Abstract
PURPOSE
The retinal pigmented epithelium (RPE) expresses many genes that play important roles in the support and maintenance of photoreceptors. The present study was conducted to develop a system amenable to the dissection of the temporal function of these genes, specifically within RPE cells. Transgenic mice were generated and characterized in which the expression of Cre recombinase could be specifically induced within the RPE.
METHODS
Transgenic mice carrying the human vitelliform macular dystrophy-2 (VMD2) promoter (PVMD2)–directed reverse tetracycline-dependent transactivator (rtTA) and the tetracycline-responsive element (TRE)–directed cre were generated. Inducible Cre expression was achieved by feeding doxycycline to these mice and was characterized by using a Cre-activatable lacZ reporter mouse strain (R26R).
RESULTS
A β-galactosidase assay of rtTA/Cre-R26R mice demonstrated that the basal level of Cre expression without doxycycline induction was negligible. Addition of doxycycline led to induction of RPE-specific Cre expression/function at least from embryonic day 9 to postnatal day 60. The highest induction occurred at approximately postnatal day 4. As measured by ERG and histology, retinal function and morphology were normal in 10-month-old rtTA/Cre mice that were treated with doxycycline at weaning age.
CONCLUSIONS
Transgenic mice were generated that express Cre recombinase in the RPE in an inducible fashion. These mice will be useful for studies of the RPE-specific role of genes that are expressed in the RPE as well as other cells, particularly for avoiding embryonic lethality and dissecting the function of genes that play dual roles in development and adulthood.
The retinal pigmented epithelium (RPE) forms the outer blood–retina barrier and serves as the gatekeeper between the choroidal blood supply and the neural retina. It also plays other important roles that are essential for the health and function of photoreceptor cells. The RPE is the site for periodic phagocytosis of photoreceptor outer segment discs and metabolism of retinoids to provide 11-cis-retinal for phototransduction. 1,2 Experimental and clinical evidence suggests that abnormality in the RPE plays an important role in some retinal and choroidal diseases, including both the dry and wet forms of age-related macular degeneration (for review, see Refs. 3–5). Some of these diseases are the result of altered expression within the RPE of genes that play essential roles elsewhere in the body.6
Genetic disruption of an essential gene expressed in multiple tissues or cell types often causes embryonic or neonatal lethality that obscures the particular role of the gene of interest in a target cell or in the adult.7,8 Such embryonic and neonatal lethality can be circumvented by using a cell-type- or tissue-specific gene-knockout strategy. Because Cre recombinase from the bacteriophage P1 is capable of catalyzing efficient site-specific DNA recombination between the 34-bp loxP sites intra- or intermolecularly,9 the Cre/lox system has become a method of choice for cell-type- or tissue-specific gene knockout in mice. In this strategy, a loxP-flanked gene or gene segment can be deleted specifically in cells that express Cre recombinase in a cell-type- or tissue-specific fashion (for review, see Ref. 10). However, tissue-specific gene disruption that occurs during early development may not yield sufficient information about a gene’s function in the adult,11 which is disadvantageous to the investigations of pathogenic mechanisms in degenerative diseases. Adding an inducible component to the Cre/lox-based gene-knockout strategy permits a temporal control on gene disruption and thus provides an ideal system for the study of postdevelopmental gene function. The tetracycline (tet)-inducible gene expression system consists of two components: the tet-dependent transactivator (tTA), which is a fusion between the tet repressor (tetR) of Escherichia coli and the activation domain of viral transcriptional regulator VP16, and the tetracycline-responsive element (TRE), which contains seven repeats of the tet operator (tetO) and the immediate early promoter of human cytomegalovirus (PhCMV). In the absence of tetracycline, the tTA binds to tetO and activates the transcription of the PhCMV-controlled gene.12 In the presence of tetracycline, binding of tTA to tetO is inhibited, and transcription is turned off. This system is named the “tet-off” system, because the presence of tetracycline turns off gene expression. A reverse version of the tet-off system, the “tet-on” system, is based on a mutant of tTA, called rtTA, that only binds to tetO and activates transcription in the presence of tetracycline. 13 The level of gene expression can be regulated up to 105 fold in a tet-off system and 103 fold in a tet-on system.13,14 Both the tet-on and tet-off systems have been successfully used for inducible gene expression in the mouse eye.15–18 To dissect the function of the RPE-expressed essential genes in the pathophysiology of retinal and choroidal diseases, we generated inducible RPE-specific Cre mice using the Cre/lox and the tet-on systems (Fig. 1A). The human VMD2 gene is associated with Best disease (vitelliform macular dystrophy; VMD) and is preferentially expressed in the RPE.19,20 Therefore, the promoter of the VMD2 gene was selected to control the inducible system in our study (Fig. 1B). This report describes the generation and characterization of the inducible RPE-specific Cre mice.
