Abstract
The structures of the genes encoding the α1 and β1 subunits of murine soluble guanylyl cyclase (sGC) were determined. Full-length cDNAs isolated from mouse lungs encoding the α1 (2.5 kb) and β1 (3.3 kb) subunits are presented in this report. The α1 sGC gene is approximately 26.4 kb and contains nine exons, whereas the β1 sGC gene spans 22 kb and consists of 14 exons. The positions of exon/intron boundaries and the sizes of introns for both genes are described. Comparison of mouse genomic organization with the Human Genome Database predicted the exon/intron boundaries of the human genes and revealed that human and mouse α1 and β1 sGC genes have similar structures. Both mouse genes are localized on the third chromosome, band 3E3-F1, and are separated by a fragment that is 2% of the chromosomal length. The 5′ untranscribed regions of α1 and β1 subunit genes were subcloned into luciferase reporter constructs, and the functional analysis of promoter activity was performed in murine neuroblastoma N1E-115 cells. Our results indicate that the 5′ untranscribed regions for both genes possess independent promoter activities and, together with the data on chromosomal localization, suggest independent regulation of both genes.
Keywords: nitric oxide, chromosomal localization
Nitric oxide-dependent signal transduction is associated with a number of important physiological processes, including smooth muscle relaxation (1), platelet aggregation (2), neurotransmission (3), cellular differentiation (4), and apoptotic cell death (5–7). Soluble guanylyl cyclase (sGC), a NO-stimulated hemoprotein, which converts GTP to cyclic guanosine monophosphate, is a key element in these processes.
sGC is a heterodimer consisting of α and β subunits (8), which are encoded by separate genes (9). A heme prosthetic group is crucial for the stimulation of the enzyme by NO (10, 11). The enzyme has been purified from various animal tissues (10, 12, 13), and corresponding cDNAs were cloned from various vertebrate species, including rat (14, 15), human (16), bovine (17, 18), and fish (19). At least two isoforms for each subunit of the enzyme have been identified in various species, prompting a recent revision of the nomenclature of sGC subunits (20). Although isoforms for both subunits were detected at the mRNA level in various tissues and were found to have an overlapping tissue distribution, until recently only the α1/β1 heterodimeric enzyme has been isolated from native sources. However, Russwurm and coworkers (21) described an additional α2/β1 heterodimer, which was shown previously to be catalytically active in vitro. Alternatively spliced transcripts for both human α (22) and β (23) subunits also have been reported. mRNA for the human α1 subunit undergoes alternative splicing, resulting in several mRNA species that are N-terminally truncated. The α2I subunit, an alternatively spliced variant of α2, has been detected in several mammalian cell lines and tissues at the mRNA level (24), and a β1 cDNA splice variant has been detected in humans (25).
Evidence that sGC activity is regulated both at the protein and mRNA levels has begun to emerge. Several groups have reported that such treatments as forskolin, dibutyryl-cAMP or 3-isobutyl-1-methyl xanthine (26), endotoxin and/or IL-1β (27), NO-donating compounds (28), and nerve growth factor (29) affect the sGC mRNA levels in various cell types. There is also evidence that levels of sGC mRNA expression are subject to developmental regulation (30). Ongoing work in our laboratory, to be reported elsewhere, clearly demonstrates hormonal regulation of the mRNA levels for α1 and β1 sGC subunits.
Despite an increasing interest in genetic aspects of sGC regulation, relatively little is known about the genes or the promoter regions of mammalian sGCs. Only the genomic organization of sGC for such distant species as the Anopheles mosquito (31) and Medaka fish (32) has recently been described.
Here we report the cDNA cloning, chromosomal localization, and structure of the mouse genes for the α1 and β1 subunits of sGC and a comparative analyses with the human sGC genes, using the Human Genome Database (National Center for Biotechnology Information, NCBI). This information furthers our understanding of the mechanisms of sGC regulation through alternative splicing and/or modulation of mRNA levels.
Materials and Methods
Isolation of a cDNA Clone for Mouse sGCα1 Subunit.
A mouse lung λ Triplex cDNA library (CLONTECH) was screened by hybridization using a 1.3-kb rat sGCα1 cDNA fragment obtained by PCR using Taq polymerase (GIBCO) and the oligonucleotide primers 5′-91TGCACTTCAGAGAACCTTG-3′ and 5′-520CTCCACCTTGTAGACATCCA-3′ (superscript indicates position of codon at which the primers start). Six positive clones were identified from approximately 1 × 106 independent phage plaques. Positive clones were subsequently purified, sequenced bidirectionally for positive clone identification, and analyzed by using dnastar software (DNAstar, Madison, WI). After analysis, the clone was defined as mouse α1 sGC and submitted to the NCBI database (accession no. AF297082). This clone was used in all subsequent experiments and alignments in this article.
