed in present work serves as a tool for further investigation in this field. The SCA rationally provides a number of hot spots in the enzyme sequence and structure which are potentially relevant to protein function and dynamics. We also expect that such analysis could be useful for large scale mutational studies by providing a list of statistically relevant positions that could be mutated in order to elucidate the details of AP function and to develop more efficient enzymes that could be useful for industrial purposes.
Materials and Methods
Protein purification and crystallization
A commercial preparation of desalted and dried culture of Trichoderma reesei was used to isolate the aspartic proteinase. The dry powder was dissolved in 1L of 20 mM sodium acetate buffer, pH 4.1 at concentration near 100 mg/mL, centrifuged to remove insoluble material (3000g, 4 °C, 40 min), then concentrated 30 times and desalted using hollow fibers with exclusion limit of 10 kDa. Resulting solution was applied on the DEAE Sepharose FF column (20×200 mm) equilibrated with 20 mM sodium acetate buffer, pH 5.0, and the protein fraction was eluted with linear gradient (0–500 mM) of sodium chloride in the same buffer. The fraction with proteinase activity with the volume of 250 mL was concentrated using Amicon PM10 membrane to 10 mL and dialyzed against two 2 L changes of 50 mM sodium acetate buffer, pH 4.1. The resulting solution was applied on the TSK CM 5PW column (21.5×150 mm) and the protein was eluted with the 300 mL linear gradient (0–300 mM) of sodium chloride in the same buffer. Purified protein was dialyzed against water and lyophilized.
The activity of the proteinase was monitored during purification by hydrolysis of a fluorogenic peptide substrate O-aminobenzoyl-Ala-Ala-Phe-Phe-Ala-p-nitroaniline. The substrate was incubated with the proteinase in 30 mM sodium phosphate/citrate buffer, pH 3.0 at 37 °C. Fluorescence was measured with Hitachi F-4000 spectrofluorimeter, λex=290 nm, λem=340 nm.41 The protein was crystallized using the hanging drop method. Five to ten µL of 20 mg/mL solution of the proteinase in water were mixed with equal volume of 15% PEG3350 solution (Sigma) in 50 mM potassium phosphate buffer, pH 6.0–7.0 and equilibrated against 1mL of 20% PEG3350 in the same buffer. Bipyramidal crystals appeared after two hours and reached maximal size of 0.3×0.3×0.6 mm after two to four days. For co-crystallization with the inhibitor, enzyme was dissolved in 1% pepstatin A solution, incubated for at least 1 hour at room temperature, centrifuged, and then mixed with the precipitant.
Enzyme-inhibitor molar ratio was approximately 1:24. Addition of the inhibitor decreased the solubility of the proteinase and caused appearance of protein precipitate during incubation. Crystals of the complex had the same shape and significantly smaller size (0.1×0.1×0.1 mm).
Data collection
For data collection, a single crystal of either the apoenzyme or the pepstatin complex was quickly frozen in gaseous nitrogen at ∼90 K (Oxford Cryosystems). X-ray data were collected by the oscillation method on a MAR345 image plate detector at the LNLS protein crystallography beam line.42 X-ray wavelength was set to 1.50 Å to maximize the signal-to-noise ratio and to optimize the speed of data collection.43 The crystal-to-detector distance was set to 250 mm and the oscillation range was equal to 1°. X-ray diffraction data were processed using programs DENZO and SCALEPACK.44 The data collection and refinement statistics are presented in Table 1.