FIGURE 1.
Tetracycline-inducible Cre expression in the RPE. (A) Cre expression using the tet-on system. (B) Diagram of transgenic constructs. RNA Pol: RNA polymerase. tetO, tet-operator; PhCMV, immediate early promoter of human cytomegalovirus; rtTA, reverse tet-inducible TransActivator; Dox, doxycycline; PVMD2, promoter of human VMD2 gene; SV40 A(n), simian virus 40 polyadenylation signal; MT-IA(n), mouse metallothionein polyadenylation signal.
MATERIALS AND METHODS
Construction of the Tetracycline-Inducible RPE-Specific Cre Transgene
The recombinant plasmid pLE119 carrying the human VMD2 promoter-controlled rtTA was constructed by replacing the promoter in the pTet-On plasmid (BD-Clontech, Mountain View, CA) with a 2.9-kb VMD2 promoter.21 The recombinant plasmid pLE116 carrying the tetO-PhCMV controlled cre was constructed by replacing the promoter for cre in the plasmid pLE10922 with the tetO-PhCMV of pTRE (Clontech).
Animal Treatment and Generation and Characterization of Transgenic Mice
The use of animals in this study followed the guidelines established by the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and was approved by the Institutional Animal Care and Use Committees at the University of Oklahoma Health Sciences Center, the Dean A. McGee Eye Institute, and the Oklahoma Medical Research Foundation. Doxycycline was administered to the mice in their drinking water by daily gavage feeding at a dose of 0.4 mg/g body weight for 2 days.
The recombinant plasmids pLE116 or pLE119 carrying the PVMD2-rtTA or tetO-PhCMV-cre transgenes were purified by using double ultracentrifugation in a cesium chloride gradient. The purified plasmids were digested with the restriction enzymes XhoI and HindIII to isolate the cre transgene and XhoI to isolate the rtTA transgene, respectively (Fig. 1B). The digested DNA fragments were purified with a gel extraction kit (Qiagen, Valencia, CA) and were co-injected into the zygotes of FVB/N background mice at the Oklahoma Medical Research Foundation Microinjection Core Facility. PCR diagnostic for the cre transgene was performed with primers a (5′-AGG TGT AGA GAA GGC ACT TAG C-3′) and b (5′-CTA ATC GCC ATC TTC CAG CAG G-3′) to detect a 411-bp product (from 621-1032 bp in the cre structure gene), according to an established procedure.10 Diagnostic PCR for the rtTA transgene was performed with primers c (5′-CGG CCT TGA ATT GAT CAT ATG CGG-3′) and d (5′-TCA AAC TCG AAG TCG GCC ATA TCC-3) to detect a 398-bp product (from 1336 to 1734 bp in the rtTA structure gene), using the following condition: 35 cycles at 94°C for 30 seconds, 64°C for 30 seconds, and 72°C for 60 seconds. PCR detection of the Cre-activatable lacZ reporter gene in R26R mice was performed with primers e (5′-GAG TTG CGT GAC TAC CTA CGG-3′) and f (5′-GGC TTC ATC CAC CAC ATA CAG G-3′), to detect a 495-bp product (bp 745-1240 in the lacZ structure gene), according to an established procedure.23
β-Galactosidase Assay
The β-galactosidase assay on retinal wholemounts and retinal sections was performed according to established methods.22,24 A liquid β-galactosidase assay was performed based on a previous method with some modifications.25 Briefly, the mice were euthanatized and their eyes were dissected and washed in cold (4°C) PBS. The lens and vitreous were removed, and the eyecup was then homogenized in 200 µL of Z-buffer containing 50 mM sodium phosphate buffer (pH 7.0), 10 mM KCl, and 1 mM MgSO4. After the cell debris was removed by centrifugation, 106 µL of the supernatant was added with 21 µL of 2-nitrophenyl β-d-galactopyranoside (ONPG; 4 mg/mL in water) and incubated at 30°C for 30 minutes. The reaction was stopped by adding 53 µL of 1 M Na2 CO3, and the optical density of the mixture was measured at 420 and 550 nm. The relative β-galactosidase activity was expressed as 1000 × [OD420 − (1.75 × OD550)]/time × protein (in milligrams).25 Protein concentrations were determined with a BCA protein assay kit (Pierce, Rockford, IL).