Isolation of Genomic Clones for Mouse sGCα1 and β1 Subunits.
A bacterial artificial chromosome (BAC) high-density membrane mouse library was purchased from Genome Systems (St. Louis). The hybridization was performed overnight at 46°C in standard hybridization solution (33). A random primer-labeled α32P-dCTP-labeled cDNA fragment (0.9 kb) for the β1 sGC probe was generated by reverse transcription–PCR from a total RNA preparation from murine neuroblastoma N1E-115 cells, using the oligonucleotides 5′--3GACACCATGTACGGTTTCGTG-3′ and 5′-243CCCTTCCTTGCTTCTCAGTAC-3′ (superscript indicates the base pairs upstream of the start codon or the position of the codon at which the primers start).
The membranes then were rehybridized with an α1 sGC cDNA probe (1.3 kb containing coding sequence) under the same conditions. Positive BAC clones were identified by using the manufacturer's procedure and purchased from Genome Systems. A BAC plasmid purification kit (CLONTECH) was used for BAC DNA isolation from bacterial culture. BAC DNA was subjected to restriction and Southern blot hybridization analysis (33) using the same hybridization probes to confirm isolation of positive clones (data not shown).
Determination of Boundaries and Sizes of Introns.
Based on the α1 and β1 sGC cDNA sequences, sequencing oligonucleotide primers were designed to determine the genomic structure of each subunit. All sequencing analyses were performed at the Molecular Core Sequencing Facility at the University of Texas-Houston Medical School on an ABI Prism 377 DNA sequencer with the DigDye Terminator cycle sequencing kit (Applied Biosystems). Primers positioned in the exons of both subunits were used to determine the intron sizes by PCR with Pfu-Turbo DNA polymerase (Stratagene) from BAC DNA templates. PCR conditions were: (i) melting step at 95°C for 1 min, (ii) primer annealing at 55°C for 1 min, and (iii) extension step at 72°C for 3 min, repeated for 35 cycles. PCR products were separated by electrophoresis on 1% agarose gels.
3′ Rapid Amplification of cDNA End (RACE) of Mouse sGCβ1 Subunit.
Poly(A)+ RNA was purified from lung tissue of CD57 mice by using an mRNA extraction kit (Dynal). Determination of the 3′ end of β1 sGC mRNA was performed by using a SMART RACE cDNA Amplification kit (CLONTECH). In brief, the first-strand cDNA synthesis was achieved by incubating the poly(A)+ RNA with a 3′ cDNA-specific primer and Superscript Reverse Transcriptase (GIBCO) for 1.5 h at 42°C. Next a “touchdown” PCR to amplify the fragment was executed by using an oligonucleotide 5′-258CTGCTACAAGCATTGCCTAGACGGACG-3′ (superscript indicates the base pairs downstream of the stop codon where the primer starts), specific to the 3′ end of the published β1 sGC sequence, and the universal primer mixture that recognizes the modified 3′ end of cDNA. PCR conditions were as suggested by the manufacturer (CLONTECH). PCR products were subcloned into pCR 2.1-Topo vector (Invitrogen), and both strands were sequenced for verification.
Chromosomal Localization of Mouse sGCα1 and β1 Subunits.
Chromosomal localization of α1 and β1 sGC genes was performed by Genome Systems using fluorescence in situ hybridization. Briefly, purified BAC DNA for each clone, containing the genomic sequence of α1 and β1 sGC, was labeled with digoxigenin dUTP by nick-translation. The labeled probe was combined with sheared mouse DNA and hybridized to normal metaphase chromosomes derived from mouse embryo fibroblast cells in a solution containing 50% formamide, 10% dextran sulfate, and 2× SSC. The hybridization was detected by using fluorescent antidigoxigenin antibodies followed by counterstaining with 4′,6-diamidino-2-phenylindole. In addition, a probe specific for the telomeric region of chromosome 3 was cohybridized with each clone to verify specific labeling of the telomere to chromosome 3. Specific measurements identifying the hybridization signal between the heterochromatic-euchromatic boundary to the telomere of chromosome 3 indicated the band location of each clone on mouse chromosome 3.
Cloning of Luciferase Plasmid Constructs.