Structure solution and refinement
The structure of the native proteinase was solved by the molecular replacement technique using the program AMORE,45 with the structure of penicillopepsin (PDB code 1BXO24) providing the search model. Location of a single molecule present in the asymmetric unit was unambiguous. Positional and temperature factor refinement was initially performed with the program REFMAC46 from CCP4 suite47 and later with PHENIX,48 following standard protocols. The program O was used to visually analyze and rebuild the model.49 Water molecules were added according to the criteria that each must make at least one stereo-chemically reasonable hydrogen bond, and that it should be well defined in the (2mFobs -DFcalc) and (mFobs - DFcalc) electron density maps. Progress of refinement was monitored by the conventional and free R-factors and by inspection of difference electron density maps. The coordinates of the native enzyme were used directly to initiate refinement of the structure of the complex. That refinement followed the same protocols as the refinement of the apoenzyme. The refinement statistics and parameters of the final models are summarized in Table 1. Figures were drawn using the program Pymol.50
Statistical coupling analysis
To perform statistical coupling analysis we applied the method described by Ranganathan and coworkers,26–30 with minor modifications. 1,337 sequences of aspartic proteinases were downloaded from the protein family (PFAM) server (http://pfam.sanger.ac.uk/) and manually adjusted to improve alignment in less conserved positions; 1,207 sequences were used in the analysis.
The conservation criteria ΔGstat for a multiple sequence alignment (MSA) are defined as:
where Pix is the binomial probability of finding a given residue x in the i position in the MSA and PMSAx is the binomial probability of finding the residue x in the MSA. Following the improvements of the method,51 the frequencies of finding each residue in the MSA were computed directly from the alignment of aspartic proteinase family of enzymes.
A minimal size in the dataset for perturbation experiments was selected to guarantee the statistical equilibrium. For that purpose, we computed averages of ΔGstat values for the five less conserved positions and stepwise reduced the working dataset by randomly excluding the sequences. Analysis of the average of ΔGstat for the least conserved positions versus dataset size, defined a minimal size for the subset for ΔΔGstat calculations to be about half of the total alignment size (600 sequences).
Perturbations were performed in every sequence position that fitted the latter criteria for the subset definition. The ΔΔGstat was computed as:
where Pi|δjx is the binomial probability of finding residue x in the i position in the subset of the alignment chosen by the perturbation in the j position. The final matrix containing all the performed perturbations was submitted to iterative cycles of cluster analysis in MATLAB. After each cycle, positions with weak signals were discarded. The final matrix included 22 columns (perturbations) and 29 rows (positions) and was used in the statistical coupling analysis. All steps of the SCA were performed using locally developed C\C++ programs.
Protein Data Bank accession codes
The atomic coordinates and structure factors of the aspartic proteinase from T. reesei and of its complex with pepstatin have been deposited in the RCSB Protein Data Bank for release upon publication. The Protein Data Bank accession codes are 3C9X for the free proteinase and 3C9Y for its complex with pepstatin A.
Acknowledgements
We thank José Ribeiro Brandão Neto for his scientific assistance during diffraction data collection and processing. We also thank Lucas Bleicher for the development and implementation of the SCA codes and also for the helpful discussions. This work was supported in part by grants 99/03387-4 and 04/08070-9 from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), and in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Blumenthal CZ. Production of toxic metabolites in Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei: justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi. Reg. Tox. Pharm. 2004;39:214–228. doi: 10.1016/j.yrtph.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Keranen S, Penttila M. Production of recombinant proteins in the filamentous fungus Trichodema reesei. Curr. Opin. Biotechnol. 1995;6:534–537. doi: 10.1016/0958-1669(95)80088-3. [DOI] [PubMed] [Google Scholar]