Electroretinography and Retinal Morphology
Mice were dark adapted overnight, and scotopic and photopic electroretinography (ERG) was performed (UTAS-E 3000 ERG system; LKC Technologies, Inc., Gaithersburg, MD), according to the conditions described previously.22,26,27 Retinal morphology was examined in hematoxylin and eosin (H & E)–stained sections according to an established method.24
RESULTS
Generation of Inducible RPE-Specific Cre Transgenic Mice
In a previous study, the promoter of the human VMD2 gene was shown to drive efficient RPE-specific transgene expression in mice.21 Therefore, this promoter was selected to direct the RPE-specific expression of the tet system transactivator gene rtTA. The rtTA is designed to activate tetO-controlled cre expression in the presence of doxycycline, through the transcription from the immediate early promoter of human cytomegalovirus (PhCMV; Figs. 1A, 1B). Our cre transgene (Fig. 1B) carried a translationally optimized cre, and the mouse metallothionein (MT-I) polyadenylation signal. The cre/MT-I polyA cassette was derived from a plasmid with a proven record of success in a transgenic study.22 During pronuclear injection, the purified PVMD2-rtTA and tetO-PhCMV-cre DNA fragments were intentionally co-injected into zygotes to result in the co-integration of both transgenes into a single chromosome. Indeed, PCR analysis of segregation patterns for several generations suggested that both transgenes were cosegregated in each transgenic founder (data not shown). This arrangement provides a distinct advantage over separate integration sites, because it will reduce the required breeding for future downstream studies using specific floxed genes of interest. After PCR analysis, the candidate transgenic mice were mated to obtain germline transmission. No abnormalities in size, morphology, or behavior were observed in any of the germline-transmitted mice. The mice were then bred with albino Cre-activatable lacZ reporter (R26R) mice.28 The resultant F1 transgenic rtTA/Cre-R26R mice did not have inherited retinal degeneration and thus were suitable for downstream characterization. Our preliminary characterization indicated that one transgenic line had productive Cre-mediated lacZ reporter expression specifically in the RPE across the retina. This mouse line was characterized further.
Localization and Functional Analysis of Inducible Cre Expression
To analyze and localize Cre expression, immunohistochemistry and immunoblotting were performed using an antibody that had a proven record of success in the characterization of photoreceptor-specific Cre mice.22,24 However, no positively stained RPE cells in retinal sections or specific immunoblotting signals from RPE extracts were identified (data not shown). These results are not inconsistent with the β-galactosidase reporter results (described later), since a productive Cre-mediated recombination requires only eight copies of Cre recombinase. 29 It is anticipated that the minimal level of Cre required for a productive Cre-mediated recombination is much lower than the threshold of detecting Cre expression by immunohistochemistry and immunoblot analysis. We took advantage of an existing Cre-activatable lacZ reporter mouse line R26R to localize Cre expression and perform functional assays.28 The R26R mice carried a loxP-flanked transcriptional “stop” DNA segment that prevents the transcription of the lacZ gene. In the transgenic Cre/rtTA-R26R mice, this transcriptional “stop” sequence can be removed only in cells expressing Cre. β-Galactosidase is then expressed under a generalized promoter ROSA26, and the Cre-expressing cells produce a blue color in the β-galactosidase assay. The expression pattern in fully induced mice is shown in Figures 2A and 2B. Our results suggested that the rtTA, Cre, and β-galactosidase were expressed exclusively in the RPE except for weak expression in the optic nerve (Fig. 2A). The Cre expression was stronger in the central retina, which is consistent with a previous transgenic study using the human VMD2 promoter.21
FIGURE 2.