To create the plasmid constructs containing the 5′ upstream regions of α1 and β1 sGC extended to the first identified exon for each gene, DNA fragments were obtained by PCR using the specific genomic clones as templates and Pfu Turbo DNA Polymerase (Stratagene). Positive-strand oligonucleotide primers for each construct were: 1.6 kb, α1 5′--1901GTCAGTGTCAGACCTGAAGATGCTG-3′ and 1.4 kb, β1 5′--1528CTCTCTGTGTGTGAGAGAGAG-3′ (superscript indicates the base pairs upstream of the start codon). Each of these positive-strand oligonucleotide primers contained a KpnI restriction site linker sequence at the 5′ end. The negative-strand primers were: 5′--104CATGATGCGATCACAGGAGGC-3′ for the α1 construct and 5′--105CGCCCGGAGCCTAGGAAGCAG-3′ for the β1 construct (superscript indicates the base pairs upstream of the start codon). Each of the negative-strand primers contained a BglII restriction site linker sequence at the 5′ end. After restriction digestion of the ends, the PCR fragments were directionally cloned into the luciferase reporter vector pGL3-Basic (Promega) between the KpnI and BglII restriction sites upstream of the luciferase gene.
Transfection and Detection of Luciferase Activity.
N1E-115 mouse neuroblastoma cells were maintained in DMEM with 4 mM l-glutamine, 4.5 g/liter glucose, 1% penicylin-streptomicyn mixture, and 10% FBS (HyClone). Cells were transiently transfected with each (α1 and β1 sGC) luciferase plasmid construct by using Fugene-6 transfection reagent (Roche Molecular Biochemicals). Cultures were incubated in the presence of Fugene and DNA (1 μg) for 48 h and assayed for luciferase activity by using a luciferase reporter assay (Promega). Cells were cotransfected with a β-galactosidase (β-gal) construct (cytomegalovirus-β-gal, 1/5 of the concentration of sGC constructs) and assayed for β-gal activity in the N1E cell lysates to normalize the transfection efficiency between cell groups (not shown).
Results
Cloning of the Mouse α1 sGC Subunit cDNA.
The cDNA for mouse α1 subunit of sGC was not previously isolated and reported. We isolated six clones by screening a mouse cDNA library (CLONTECH) using a rat cDNA sequence as a probe. The clone containing the longest insertion was sequenced and analyzed for the presence of the ORF encoding the α1 sGC subunit. The sequence comparison of isolated cDNA demonstrated 93.3% and 83.9% homology with rat and human α1 cDNA, respectively, confirming that the isolated clone indeed encodes mouse α1 sGC.
Isolation of 3′ End Fragment of β1 sGC cDNA.
The NCBI database contains a 2.3-kb cDNA sequence for mouse β1 sGC (GenBank accession no. AF020339). Northern analysis of mouse lung total RNA performed in our laboratory showed a 4-kb transcript for β1 sGC (data not shown). To find the missing portion of the mRNA for β1 sGC, a 3′ rapid amplification of cDNA end analysis was performed on the mouse lung mRNA preparation (see Materials and Methods). The first cDNA strand was generated by using a primer located upstream of the known 3′ end of mouse cDNA and the oligo(dT) adaptor primer. A 1-kb fragment was successfully isolated. Sequence analysis of this fragment indicated that it contained the expected 70-bp region identical to the known mouse 3′ end of β1 cDNA followed by a 956-bp novel sequence containing a conservative consensus for the polyadenylation signal and poly(A) stretch. The sequence was highly homologous to the rat 3′ end of β1 cDNA (data not shown). This allowed us to conclude that the complete 3′ untranslated region for β1 sGC cDNA had been isolated. The full cDNA sequence of mouse β1 sGC subunit was submitted to the NCBI database (accession no. AF297083).
Genomic Organization of Mouse α1 and β1 sGC Genes.
Three overlapping BAC clones were isolated for each of the sGC genes by separate screening of a BAC mouse genomic library (Genome Systems) by using probes containing a 1.3-kb fragment of the coding sequence for mouse α1 sGC and a 0.9-kb N-terminal fragment of the coding mouse β1 sGC sequence (see Materials and Methods). Southern analysis of isolated clones with probes specific for the 5′ and 3′ cDNA fragments for both sGC subunits confirmed that at least two of three clones for each subunit contained a genomic fragment that hybridized with both 5′ and 3′ probes from the α1 and β1 cDNAs (data not shown). Genomic sequences isolated during screening have 99% and 100% homology in coding regions with the murine α1 and β1 cDNAs, respectively, confirming successful isolation of the genes for these two isoforms and subunits. Comparison of the coding sequences for the α1 sGC subunit gene with previously cloned cDNA revealed seven mismatches in codons 49 (TAC→ GAC), 52 (GAG→GAA), 319 (AAC→AGC), 343 (AAC→AAT), 445 (GAA→GAG), 487 (ATC→ACC), and 690 (GTA→ATA). Of the seven codons, four of the replacements (codons 49, 319, 487, and 690) introduce different amino acid residues in the final protein sequence. The source for the BAC genomic library (Genomic Systems) used in our analyses differed from that of the cDNA library (CLONTECH), suggesting that the inconsistency in these sequences reflect DNA polymorphism between different strains of mice (i.e., 129/SvJ I vs. 200 BALB/c, respectively).