- 3.Neustroev KN, Firsov LM. Acid proteinase and multiplicity of forms glucoamylase from Aspergillus awamori. Biokhimiya. 1990;55:776–785. [Google Scholar]
- 4.Mischak H, Hofer F, Messner R, Weissinger E, Hayn M, Tomme P, Esterbauer H, Kuchler E, Claeyssens M, Kubicek CP. Monoclonal antibodies against different domains of cellobiohydrolase I and II from Trichoderma reesei. Biochim. Biophys. Acta. 1989;990:1–7. doi: 10.1016/s0304-4165(89)80003-0. [DOI] [PubMed] [Google Scholar]
- 5.Andreeva NS, Fedorov AA, Gushchina AE, Shutskever NE. X-ray structural analysis of pepsin. V. Conformational of the main chain of the enzyme. Mol Biol (Moscow) 1978;12:922–936. [PubMed] [Google Scholar]
- 6.Andreeva N. A consensus template of the aspartic proteinase fold. In: Dunn B, editor. Structure and Function of the Aspartic Proteinases. New York, NY: Plenum Press; 1991. pp. 559–572. [DOI] [PubMed] [Google Scholar]
- 7.Andreeva NS, Rumsh LD. Analysis of crystal structures of aspartic proteinases: On the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes. Prot. Sci. 2001;10:2439–2450. doi: 10.1110/ps.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 9.Miller M, Jaskólski M, Rao JKM, Leis J, Wlodawer A. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature. 1989;337:576–579. doi: 10.1038/337576a0. [DOI] [PubMed] [Google Scholar]
- 10.Illany-Feigenbaum J, Netzer A. Milk-clotting activity of proteolytic enzymes. J. Dairy Sci. 1969;52:43–50. [Google Scholar]
- 11.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 12.Bailey D, Cooper JB, Veerapandian B, Blundell TL, Atrash B, Jones DM, Szelke M. X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin. Biochem. J. 1993;289:363–371. doi: 10.1042/bj2890363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krissiel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 14.James MNG, Sielecki A, Salituro F, Rich DH, Hofmann T. Conformational flexibility in the active sites of aspartyl proteinases revealed by a pepstatin fragment binding to penicillopepsin. Proc. Natl. Acad. Sci. USA. 1982;79:6137–6141. doi: 10.1073/pnas.79.20.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sali A, Veerapandian B, Cooper JB, Moss DS, Hofmann T, Blundell TL. Domain flexibility in aspartic proteinases. Proteins. 1992;12:158–170. doi: 10.1002/prot.340120209. [DOI] [PubMed] [Google Scholar]
- 16.Allen B, Blum M, Cunningham A, Tu G-G, Hofmann T. A ligand-induced, temperature dependent conformational change in penicillopepsin: Evidence from non-linear Arrhenius plots and from circular dichroism studies. J. Biol. Chem. 1990;265:5060–5065. [PubMed] [Google Scholar]
- 17.Fruton JS. Fluorescence studies on the active sites of proteinases. Mol. Cell. Biol. 1980;32:105–114. doi: 10.1007/BF00227803. [DOI] [PubMed] [Google Scholar]
- 18.Veerapandian B, Cooper JB, Sali A, Blundell TL, Rosatti RL, Dominy BW, Damon DB, Hoover D. Direct observation by X-ray analysis of tetrahedral “intermediate” of aspartic proteinases. Protein Sci. 1992;1:322–328. doi: 10.1002/pro.5560010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamitori S, Ohtaki A, Ino H, Takeuchi M. Crystal structures of Aspergillus oryzae aspartic proteinase and its complex with an inhibitor pepstatin at 1.9 Å resolution. J. Mol. Biol. 2003;326:1503–1511. doi: 10.1016/s0022-2836(03)00078-0. [DOI] [PubMed] [Google Scholar]
- 20.Asojo OA, Afonina E, Gulnik SV, Yu B, Erickson JW, Randad R, Mehadjed D, Silva AM. Structures of Ser205 mutant plasmepsin II from Plasmodium falciparum at 1.8 Å in complex with the inhibitors RS367 and RS370. Acta Crystallogr. 2002;D58:2001–2008. doi: 10.1107/s0907444902014695. [DOI] [PubMed] [Google Scholar]
- 21.Coates L, Erskine PT, Wood SP, Myles DAA, Cooper JB. A neutron Laue diffraction study of endothiapepsin: implications for the aspartic proteinase mechanism. Biochemistry. 2001;40:13149–13157. doi: 10.1021/bi010626h. [DOI] [PubMed] [Google Scholar]
- 22.Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J. Biol. Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 23.Pearl L, Blundell T. The active site of aspartic proteinases. FEBS Lett. 1984;174:96–101. doi: 10.1016/0014-5793(84)81085-6. [DOI] [PubMed] [Google Scholar]