Representative results of β-galactosidase staining assay. F1 albino rtTA/Cre-R26R mice were used. Only Cre-expressing cells are blue. (A, B) Localization of Cre-activated β-galactosidase expression in a retinal section and an RPE/choroid flat-mount, respectively. (C–F) Induction initiated on E9, -13, -17, or P5 by feeding doxycycline to the mother for 2 days. Assays were performed 10 days after induction. (G–I) direct feeding of mice with doxycycline on P14, -25, or -60. Assays were performed 10 days after induction. (J) No induction to P35 (control). Cre-activated β-galactosidase expression was induced from E9 to P60 and was stronger from E17 to P25. RPE, retinal pigmented epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GC, ganglion cells. Scale bar, 50 µm.
To examine the leakiness of the system without induction, all mice were placed on an autoclaved diet (LabDiet 5010; Statewide Supply, Oklahoma City, OK). The trace amount of tetracycline derivatives in the diet is thought to be heat-inactivated by autoclaving. The expression of the Cre-activated β-galactosidase in the RPE of the noninduced control animals demonstrated the presence of the inherent tet-system leakage in our mice; however, the majority of the noninduced RPE cells did not express Cre (Fig. 2J), suggesting that the leakage will not likely be a serious issue in the utilization of the system. The induction of Cre-activated β-galactosidase during embryonic and early neonatal development was achieved by feeding mothers with doxycycline. Since induction at embryonic day (E)7 to -8 did not yield productive Cre-mediated recombination (data not shown) and induction at E9 to -10 resulted in homogenous Cre-activated β-galactosidase expression in the RPE (Fig. 2C), our result suggests that the initiation of Cre-mediated recombination and the onset of rtTA expression occurred around E9. Our results also indicated that Cre-activated β-galactosidase expression in the RPE could be induced in the inducible RPE-specific Cre mice up to at least 2 months of age (Fig. 2I), suggesting that the rtTA was expressed or functional at least until then. However, the intensity of Cre-activated β-galactosidase staining was stronger from E17 to P5 (Figs. 2E, 2F) in mice induced through feeding doxycycline to their mothers. To prevent the reduced induction of Cre-activated β-galactosidase in older neonates through doxycycline feeding to their mothers, gavage feeding of the neonates was performed to achieve productive induction after P3. Mice induced in this manner between P14 and −25 also demonstrated relatively high levels of Cre-activated β-galactosidase activity (Figs. 2G, 2H). The intensity of Cre-activated β-galactosidase staining was relatively low and more mosaic at P60 (Fig. 2I). These results suggest that the highest level of rtTA expression occurred between E17 and P25.
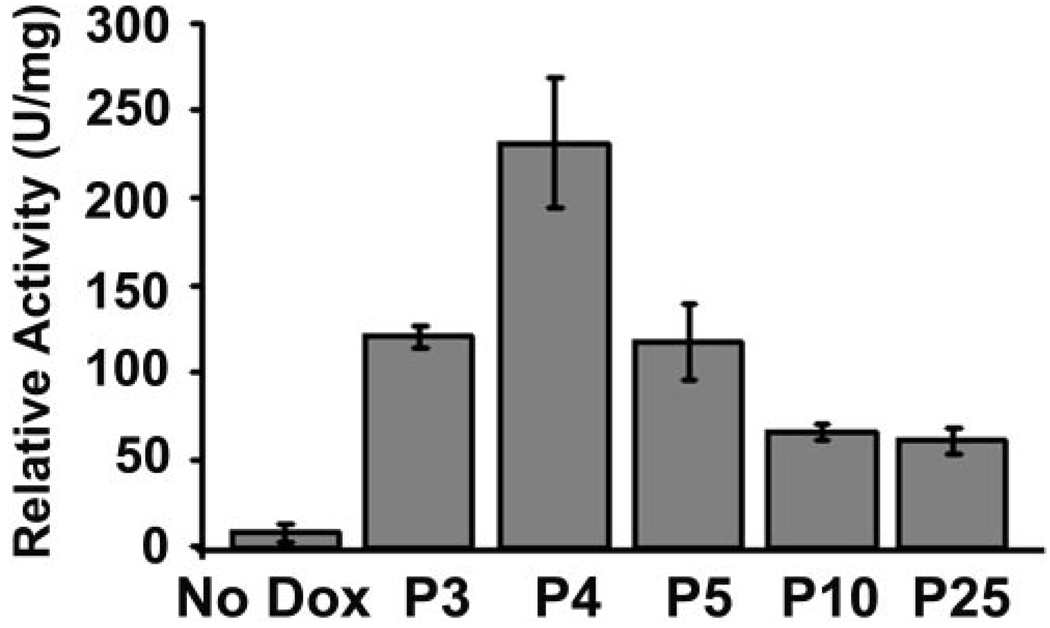
To identify the conditions for the maximum induction of Cre-activated β-galactosidase or expression of VMD2 promoter-controlled rtTA, we performed a quantitative β-galactosidase activity assay. Although it would be ideal to include all aged mice in the experiment, the inability to feed embryos and neonates younger than P3 directly with doxycycline made it difficult to perform a direct comparison. Since a previous study demonstrated that the highest level of VMD2 mRNA expression occurred around P4,30 the assay was performed using homogenized retinal extracts from mice that were fed doxycycline by gavage at various time points from P3 to −25. Our results demonstrate that the maximum Cre-activated β-galactosidase activity was 28-fold greater than the control when induction occurred on P4 (Fig. 3). This result, along with the result from the β-galactosidase staining assay (Fig. 2), suggests that VMD2 promoter-controlled rtTA expression or Cre-activated β-galactosidase expression increased gradually from E9, reached a peak at approximately P4, and gradually declined afterward. This finding is consistent with the expression pattern of mouse bestrophin, the counterpart of human VMD2.30
FIGURE 3.