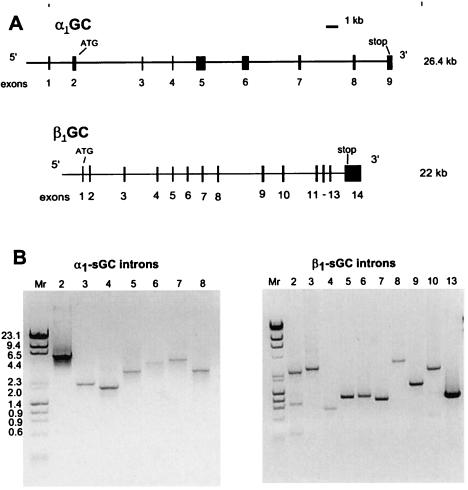
The positions of the exon/intron boundaries were identified by sequencing using oligonucletide primers located within the coding sequences of each gene (Table 1). Intron sizes were estimated by using PCR (see Table 1, Fig. 1) and for introns 1, 2, 10, 11, and 12 of β1 sGC by complete sequencing. Although the size of intron 1 for sGCα1 was not determined, partial sequencing suggests it is more than 2 kb.
Table 1.
Exon-intron splice junctions of the α1 and β1 sGC genes
| Splice donor* | Size of intron, kb | Splice acceptor | |
|---|---|---|---|
| α1 sGC | |||
| Intron 1 | cat−103g/GTGGGTTCGCTCAGC | >2.0 | TCCACTGCTCATAG/gt gct |
| Intron 2 | cca85gag/GTGAGTGTTCTCCC | 5.5 | TCTTTTTCTTTCCAG/tgt gag |
| Intron 3 | aac106ag/GTAAGCTAAGTTACC | 2.2 | TTAATTATTCCCAG/g aaa |
| Intron 4 | gca126g/GTAATAAATAAAACT | 1.9 | CTGTGTGCTTGCAG/gtg ccc |
| Intron 5 | tca362agg/GTAAGGAAAATGTAA | 3.0 | CCTTTCCTTTGCAG/gtt atg |
| Intron 6 | tac524aag/GTAGGGAAGGTGGAA | 4.2 | TATATTGTATCTAG/gtg gag |
| Intron 7 | atc572aag/GTA AGGCCGTGACTT | 4.4 | TGTTTTGCCTTCAG/atg cga |
| Intron 8 | tac624ag/GTATGGATGGCACTA | 2.8 | TAAATTGTTCTCAG/g tta |
| β1 sGC | |||
| Intron 1 | acc1atg/GTGAGTGCTGTCAG | 0.5 | TCTCTGCCCTTCAG/tac ggt |
| Intron 2 | atc26aa/GTAAGTGAACAGCC | 2.5 | TCCATTTTCTTTCAG/a aaa |
| Intron 3 | ctc60a/GTAGGTTGAAAAC | 2.4 | CATCTACAAAACAG/ac ctc |
| Intron 4 | tttg98cag/GTGAGATGTTCGAG | 0.8 | CTGCTGCACTACAG/aac ctc |
| Intron 5 | atg164aag/GTAGTGTTCACCCG | 1.1 | CCATTGACATCTAG/gtg att |
| Intron 6 | ccc241cag/GTAAAATGCACAG | 1.1 | TTTCTGTGTCTTAG/ctc cag |
| Intron 7 | agc280aag/GTAAGCAAGAACC | 1.0 | CTTTCCTGTTTAAG/gaa ggg |
| Intron 8 | cca325ag/GTAACAACTTTTAA | 3.0 | CTCTGTGTGACAG/t gtg |
| Intron 9 | gac391ac/GTAAGCAAGGGAG | 1.5 | CTAATTCCCACAG/a ttg |
| Intron 10 | tac470aag/GCAAGTCTTCATGG | 2.5 | TGTGTCACCCTAG/gtg gaa |
| Intron 11 | gtt517cag/GTGAGTAAATAAAT | 0.4 | CTTTGCTTCTGCAG/ata aca |
| Intron 12 | tac569ag/GTGAGGAGGGAAAT | 0.3 | CTCATGACTTTCAG/g tgt |
| Intron 13 | acg611gag/GTATGGCTCATTAG | 1.1 | TCGACCCATTTAAG/gaa aca |
The positions of codons at which the introns interrupt the coding sequence are indicated, except for the first intron of α1 sGC gene, where it indicates the base pairs upstream of the start codon.