- 24.Khan AR, Parrish JC, Fraser ME, Smith WW, Bartlett PA, James MN. Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes. Biochemistry. 1998;37:16839–16845. doi: 10.1021/bi9821364. [DOI] [PubMed] [Google Scholar]
- 25.Fujinaga M, Cherney MM, Tarasova NI, Bartlett PA, Hanson JE, James MN. Structural study of the complex between human pepsin and a phosphorus-containing peptidic-transition-state analog. Acta Crystallogr. 2000;D56:272–279. doi: 10.1107/s0907444999016376. [DOI] [PubMed] [Google Scholar]
- 26.Lockless SW, Ranganathan R. Evolutionary conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 27.Süel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 28.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Naturallike function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 29.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 30.Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R. Allosteric determinants in guanine nucleotide-binding proteins. Proc. Natl. Acad. Sci. USA. 2003;100:14445–14450. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi H, Takahashi T, Momose H, Inouye M, Maeda Y, Matsuzawa H, Ohta T. Enhancement of the thermostability of subtilisin E by introduction of a disulfide bond engineered on the basis of structural comparison with a thermophilic serine protease. J. Biol. Chem. 1990;265:6874–6878. [PubMed] [Google Scholar]
- 32.Ikegaya K, Ishida Y, Murakami K, Masaki A, Sugio N, Takechi K, Murakami S, Tatsumi H, Ogawa Y, Nakano E, Motai H, Kawabe H. Enhancement of the thermostability of the alkaline protease from Aspergillus oryzae by introduction of a disulfide bond. Biosci. Biotechnol. Biochem. 1992;56:326–327. doi: 10.1271/bbb.56.326. [DOI] [PubMed] [Google Scholar]
- 33.Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Structure, Function, and Bioinformatics. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz DA, Baldwin RL. Cis proline mutants of ribonuclease A. I. Thermal stability and function. Protein Science. 1992;1:910–916. doi: 10.1002/pro.5560010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nathaniel C, Wallace LA, Burke J, Dirr HW. The role of an evolutionarily conserved cis-proline in the thioredoxin-like domain of human class Alpha glutathione transferase A1-1. Biochem. J. 2003;372:241–246. doi: 10.1042/BJ20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal D, Chakrabarti P. Cis Peptide Bonds in Proteins: Residues Involved, their Conformations, Interactions and Locations. J. Mol. Biol. 1999;294:271–288. doi: 10.1006/jmbi.1999.3217. [DOI] [PubMed] [Google Scholar]
- 39.Foulkes-Murzycki JE, Scott WRP, Schiffer C. Hydrophobic sliding: A possible mechanism for drug resistance in Human Immunodefiiciency virus type 1 protease. Structure. 2007;15:223–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishima R, Louis JM, Torchia DA. Characterization of two hydrophobic methyl clusters in HIV-1 protease by NMR Spin Relaxation in solution. J. Mol Biol. 2001;305:515–521. doi: 10.1006/jmbi.2000.4321. [DOI] [PubMed] [Google Scholar]
- 41.Filippova IY, Lysogorskaya EN, Anisimova VV, Suvorov LI, Oksenoit ES, Stepanov VM. Fluorogenic peptide substrates for assay of aspartyl proteinases. Anal. Biochem. 1996;234:113–118. doi: 10.1006/abio.1996.0062. [DOI] [PubMed] [Google Scholar]
- 42.Polikarpov I, Oliva G, Castellano EE, Garratt R, Arruda P, Leite A, Craievich A. Protein crystallography station at LNLS, The Brazilian National Synchrotron Light Source. Nucl. Instr. Methods A. 1998;405:159–164. [Google Scholar]
- 43.Polikarpov I, Teplyakov A, Oliva G. The ultimate wavelength for protein crystallography. Acta Crystallogr. 1997;D53:734–737. doi: 10.1107/S0907444997007233. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Afonine PV, Grosse-Kunstleve RW, Adams PD. The phoenix refinement framework. CCP4 Newsl. 2005;42 contribution 8. [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard G. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- 51.Dima RI, Thirumalai D. Determination of network of residues that regulate allostery in protein families using sequence analysis. Prot. Sci. 2006;15:258–268. doi: 10.1110/ps.051767306. [DOI] [PMC free article] [PubMed] [Google Scholar]