In vivo induction of Cre-activated β-galactosidase activity in the F1 rtTA/Cre-R26R mice. Doxycycline (0.4 mg/g body weight) was fed to mice on P3, -4, -5, -10, or -25 by gavage for 2 days. Five days after induction was initiated, the β-galactosidase assay was performed. No dox: P21 control (rtTA-Cre/R26R) mice without doxycycline induction. Each data point was derived from at least eight retinas. The strongest Cre-activated β-galactosidase activity was observed with induction at P4.
RPE Integrity
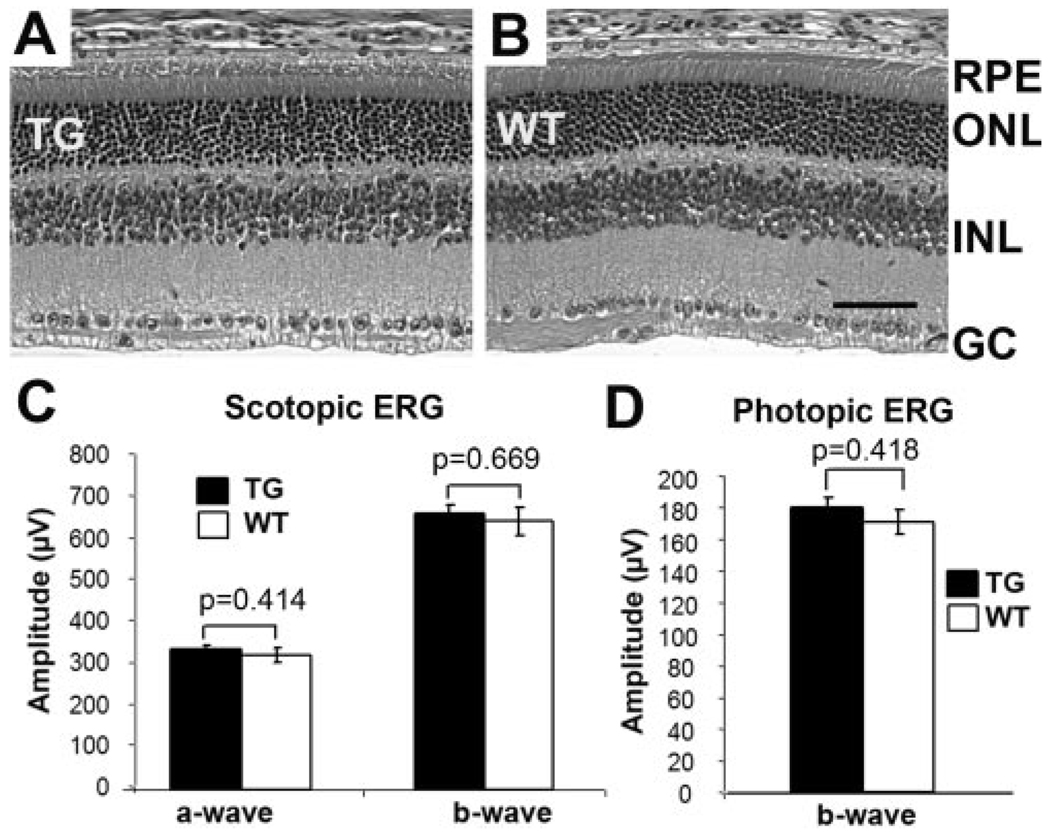
Studies have demonstrated that overexpression of Cre in rod photoreceptors causes retinal degeneration.24,31 Although the mechanism is still unclear, it is known that overexpression of Cre can cause unwanted chromosomal rearrangement in mammals. 32,33 Because the goal of our study was to establish a useful system for gene-function studies, it was necessary to determine whether there was any RPE degeneration in the inducible RPE-specific Cre mice. Therefore, the number of RPE cells was quantified in the transgenic mice to determine whether RPE cell loss occurred over time, by counting the number of the RPE nuclei across the vertical meridian in the H&E-stained retinal sections. Our results indicated that there was no detectable loss of the RPE in the 10-month-old inducible RPE-specific Cre mice that had a brief doxycycline treatment at the weaning age (Fig. 4). Because the loss of RPE causes photoreceptor degeneration, the retinal morphology and function in these mice were also examined. Our results demonstrated that there was no detectable photoreceptor degeneration in the inducible RPE-specific Cre mice treated with doxycycline at the weaning age (Figs. 5A, 5B). Likewise, the scotopic and photopic ERG analyses showed that these transgenic mice had normal photoreceptor function (Figs. 5C, 5D). In short, our results suggest that Cre expression did not cause any detectable RPE or photoreceptor degeneration in the inducible RPE-specific Cre mice.
FIGURE 4.
Relative number of RPE cells in H&E-stained retinal sections of 10 month-old inducible RPE-specific Cre mice. The value was the average number of total RPE nuclei along the vertical meridian in H&E-stained retinal sections. No significant loss of the RPE cells was detected in the inducible RPE-specific Cre mice. WT, wild-type; TG, transgenic.
FIGURE 5.
Retinal morphology and function of 10-month-old inducible RPE-specific Cre mice. (A, B) Representative H&E-stained retinal sections of transgenic (TG) and wild-type (WT) mice. RPE, retinal pigmented epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GC, ganglion cells. Scale bar, 40 µm. (C, D) Scotopic and photopic ERGs. At least eight mice were included in each group. No significant difference in retinal morphology and function was detected in the inducible RPE-specific Cre mice.