Figure 1.
Genomic structure and size of introns of murine α1 and β1 sGC. (A) Exon-intron distribution of murine α1 and β1 sGC genes (drawn to scale). Exons are represented by black boxes. (B) Estimation of the intron sizes by PCR using primers located in cDNA sequences flanking the introns (see Materials and Methods). The amplified DNA products were separated on 1% agarose gel and stained with ethidium bromide. The molecular mass markers (mixture of Lambda DNA–HindIII digest and φX174 DNA–HaeIII digest, New England Biolabs) are shown at the left of each panel.
The α1 sGC gene encompasses at least 26.4 kb and includes nine exons and eight introns, whereas the β1 sGC gene contains 14 exons and 13 introns and spans 22 kb (see Fig. 1). Start codons are positioned in the second and first exons of the α1 and β1 genes, respectively. The GT/AG donor/acceptor consensus was maintained in all introns for both genes, except for intron 10 in β1 sGC, where the donor site was GC. The sequences that flank the exon/intron boundaries in the α1 and β1 sGC genes are presented in Table 1.
Chromosomal Localization.
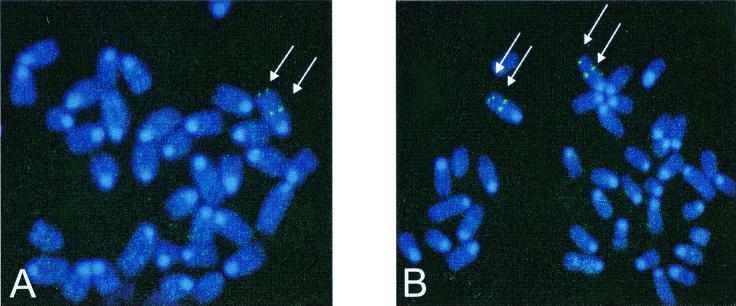
BAC clones containing α1 and β1 genes were used for chromosomal localization by fluorescence in situ hybridization analysis as described in Materials and Methods. A total of 80 metaphase cells were analyzed for each genomic clone with 71 and 72 chromosomes exhibiting specific labeling for α1 and β1 sGC genes, respectively (Fig. 2). Both genes colocalized to mouse chromosome 3. The α1 sGC gene positioned at 44% and β1 sGC at 46% of the distance from the heterochromatic-euchromatic boundary to the telomere of chromosome 3, which corresponds to band 3E3-F1.
Figure 2.
Chromosomal localization of murine α1 and β1 sGC genes. DNA from BAC clones containing genomic regions of α1 and β1 sGC genes was labeled with digoxigenin dUTP and hybridized to normal metaphase chromosomes derived from mouse embryo fibroblast cells (for details see Materials and Methods). Cohybridization of α1 sGC and β1 sGC probe with a probe specific for telomeric region on chromosome 3 is shown on A and B, respectively. Arrows indicate the sites of specific hybridizations.
Analysis of the Promoter Activity of 5′ Regions for α1 and β1 sGC Genes in the N1E-115 Cell Line.
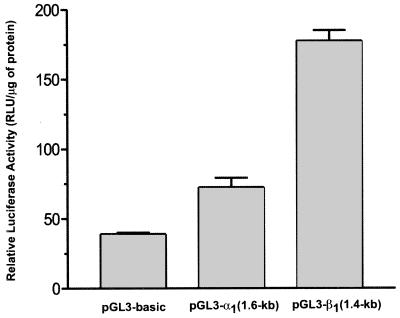
Murine neuroblastoma N1E-115 cells were selected for promoter analysis as host cells because expression of sGC was previously shown in this cell line (34); 1.6 kb and 1.4 kb of the 5′-flanking regions extended until the first identified exons of the α1 and β1 sGC subunit genes were subcloned upstream of the luciferase gene of the pGL3-Basic luciferase reporter vector (Promega). The constructs were transiently transfected in N1E-115 cells (see Materials and Methods). Luciferase activity generated by each of the constructs was compared with the activity of the promotorless pGL3-basic plasmid. The upstream regions of the α1 and β1 sGC genes demonstrated different transcriptional activity in N1E-115 cells (Fig. 3). The relative level of luciferase activity for the β1 sGC construct was 4.6-fold higher when compared with the control, whereas the α1 sGC construct activity was only 2-fold higher than the control promoterless plasmid.
Figure 3.
Promoter activity of 5′ flanking regions of α1 and β1 sGC in N1E-115 cells. Summary of luciferase expression assays of α1 and β1 sGC putative promoter regions in murine neuroblastoma N1E-115 cells. Luciferase activity was assayed as described in Materials and Methods. Values were normalized by using a β-gal construct (cytomegalovirus-β-gal) cotransfections and the total protein concentration of each group. The values represent the normalized means ± SD of three different experiments.
Prediction of the Organization of the Human sGC Genes.
cDNAs for mouse and human α1 and β1 subunits were compared with the Human Genome Database. We identified a clone containing putative genomic regions for both human α1 and β1 subunits (clone AC021433), which is ascribed to chromosome 2. Exons 3–9 of the human α1 gene and all exons of the human β1 gene were identified in this genomic fragment. The comparison of predicted exon-intron boundaries and donor/acceptor sites from the human genes with those of the mouse genes revealed that they are identical in both species with the exception of intron 4 (donor and acceptor sites) and intron 9 (acceptor site) of the β1 subunit.
Discussion
Despite the significant role sGC plays in numerous physiological processes, little is known about the genomic organization of the genes for this enzyme in mammalian species. Recently, the genomic organization for the α1 and β1 subunits of sGC in Medaka fish (32) and the β subunit of sGC in mosquito (Anopheles gambiae) were reported (31). Tandem genomic organization and evidence of directly coordinated transcription for α1 and β1 subunits in Medaka fish suggested the possibility of a similar mechanism of expression of sGC subunts in all vertebrates. Coexpression of both α and β subunits in transfected cells is required for enzyme activity (15). Furthermore, the α1 and β1 sGC genes have been localized to the same chromosome in rat and human (35, 36). Coexpression of both α1 and β1 subunits is obligatory for enzyme activity in rat lung (8, 9, 14, 15) and human cerebral cortex, cerebellum, and lung (20). However, the ratio of expression levels for both subunits varies in a tissue-dependent manner (20), suggesting that the regulation of expression for these subunits is not tightly coordinated as was suggested for Medaka fish. Here, we report that mouse α1 and β1 sGC genes map to the third chromosome (see Fig. 2). However, they are separated from each other by an extended region comprising 2% of the total chromosomal length. This finding excludes the possibility of tandem organization and directly coordinated transcription as proposed for the fish genes. This conclusion is supported by the independent ability of the 5′ flanking regions of the α1 and β1 sGC genes to drive transcription (see Fig. 3). However, the trans-coordinated transcriptional regulation by the same factors is possible.
The rat α1 and β1 genes map to chromosome 2 and are closely linked to the particulate guanylyl cyclase isoform locus and quantitative trait locus, which have been associated with salt-sensitive hypertension in Dahl rats (36). Thus far, no direct connection of the genomic loci identified for α1 and β1 sGC to hypertension in mouse or human has been demonstrated (37). Here we show that both mouse genes colocalize on chromosome 3 in the area corresponding to band 3E3-F1. The α1 and β1 subunits are colocalized with Muc1, Pklr, Ntrk1, CD1, and Fcfr1 loci located at 44.8–46.5 positions of chromosome 3. The chromosomal positioning of mouse sGC genes will stimulate the search for diseases associated with genomic aberrations in this region in mice.
Probing of the Human Genome Database with the mouse and human cDNA sequences identified the clone AC021433, which contains eight exons of the human α1 gene and 14 exons of the human β1 gene. This fragment is ascribed to human chromosome 2. Although this fragment is missing the first two exons of the α1 gene, this is the most probable candidate for the locus of the human α1 and β1 genes. In a previous report (35), α3 and β3 subunits of human sGC (later confirmed to be α1 and β1 sGC genes; ref. 20) were colocalized to human chromosome 4 at q31.1-q33, which represents some discrepancy with recent results. However, previously reported mapping of the chromosomal position for α1 and β1 of sGC were made by using cDNA for corresponding isoforms as probes for the localization (35). Considering the high level of homology between coding regions of both sGC subunits in various isoforms and, in addition, the existence of regions of extensive homologies (in the catalytic domain, for example) to the particulate guanylyl cyclase family, the use of cDNA for chromosomal localization could result in ambiguities and errors.