DISCUSSION
To investigate the function of widely expressed genes in the RPE, several laboratories have made extensive efforts to generate RPE-specific Cre mice. Although lines of mice that have Cre-mediated recombination in the RPE were established,34–36 the RPE-specific Cre expression in these mice occurs during early embryogenesis and could cause developmental defects in gene-knockout studies.11,37 This limits the usefulness of these Cre mice for functional studies of genes that play roles in both development and adulthood. To overcome this problem and add a temporal control to the system, we generated transgenic mice that express Cre recombinase in the RPE in a doxycycline-inducible fashion. The expression of the reverse tetracycline transactivator (rtTA) was directed by the promoter of the human VMD2 gene, which is preferentially expressed in the RPE.21 In the presence of the doxycycline, the rtTA activated the transcription of Cre through the TRE. Indeed, functional expression of Cre recombinase was restricted to the RPE in the retina, with relatively weak expression detected in the optic nerve. Regulation of Cre expression with variable concentrations of doxycycline was achievable (data not shown); however, the control of induction time resulted in better reproducibility. Because Cre-mediated excisive recombination is usually permanent, induction of Cre expression at various times using a sufficient amount of doxycycline provides a regulatable phenotype, as shown in Figure 2 and Figure 3. The highest Cre-activated β-galactosidase reporter expression occurred after birth, which is similar to that of mouse bestrophin, the counterpart of human VMD2,30 suggesting that the highest level of rtTA and Cre expression can be induced after birth. The novelty of our system is that the time of the RPE-specific Cre expression can be regulated. Therefore, this system can be used to determine the temporal function of a gene, a unique advantage over other published mouse lines that cannot control the time of Cre expression in the RPE.34–36
Our inducible RPE-specific Cre mice bypass the early development and therefore can be useful for temporal gene activation and inactivation in the RPE, particularly for genes that are implicated in both development and degenerative diseases. We are interested in the function of RPE-expressed growth factors, their receptors, and downstream targets in the maintenance of the outer blood–retina barrier function and RPE survival. The inducible RPE-specific Cre mice will be invaluable in these studies. In addition to inducible gene activation and inactivation, our transgenic mice also provide a tool for inducible RPE-specific expression of other gene of interest, because the VMD2 promoter-controlled rtTA is functional in the RPE.