The transcriptional regulation of the expression of sGC has not been previously examined. Recently, evidence to support altered expression of mRNA of sGC subunits has emerged. In primary rat pulmonary artery smooth muscle cells, prolonged NO treatment leads to decreased NO-stimulated sGC activity and mRNA levels (28). sGC levels rise in unborn rat pulmonary artery, beginning at approximately 20 days of gestation, and mRNA, protein, and activity remain elevated at least 8 days after birth (30). Decreased rates of sGC transcription also have been suggested in other models after NO treatment and administration of cAMP-elevating agents (24, 25). Furthermore, nerve growth factor administration to rat PC-12 cells results in decreased steady-state levels of sGC α1 and β1 mRNA. In a related study in our laboratory, we found that estrogen treatment decreases α1 and β1 sGC mRNA levels in rat uterus (to be reported elsewhere). The precise mechanisms underlying these effects on sGC in specific tissues are largely unknown. Our findings on the activity of putative promoter regions demonstrate different transcriptional activity for both subunits, suggesting the potential for finely tuned regulation. These data represent the initial studies on the transcriptional regulation of mouse sGC genes and will be continued in our laboratory. Considering the increased interest in sGC function and multiple reports concerning the functional significance of the splice variants for both subunits (ref. 22 and references therein), our results should help to gain insights on the genetic basis of regulation of this important enzyme.
Acknowledgments
Research support was provided by the John S. Dunn Foundation, the Harold and Leila Y. Mathers Foundation, and the University of Texas.
Abbreviations
- sGC
soluble guanylyl cyclase
- NCBI
National Center for Biotechnology Information
- BAC
bacterial artificial chromosome
- β-gal
β-galactosidase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the NCBI database (accession nos. AF297082 and AF297083).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190331697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190331697
References
- 1.Huang P L. Semin Perinatol. 2000;24:87–90. doi: 10.1016/s0146-0005(00)80064-6. [DOI] [PubMed] [Google Scholar]
- 2.Severina I S. Biochemistry (Moscow) 1998;63:794–801. [PubMed] [Google Scholar]
- 3.Dawson V L, Dawson T M. Prog Brain Res. 1998;118:215–229. doi: 10.1016/s0079-6123(08)63210-0. [DOI] [PubMed] [Google Scholar]
- 4.Boss G R. Proc Natl Acad Sci USA. 1989;86:7174–7178. doi: 10.1073/pnas.86.18.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thippeswamy T, Morris R. Brain Res. 1997;774:116–122. doi: 10.1016/s0006-8993(97)81694-0. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Maher P, Schubert D. J Cell Biol. 1997;139:1317–1324. doi: 10.1083/jcb.139.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Stamler J S. Cell Death Differ. 1999;6:937–942. doi: 10.1038/sj.cdd.4400578. [DOI] [PubMed] [Google Scholar]
- 8.Kamisaki Y, Saheki S, Nakane M, Palmieri J A, Kuno T, Chang B Y, Waldman S A, Murad F. J Biol Chem. 1986;261:7236–7241. [PubMed] [Google Scholar]
- 9.Nakane M, Murad F. Adv Pharmacol. 1994;26:7–18. doi: 10.1016/s1054-3589(08)60048-4. [DOI] [PubMed] [Google Scholar]
- 10.Garbers D L. J Biol Chem. 1979;254:240–243. [PubMed] [Google Scholar]
- 11.Ignarro L J, Degnan J N, Baricos W H, Kadowitz P J, Wolin M S. Biochim Biophys Acta. 1982;718:49–59. doi: 10.1016/0304-4165(82)90008-3. [DOI] [PubMed] [Google Scholar]
- 12.Gerzer R, Bohme E, Hofmann F, Schultz G. FEBS Lett. 1981;132:71–74. doi: 10.1016/0014-5793(81)80429-2. [DOI] [PubMed] [Google Scholar]