A previous study demonstrated that doxycycline induction does not result in detectable changes in the mouse retina and is unlikely to present a problem in the use of our mice for gene functional studies.16 Transgene insertion and Cre overexpression, however, could cause defects in the RPE and its secondary effect in the retina and prevent the rtTA/Cre mice from becoming a useful genetic tool. Thus, it was important to determine whether the retina is normal in the transgenic mice, as we have examined with all our retinal cell-specific Cre mice.24,38 There was no detectable loss of RPE cells, rod photoreceptors, or photoreceptor function in the mice until at least 10 months of age, which provides a crucial window to ensure the usefulness of the rtTA/Cre mice in gene function studies.
Although our inducible RPE-specific Cre mice carry two transgenes, results from PCR analysis of rtTA and cre for several generations indicated that these transgenes cosegregate. This feature is attributed to the use of a co-injection strategy during the generation of the mice. Because the major goal of our study was to develop a genetic system to induce gene activation or inactivation with a variety of genes, substantial breeding of mice carrying modified genes on different chromosomes would be required. The presence of rtTA and cre on the same chromosome will permit a significant reduction of breeding effort for these types of studies.
Acknowledgments
The authors thank the Microinjection Core Facility at the Oklahoma Medical Research Foundation for the generation of the transgenic mice, John D. Ash for helpful suggestions, and Yuwei Le for technical assistance.
Supported by National Institutes of Health Grant P20 RR17703 and National Eye Institute Grant P30 EY12190; Oklahoma Center for the Advancement of Science and Technology (OCAST) contract HR05-133; American Diabetes Association (ADA) Grant 1-06-RA-76; and unrestricted research awards from Hope for Vision and Research to Prevent Blindness, Inc.
Footnotes
Disclosure: Y.-Z. Le, None; W. Zheng, None; P.-C. Rao, None; L. Zheng, None; R.E. Anderson, None; N. Esumi, None; D.J. Zack, None; M. Zhu, None
References
- 1.Young RW, Bok D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saari JC. Biochemistry of visual pigment regeneration: the Friedenwald lecture. Invest Ophthalmol Vis Sci. 2000;41:337–348. [PubMed] [Google Scholar]
- 3.Sunness JS. The natural history of geographic atrophy, the advanced atrophic form of age-related macular degeneration. Mol Vis. 1999;5:25. [PubMed] [Google Scholar]
- 4.Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- 5.Lutty G, Grunwald J, Majji AB, Uyama M, Yoneya S. Changes in choriocapillaris and retinal pigment epithelium in age-related macular degeneration. Mol Vis. 1999;5:35. [PubMed] [Google Scholar]
- 6.Lopez PF, Sippy BD, Lambert HM, Thach AB, Hinton DR. Transdifferentiated retinal pigment epithelial cells are immunoreactive for vascular endothelial growth factor in surgically excised age-related macular degeneration-related choroidal neovascular membranes. Invest Ophthalmol Vis Sci. 1996;37:855–868. [PubMed] [Google Scholar]
- 7.Ferrara N, Carver-Moore K, Chen H, et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 8.Rucker EB, 3rd, Dierisseau P, Wagner KU, et al. Bcl-x and Bax regulate mouse primordial germ cell survival and apoptosis during embryogenesis. Mol Endocrinol. 2000;14:1038–1052. doi: 10.1210/mend.14.7.0465. [DOI] [PubMed] [Google Scholar]
- 9.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Y, Sauer B. Conditional gene knockout using cre recombinase. Methods Mol Biol. 2000;136:477–485. doi: 10.1385/1-59259-065-9:477. [DOI] [PubMed] [Google Scholar]
- 11.Marneros AG, Fan J, Yokoyama Y, et al. Vascular endothelial growth factor expression in the retinal pigment epithelium is essential for choriocapillaris development and visual function. Am J Pathol. 2005;167:1451–1459. doi: 10.1016/S0002-9440(10)61231-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 14.Kistner A, Gossen M, Zimmermann F, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeletti B, Loster J, Auricchio A, et al. An in vivo doxycycline-controlled expression system for functional studies of the retina. Invest Ophthalmol Vis Sci. 2003;44:755–760. doi: 10.1167/iovs.02-0340. [DOI] [PubMed] [Google Scholar]
- 16.Chang MA, Horner JW, Conklin BR, DePinho RA, Bok D, Zack DJ. Tetracycline-inducible system for photoreceptor-specific gene expression. Invest Ophthalmol Vis Sci. 2000;41:4281–4287. [PubMed] [Google Scholar]
- 17.Kerrison JB, Duh EJ, Yu Y, Otteson DC, Zack DJ. A system for inducible gene expression in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2005;46:2932–2939. doi: 10.1167/iovs.04-1237. [DOI] [PubMed] [Google Scholar]
- 18.Chikama T, Hayashi Y, Liu CY, et al. Characterization of tetracycline-inducible bitransgenic Krt12rtTA/LacZ mice. Invest Ophthalmol Vis Sci. 2005;46:1966–1972. doi: 10.1167/iovs.04-1464. [DOI] [PubMed] [Google Scholar]
- 19.Petrukhin K, Koisti MJ, Bakall B, et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 20.Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease) Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- 21.Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem. 2004;279:19064–19073. doi: 10.1074/jbc.M309881200. [DOI] [PubMed] [Google Scholar]
- 22.Le Y, Ash JD, Al-Ubaidi MR, Chen Y, Ma J, Anderson RE. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004;10:1011–1018. [PubMed] [Google Scholar]
- 23.Le Y, Gagneten S, Larson T, et al. Far-upstream elements are dispensable for tissue-specific proenkephalin expression using a Cre-mediated knock-in strategy. J Neurochem. 2003;84:689–697. doi: 10.1046/j.1471-4159.2003.01573.x. [DOI] [PubMed] [Google Scholar]
- 24.Le Y, Zheng L, Zheng W, et al. Mouse opsin promoter controlled expression of Cre recombinase in transgenic mice. Mol Vis. 2006;12:389–398. [PubMed] [Google Scholar]
- 25.Rajala RV, McClellan ME, Chan MD, Tsiokas L, Anderson RE. Interaction of the retinal insulin receptor beta-subunit with the p85 subunit of phosphoinositide 3-kinase. Biochemistry. 2004;43:5637–5650. doi: 10.1021/bi035913v. [DOI] [PubMed] [Google Scholar]
- 26.Quiambao AB, Peachey NS, Mangini NJ, Rohlich P, Hollyfield JG, al-Ubaidi MR. A 221-bp fragment of the mouse opsin promoter directs expression specifically to the rod photoreceptors of transgenic mice. Vis Neurosci. 1997;14:617–625. doi: 10.1017/s095252380001258x. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Quiambao AB, Roveri L, et al. Degeneration of cone photoreceptors induced by expression of the Mas1 protooncogene. Exp Neurol. 2000;163:207–219. doi: 10.1006/exnr.2000.7370. [DOI] [PubMed] [Google Scholar]
- 28.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 29.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 30.Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD. Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci. 2003;44:3622–3628. doi: 10.1167/iovs.03-0030. [DOI] [PubMed] [Google Scholar]
- 31.Jimeno D, Feiner L, Lillo C, et al. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest Ophthalmol Vis Sci. 2006;47:5039–5046. doi: 10.1167/iovs.06-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loonstra A, Vooijs M, Beverloo HB, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guyonneau L, Rossier A, Richard C, Hummler E, Beermann F. Expression of Cre recombinase in pigment cells. Pigment Cell Res. 2002;15:305–309. doi: 10.1034/j.1600-0749.2002.02039.x. [DOI] [PubMed] [Google Scholar]
- 35.Mori M, Metzger D, Garnier JM, Chambon P, Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2002;43:1384–1388. [PubMed] [Google Scholar]
- 36.Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–1939. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
- 37.Mori M, Metzger D, Picaud S, et al. Retinal dystrophy resulting from ablation of RXR alpha in the mouse retinal pigment epithelium. Am J Pathol. 2004;164:701–710. doi: 10.1016/s0002-9440(10)63157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le YZ, Ash JD, Al-Ubaidi MR, Chen Y, Ma JX, Anderson RE. Conditional gene knockout system in cone photoreceptors. Adv Exp Med Biol. 2006;572:173–178. doi: 10.1007/0-387-32442-9_26. [DOI] [PubMed] [Google Scholar]