- 13.Ohlstein E H, Wood K S, Ignarro L J. Arch Biochem Biophys. 1982;218:187–198. doi: 10.1016/0003-9861(82)90335-6. [DOI] [PubMed] [Google Scholar]
- 14.Nakane M, Saheki S, Kuno T, Ishii K, Murad F. Biochem Biophys Res Commun. 1988;157:1139–1147. doi: 10.1016/s0006-291x(88)80992-6. [DOI] [PubMed] [Google Scholar]
- 15.Nakane M, Arai K, Saheki S, Kuno T, Buechler W, Murad F. J Biol Chem. 1990;265:16841–16845. [PubMed] [Google Scholar]
- 16.Giuili G, Scholl U, Bulle F, Guellaen G. FEBS Lett. 1992;304:83–88. doi: 10.1016/0014-5793(92)80594-7. [DOI] [PubMed] [Google Scholar]
- 17.Koesling D, Herz J, Gausepohl H, Niroomand F, Hinsch K D, Mulsch A, Bohme E, Schultz G, Frank R. FEBS Lett. 1988;239:29–34. doi: 10.1016/0014-5793(88)80539-8. [DOI] [PubMed] [Google Scholar]
- 18.Koesling D, Harteneck C, Humbert P, Bosserhoff A, Frank R, Schultz G, Bohme E. FEBS Lett. 1990;266:128–132. doi: 10.1016/0014-5793(90)81523-q. [DOI] [PubMed] [Google Scholar]
- 19.Mikami T, Kusakabe T, Suzuki N. Eur J Biochem. 1998;253:42–48. doi: 10.1046/j.1432-1327.1998.2530042.x. [DOI] [PubMed] [Google Scholar]
- 20.Zabel U, Weeger M, La M, Schmidt H H. Biochem J. 1998;335:51–57. doi: 10.1042/bj3350051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russwurm M, Behrends S, Harteneck C, Koesling D. Biochem J. 1998;335:125–130. doi: 10.1042/bj3350125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritter D, Taylor J F, Hoffmann J W, Carnaghi L, Giddings S J, Zakeri H, Kwok P Y. Biochem J. 2000;346:811–816. [PMC free article] [PubMed] [Google Scholar]
- 23.Behrends S, Steenpass A, Porst H, Scholz H. Biochem Pharmacol. 2000;59:713–717. doi: 10.1016/s0006-2952(99)00381-0. [DOI] [PubMed] [Google Scholar]
- 24.Behrends S, Harteneck C, Schultz G, Koesling D. J Biol Chem. 1995;270:21109–21113. doi: 10.1074/jbc.270.36.21109. [DOI] [PubMed] [Google Scholar]
- 25.Chhajlani V, Frandberg P A, Ahlner J, Axelsson K L, Wikberg J E. FEBS Lett. 1991;290:157–158. doi: 10.1016/0014-5793(91)81248-7. [DOI] [PubMed] [Google Scholar]
- 26.Papapetropoulos A, Marczin N, Mora G, Milici A, Murad F, Catravas J D. Hypertension. 1995;26:696–704. doi: 10.1161/01.hyp.26.4.696. [DOI] [PubMed] [Google Scholar]
- 27.Papapetropoulos A, Abou-Mohamed G, Marczin N, Murad F, Caldwell R W, Catravas J D. Br J Pharmacol. 1996;118:1359–1366. doi: 10.1111/j.1476-5381.1996.tb15545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippov G, Bloch D B, Bloch K D. J Clin Invest. 1997;100:942–948. doi: 10.1172/JCI119610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Force T, Bloch K D. J Biol Chem. 1997;272:6038–6043. doi: 10.1074/jbc.272.9.6038. [DOI] [PubMed] [Google Scholar]
- 30.Bloch K D, Filippov G, Sanchez L S, Nakane M, de la Monte S M. Am J Physiol. 1997;272:L400–L406. doi: 10.1152/ajplung.1997.272.3.L400. [DOI] [PubMed] [Google Scholar]
- 31.Caccone A, Garcia B A, Mathiopoulos K D, Min G S, Moriyama E N, Powell J R. Insect Mol Biol. 1999;8:23–30. doi: 10.1046/j.1365-2583.1999.810023.x. [DOI] [PubMed] [Google Scholar]
- 32.Mikami T, Kusakabe T, Suzuki N. J Biol Chem. 1999;274:18567–18573. doi: 10.1074/jbc.274.26.18567. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Arroyo C M, Forray C. Eur J Pharmacol. 1991;208:157–161. doi: 10.1016/0922-4106(91)90066-q. [DOI] [PubMed] [Google Scholar]
- 35.Giuili G, Roechel N, Scholl U, Mattei M G, Guellaen G. Hum Genet. 1993;91:257–260. doi: 10.1007/BF00218267. [DOI] [PubMed] [Google Scholar]
- 36.Azam M, Gupta G, Chen W, Wellington S, Warburton D, Danziger R S. Hypertension. 1998;32:149–154. doi: 10.1161/01.hyp.32.1.149. [DOI] [PubMed] [Google Scholar]
- 37.Danziger R S, Pappas C, Barnitz C, Varvil T, Hunt S C, Leppert M F. J Hypertens. 2000;18:263–266. doi: 10.1097/00004872-200018030-00004. [DOI] [PubMed] [Google Scholar]